Intergenomic Crossover Formation in Newly Synthesized Trigeneric Hybrids Involving Wheat, Rye and Barley †
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
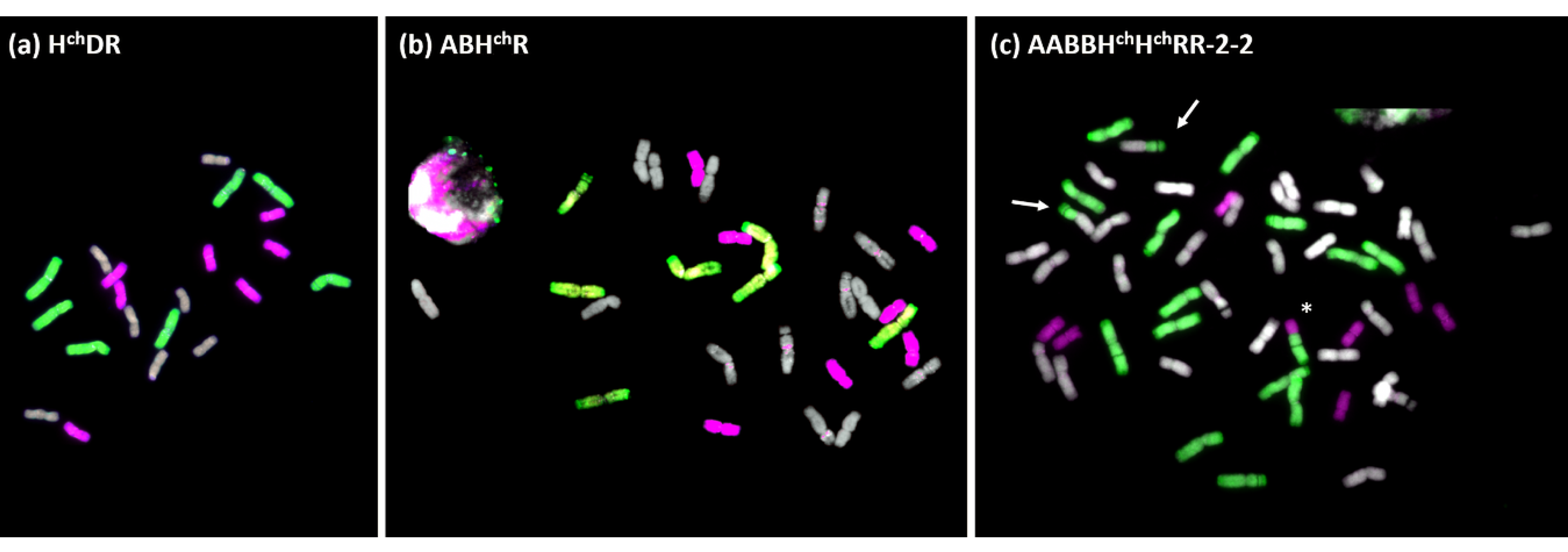
3.1. Production and Chromosome Constitution of the Trigeneric Hybrids and Amphyploids
3.1.1. Hybrid HchDR
3.1.2. Hybrid ABHchR
3.1.3. Amphiploid AABBHchHchRR
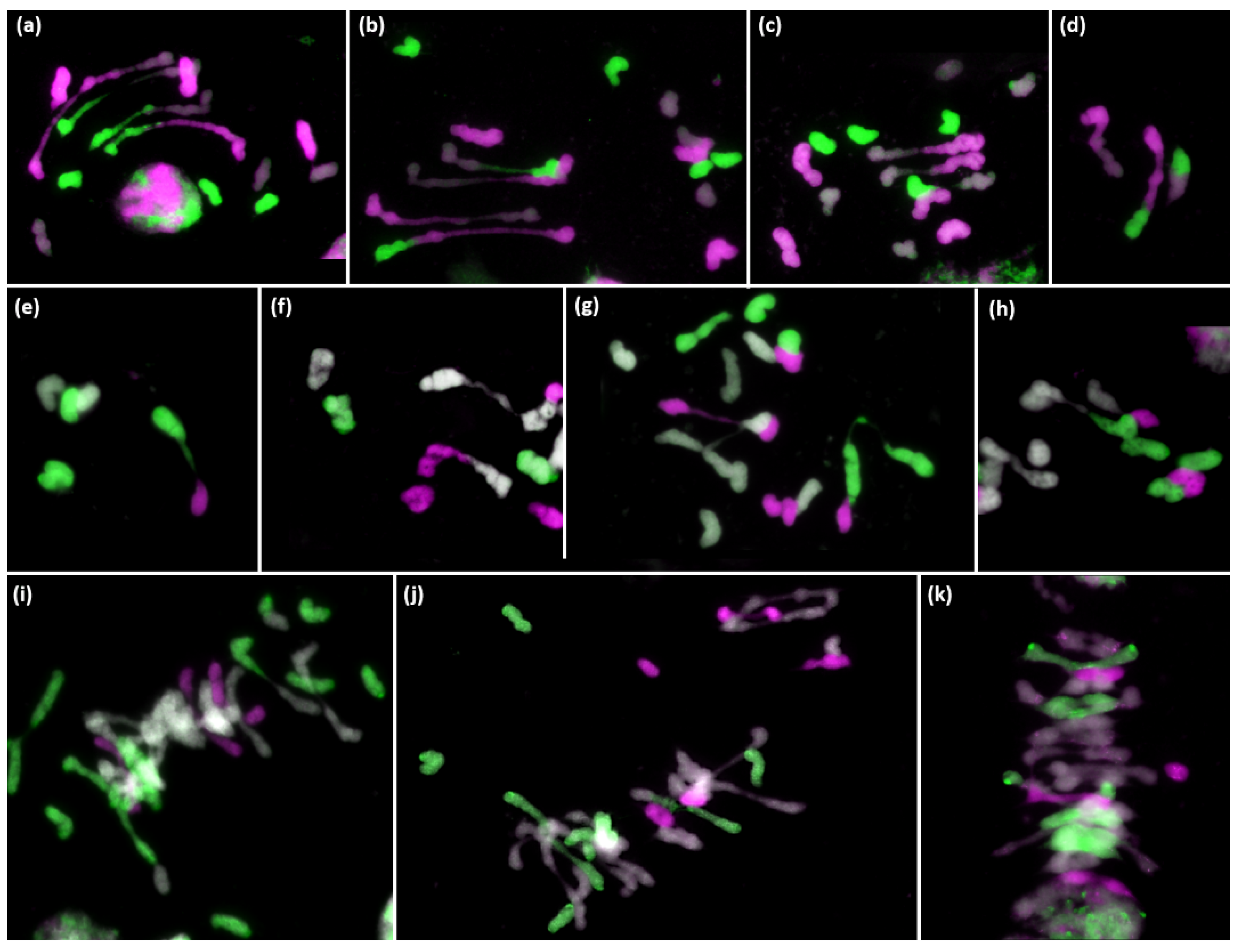
3.2. Analysis of Meiotic Metaphase I Configuration in the Trigeneric Hybrid HchDR
Analysis of Meiotic Metaphase I Configuration in the Trigeneric Hybrid ABHchR and Its Corresponding Amphiploid AABBHchHchRR
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, A.C.; Borrill, P.; Higgins, J.; Alabdullah, A.; Ramírez-González, R.H.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Genome-wide transcription during early wheat meiosis is independent of synapsis, ploidy level, and the Ph1 Locus. Front. Plant Sci. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Gupta, P.K.; Fedak, G. Hybrids of bread wheat (Triticum aestivum) with Thinopyrum scirpeum (4×) and Thinopyrum junceum (6×). Plant Breed. 1986, 97, 107–111. [Google Scholar] [CrossRef]
- Orellana, J.; Vazquez, J.F.; Carrillo, J.M. Genome analysis in wheat–rye–Aegilops caudata trigeneric hybrids. Genome 1989, 32, 169–172. [Google Scholar] [CrossRef]
- Fernandez-Escobar, J.; Martin, A. A self-fertile trigeneric hybrid in the Triticeae involving Triticum, Hordeum, and Secale. Euphytica 1989, 42, 291–296. [Google Scholar] [CrossRef]
- Yu, C.; Jia, X.; Hu, S.; Zhuang, J. Cytogenetics of the hybrids and their pollen plants of three genera Triticum secale and Thinopyrum. Acta Genet. Sin. 1993, 21, 447–452. [Google Scholar]
- Li, X.F.; Song, Z.Q.; Liu, S.B.; Gao, J.R.; Wang, H.G. Cytogenetic study of a trigeneric (triticale× trileymus) hybrid. Euphytica 2006, 150, 117–122. [Google Scholar] [CrossRef]
- Sears, E.R.; Okamoto, M. Intergenomic chromosome relationship in hexaploidy wheat. In Proceedings of the 10th International Congress of Genetics, Montreal, QC, Canada, 17–25 August 1958; Volume 2, pp. 258–259. [Google Scholar]
- Riley, R.; Chapman, V. Genetic control of the cytologically diploid behavior of hexaploidy wheat. Nature 1958, 182, 713–715. [Google Scholar] [CrossRef]
| No. and Type of Chromosome Associations | Total % of Associations | |||
|---|---|---|---|---|
| Rod Bivalent | Ring Bivalent | Trivalent | ||
| Hordeum-Hordeum | 3 | 0 | 0 | 1.7 |
| Aegilops-Aegilops | 1 | 0 | 0 | 0.6 |
| Secale-Secale | 15 | 0 | 0 | 8.5 |
| Hordeum-Aegilops | 73 | 5 | 0 | 47.2 |
| Hordeum-Secale | 13 | 0 | 0 | 7.4 |
| Aegilops-Secale | 51 | 0 | 0 | 29.0 |
| Hordeum-Aegilops-Secale | 0 | 0 | 4 | 4.5 |
| Hordeum-Aegilops-Aegilops | 0 | 0 | 1 | 1.1 |
| Total No. of associations | 156 | 10 | 10 | |
| Rod Bivalent | Total % of Associations | |
|---|---|---|
| Triticum-Triticum | 4 | 8.5 |
| Hordeum-Hordeum | 5 | 11.9 |
| Secale-Secale | 4 | 8.5 |
| Triticum-Hordeum | 15 | 35.7 |
| Triticum-Secale | 10 | 23.8 |
| Hordeum-Secale | 4 | 8.5 |
| Total No. of associations | 42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey, M.-D.; Martín, A.C. Intergenomic Crossover Formation in Newly Synthesized Trigeneric Hybrids Involving Wheat, Rye and Barley. Biol. Life Sci. Forum 2021, 4, 24. https://doi.org/10.3390/IECPS2020-08747
Rey M-D, Martín AC. Intergenomic Crossover Formation in Newly Synthesized Trigeneric Hybrids Involving Wheat, Rye and Barley. Biology and Life Sciences Forum. 2021; 4(1):24. https://doi.org/10.3390/IECPS2020-08747
Chicago/Turabian StyleRey, María-Dolores, and Azahara C. Martín. 2021. "Intergenomic Crossover Formation in Newly Synthesized Trigeneric Hybrids Involving Wheat, Rye and Barley" Biology and Life Sciences Forum 4, no. 1: 24. https://doi.org/10.3390/IECPS2020-08747
APA StyleRey, M.-D., & Martín, A. C. (2021). Intergenomic Crossover Formation in Newly Synthesized Trigeneric Hybrids Involving Wheat, Rye and Barley. Biology and Life Sciences Forum, 4(1), 24. https://doi.org/10.3390/IECPS2020-08747







