Characterization of Olive-Associated Fungi of Cultivars with Different Levels of Resistance to Anthracnose †
Abstract
:1. Introduction
2. Experiments
2.1. Sample Collection
2.2. Fungal Isolation and Enumeration
2.3. Fungal Identification
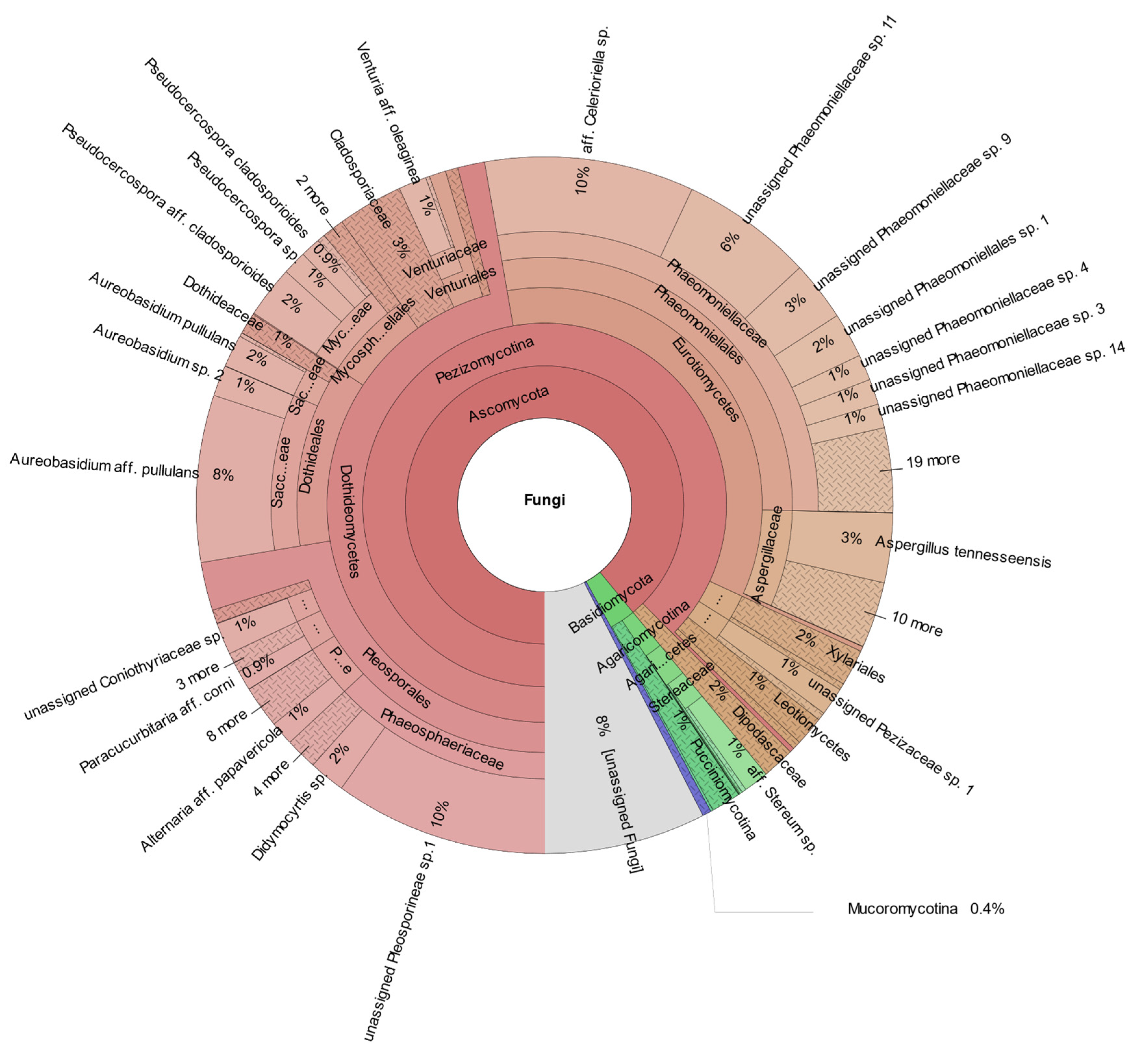
3. Results
General Description of the Fungal Community
4. Discussion and Conclusions
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cacciola, S.O.; Faedda, R.; Sinatra, F.; Agosteo, G.E.; Schena, L.; Frisullo, S.; di San Lio, G.M. Olive anthracnose. J. Plant Pathol. 2012, 94, 29–44. [Google Scholar]
- Besnard, G.; Terral, J.F.; Cornille, A. On the origins and domestication of the olive: A review and perspectives. Ann. Bot. 2018, 121, 385–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Olive Council. Available online: https://www.internationaloliveoil.org/#newsletter (accessed on 10 September 2020).
- Talhinhas, P.; Loureiro, A.; Oliveira, H. Olive anthracnose: A yield-and oil quality-degrading disease caused by several species of Colletotrichum that differ in virulence, host preference and geographical distribution. Mol. Plant Pathol. 2018, 19, 1797–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhinhas, P.; Mota-Capitão, C.; Martins, S.; Ramos, A.P.; Neves-Martins, J.; Guerra-Guimarães, L.; Várzea, V.; Silva, M.C.; Sreenivasaprasad, S.; Oliveira, H. Epidemiology, histopathology and aetiology of olive anthracnose caused by Colletotrichum acutatum and C. gloeosporioides in Portugal. Plant Pathol. 2011, 60, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Moral, J.; Xaviér, C.J.; Viruega, J.R.; Roca, L.F.; Caballero, J.; Trapero, A. Variability in susceptibility to anthracnose in the world collection of olive cultivars of Cordoba (Spain). Front. Plant Sci. 2017, 8, 1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simeone, V.; Baser, N.; Perrelli, D.; Cesari, G.; El Bilali, H.; Natale, P. Residues of rotenone, azadirachtin, pyrethrins and copper used to control Bactrocera oleae (Gmel.) in organic olives and oil. Food Addit. Contam. 2009, 26, 475–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacon, C.W.; White, J.F. Functions, mechanisms and regulation of endophytic and epiphytic microbial communities of plants. Symbiosis 2016, 68, 87–98. [Google Scholar] [CrossRef]
- Landum, M.C.; do Rosário Félix, M.; Alho, J.; Garcia, R.; Cabrita, M.J.; Rei, F.; Varanda, C.M. Antagonistic activity of fungi of Olea europaea L. against Colletotrichum acutatum. Microbiol. Res. 2016, 183, 100–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preto, G.; Martins, F.; Pereira, J.A.; Baptista, P. Fungal community in olive fruits of cultivars with different susceptibilities to anthracnose and selection of isolates to be used as biocontrol agents. Biol. Control 2017, 110, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Martins, F.; Pereira, J.A.; Bota, P.; Bento, A.; Baptista, P. Fungal endophyte communities in above-and belowground olive tree organs and the effect of season and geographic location on their structures. Fungal Ecol. 2016, 20, 193–201. [Google Scholar] [CrossRef]
- Gomes, T.; Pereira, J.A.; Benhadi, J.; Lino-Neto, T.; Baptista, P. Endophytic and epiphytic phyllosphere fungal communities are shaped by different environmental factors in a Mediterranean ecosystem. Microb. Ecol. 2018, 76, 668–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Federhen, S. Type material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015, 43, D1086–D1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahri, H.; Ramos, V.; Mina, D.; Pereira, J.A.; Baptista, P. Characterization of Olive-Associated Fungi of Cultivars with Different Levels of Resistance to Anthracnose. Biol. Life Sci. Forum 2021, 4, 60. https://doi.org/10.3390/IECPS2020-08878
Bahri H, Ramos V, Mina D, Pereira JA, Baptista P. Characterization of Olive-Associated Fungi of Cultivars with Different Levels of Resistance to Anthracnose. Biology and Life Sciences Forum. 2021; 4(1):60. https://doi.org/10.3390/IECPS2020-08878
Chicago/Turabian StyleBahri, Hamdi, Vitor Ramos, Diogo Mina, José A. Pereira, and Paula Baptista. 2021. "Characterization of Olive-Associated Fungi of Cultivars with Different Levels of Resistance to Anthracnose" Biology and Life Sciences Forum 4, no. 1: 60. https://doi.org/10.3390/IECPS2020-08878
APA StyleBahri, H., Ramos, V., Mina, D., Pereira, J. A., & Baptista, P. (2021). Characterization of Olive-Associated Fungi of Cultivars with Different Levels of Resistance to Anthracnose. Biology and Life Sciences Forum, 4(1), 60. https://doi.org/10.3390/IECPS2020-08878







