Naturally Occurring Green Tea Polyphenols as Anti-Mycobacterial Agents †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Docking Analysis
2.2. In Silico Drug-Likeness and ADMET (Absorption, Distribution, Metabolism, Excretion, Toxicity) Analysis
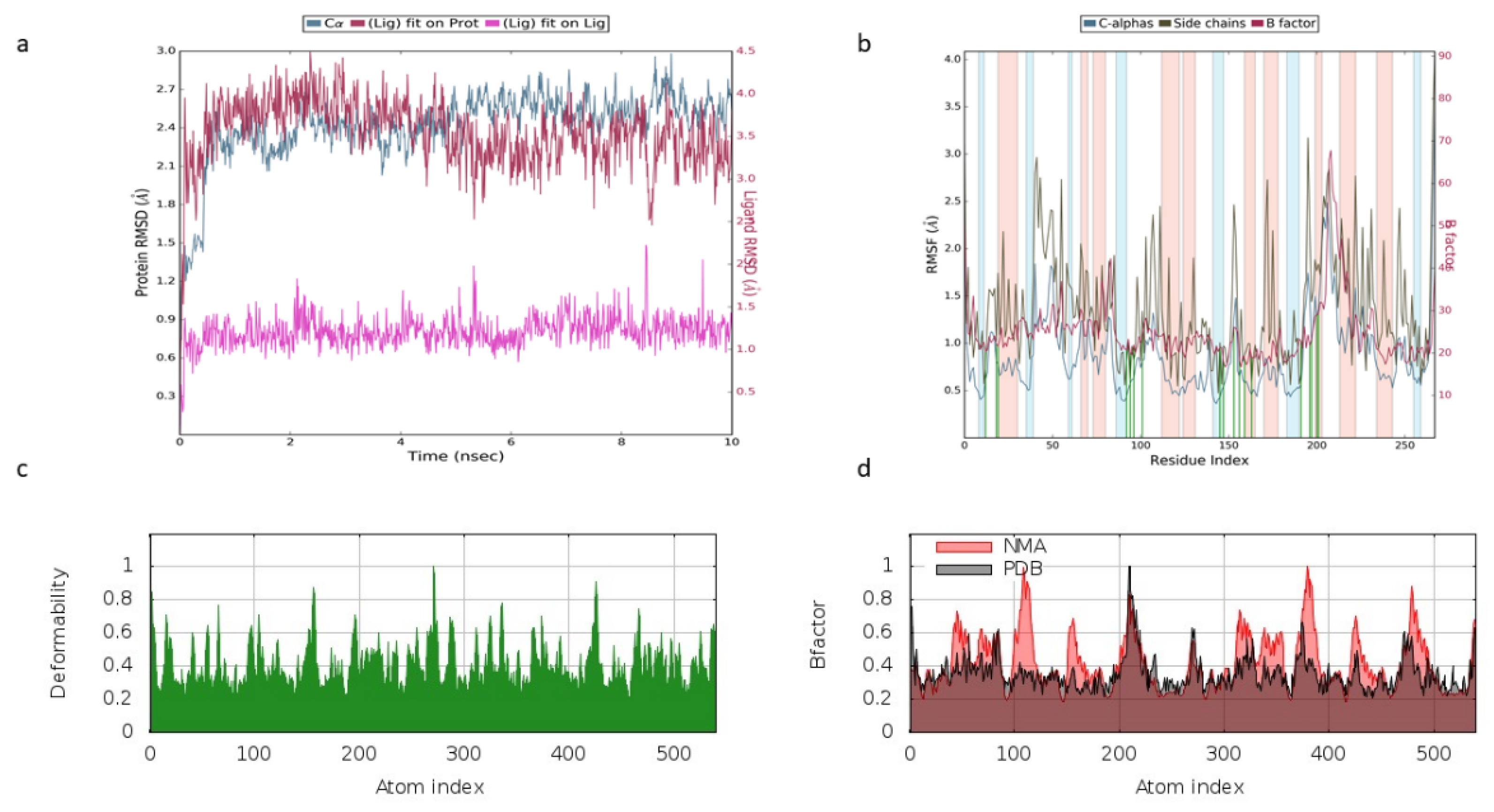
2.3. Normal Mode Analysis
2.4. Molecular Dynamics Analysis
3. Results and Discussion
3.1. Molecular Docking Simulations
3.2. Molecular Dynamics Simulation and Normal Mode Analysis
3.3. In Silico ADME Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poro, K.E.; Hoekou, Y.; Pissang, P.; Kpabi, I.; Novidzro, K.M.; Dagnra, A.Y.; Tchacondo, T.; Batawila, K. In vitro Antimycobacterial Activity of Selected Medicinal Plants against Mycobacterium tuberculosis. Int. J. Curr. Microbiol. Appl. Sci. 2021, 10, 3201–3208. [Google Scholar]
- WHO TB Factsheet. Available online: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed on 17 July 2021).
- Banerjee, A.; Dubnau, E.; Quemard, A.; Balasubramanian, V.; Um, K.S.; Wilson, T.; Collins, D.; de Lisle, G.; Jacobs, W.R., Jr. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 1994, 263, 227–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henss, L.; Auste, A.; Schürmann, C.; Schmidt, C.; von Rhein, C.; Mühlebach, M.D.; Schnierle, B.S. The green tea catechin epigallocatechin gallate inhibits SARS-CoV-2 infection. J. Gen. Virol. 2021, 102, 001574. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.K.; Kaul, D.; Sharma, M. Green tea polyphenol inhibits Mycobacterium tuberculosis survival within human macrophages. Int. J. Biochem. Cell Biol. 2006, 38, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Ramesh, K.V. Epigallocatechin gallate, a green tea polyphenol inhibits Mycobacterium smegmatis: In silico and in vitro studies. Indian J. Pharm. Sci. 2017, 79, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, V.K.; Singh, R.; Sharma, J.; Rajendran, V.; Purohit, R.; Kumar, S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021, 39, 3449–3458. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, J.; Chacón, P.; Abagyan, R. Predictions of Protein Flexibility: First Order Measures. PROTEINS: Structure, Function, and Bioinformatics. Proteins 2004, 56, 661–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, S.N.; Chaudhari, H.K. Computational studies on imidazo [1,2-a] pyridine-3-carboxamide analogues as antimycobacterial agents: Common pharmacophore generation, atom-based 3D-QSAR, molecular dynamics simulation, QikProp, molecular docking and prime MMGBSA approaches. Open Pharm. Sci. J. 2018, 5, 12–23. [Google Scholar] [CrossRef] [Green Version]
- Hsu, K.C.; Chen, Y.F.; Lin, S.R.; Yang, J.M. iGEMDOCK: A graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform. 2011, 12, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, S.N.; Pandey, A.; Thorat, B.R. Multiple 3D- and 2D-quantitative structure–activity relationship models (QSAR), theoretical study and molecular modeling to identify structural requirements of imidazopyridine analogues as anti-infective agents against tuberculosis. Struct. Chem. 2022. [Google Scholar] [CrossRef]
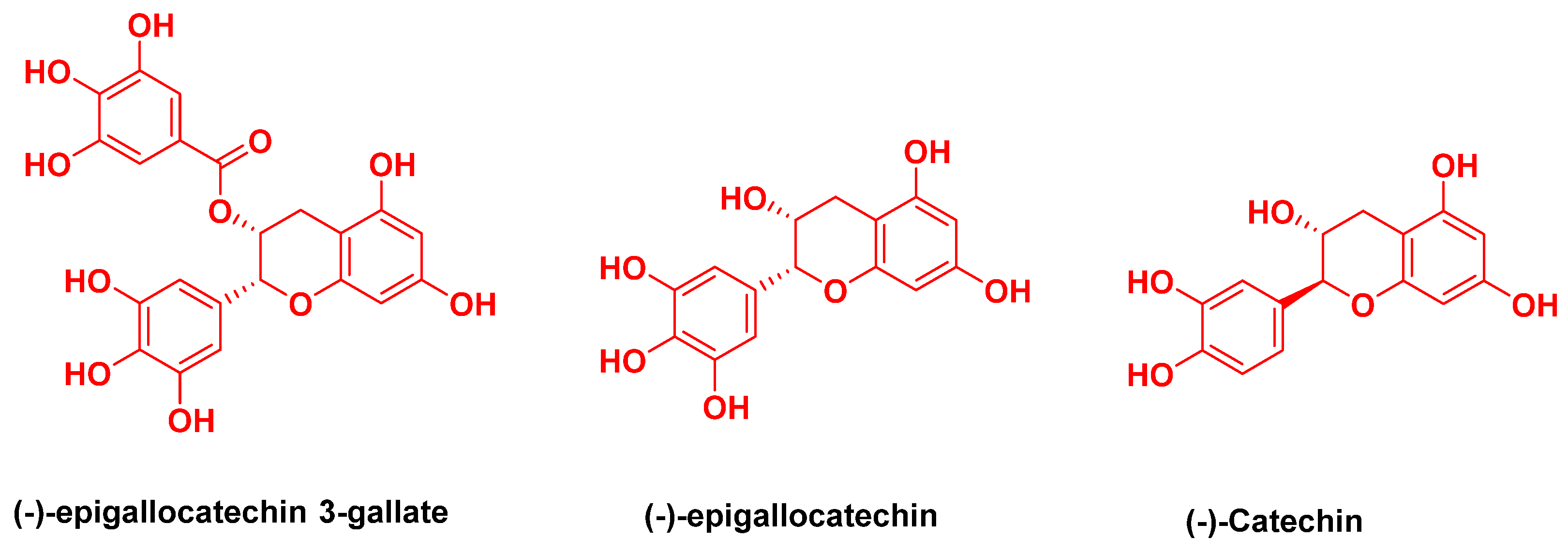
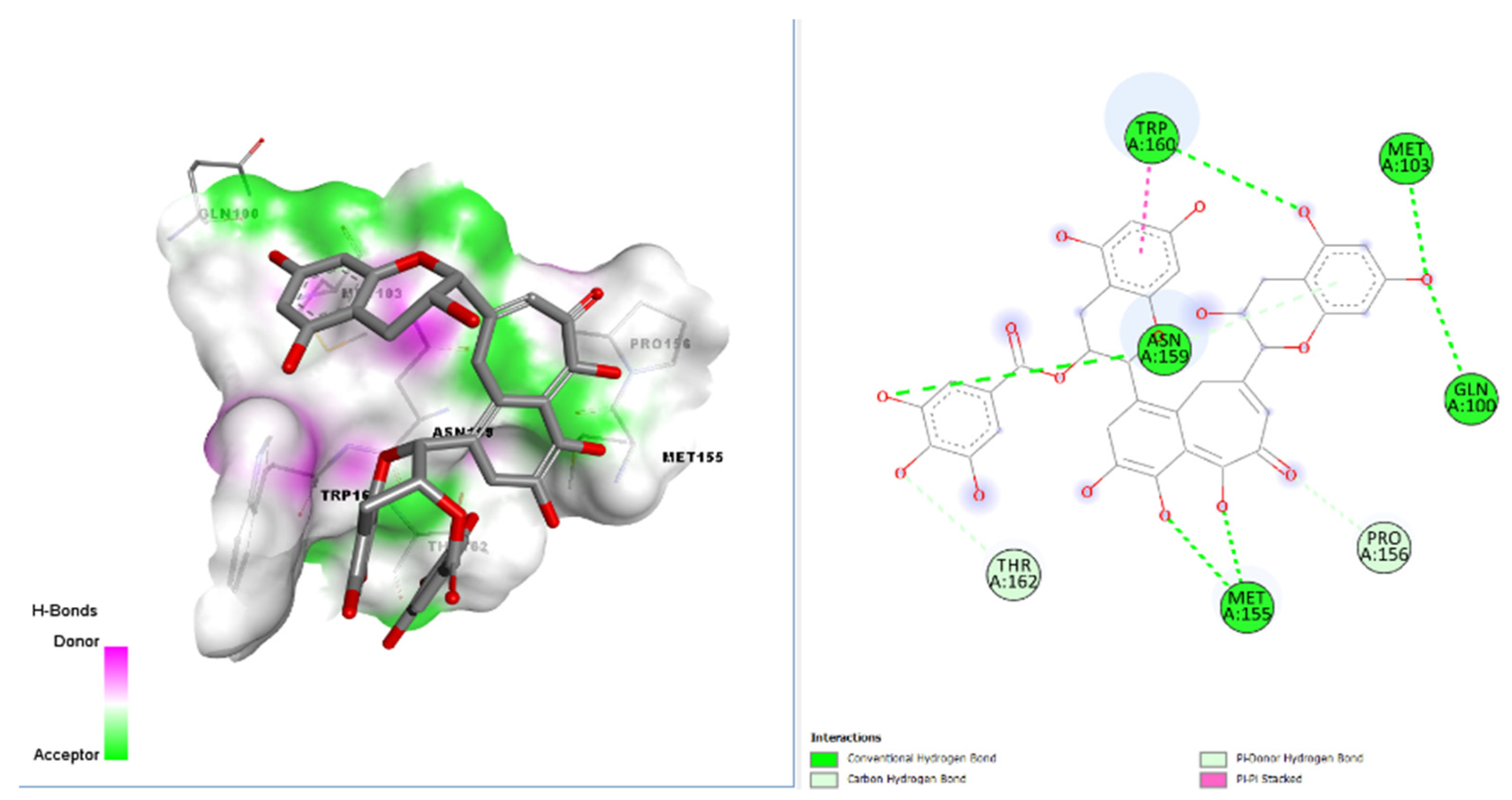
| Molecules | iGemDock Interaction Energy | Molecules | -iGemDock Interaction Energy |
|---|---|---|---|
| Oolonghomobisflavan A | −66.2219 | Theaflavic Acid | −84.4934 |
| Theasinensin D | −72.1619 | Barrigenol R1 | −86.4843 |
| Theaflavin-3-gallate | −134.13 | Barringtogenol | −89.0693 |
| Isotheaflavin | −72.621 | Camelliagenin | −95.1799 |
| Epigallocatechin-3,5-Di-O-Gallate | −72.0176 | Gallocatechin | −86.7374 |
| Oolonghomobisflavan B | −75.4779 | Catechin | −102.992 |
| Cis-3-Hexenol | −63.5566 | Epicatechin | −98.6033 |
| Epigallocatechin-3,4-Di-O-Gallate | −92.6784 | Epiafzelechin | −91.5357 |
| Vicenin 2 | −96.9806 | Quercetin | −102.834 |
| Epicatechin-3,5-Di-O-Gallate | −101.495 | Cryptoxanthin | −95.1799 |
| Rutin | −87.1416 | Myricetin | −83.5936 |
| Proanthocyanidin | −84.8129 | Apigenin | −83.6163 |
| Pheophytin | −90.2865 | Nerolidol | −84.584 |
| Benzaldehyde | −91.9877 | Kaempferol | −89.1838 |
| Epitheaflavic Acid 3’-Gallate | −65.361 | Theanine | −83.9851 |
| Epigallocatechin Gallate | −122.3403 | Ascorbic Acid | −80.1271 |
| Theasinensin E | −62.6409 | Quinic Acid | −85.3299 |
| Myricitrin | −61.915 | Succinic Acid | −85.5696 |
| Theaflavin | −65.9704 | Methyl Salicylate | −81.1848 |
| Epicatechin Gallate | −75.5287 | Theobromine | −84.7269 |
| Kaempferitrin | −72.7401 | Caffeine | −84.4502 |
| Isoquercetin | −89.9058 | Xanthine | −86.7595 |
| Epiafzelechin 3-O-Gallate | −79.4119 | Linalool Oxide | −83.9907 |
| Pheophorbide | −71.1657 | Phenylacetaldehyde | −87.8044 |
| Epigallocatechin 3-O-P-Coumarate | −78.8643 | Methylxanthine | −79.6185 |
| Pheophorbide | −68.9266 | Theophylline | −88.1319 |
| Oxalic Acid | −87.9277 | Geraniol | −95.2378 |
| Cryptoxanthin | −81.2634 | Hexanal | −95.8974 |
| Isovitexin | −82.924 | Diphenylamine | −93.4455 |
| Vitexin | −85.6638 | Trans−2-Hexenal | −94.076 |
| Chlorogenic Acid | −89.7604 | Linalool | −86.4307 |
| Coumaroyl Quinic Acid | −94.7189 | Phenylethanol | −101.468 |
| Epigallocatechin | −115.6776 | Ciprofloxacin * | −108.9558 |
| Sr. No. | Molecules | Residues with Contribution Energy (kcal/mol) |
|---|---|---|
| 1 | Isoniazide | TYR A:158 (PI-PI STACKING); VAL A:203; MET A:199; LYS A:165 |
| 2 | Pyrazinamide | TYR A:158; MET A:161; ALA A:198 |
| 3 | Ciprofloxacin | PRO A:156; MET A:199; TYR A:158; VAL A:203 |
| 4 | Theaflavin-3-gallate (Best docked) | TRP A:160; MET A:103; GLN A:100; ASN A:159; MET A:155; THR A:162; PRO A:156 |
| 5 | Epigallocatechin | ALA A:198; MET A:162; PRO A:193; PHE A:149; MET A:199; TYR A:158 |
| 6 | Epigallocatechin Gallate (EGCG) | ALA A:198; MET A:162; PRO A:193; PHE A:149; MET A:199; TYR A:158 |
| 7 | Inbound ligand | ALA A:198; MET A:162; PRO A:193; PHE A:149; MET A:199; TYR A:158 |
| Properties | Theaflavin-3-Gallate | Epigallocatechin | Epigallocatechin Gallate (EGCG) |
|---|---|---|---|
| CYP450 2C9 Substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 2D6 Substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 3A4 Substrate | Non-substrate | Non-substrate | Non-substrate |
| Human Ether-a-go-go-Related Gene Inhibition | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| AMES Toxicity | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic |
| Carcinogens | None | None | None |
| Acute Oral Toxicity | IV | IV | IV |
| P-glycoprotein Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Rat Acute Toxicity (LD50, mol/kg) | 2.6693 | 1.8700 | 2.6643 |
| Human Intestinal Absorption | + | + | + |
| Blood–brain barrier | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mali, S.N.; Pandey, A. Naturally Occurring Green Tea Polyphenols as Anti-Mycobacterial Agents. Med. Sci. Forum 2021, 7, 5. https://doi.org/10.3390/ECMS2021-10844
Mali SN, Pandey A. Naturally Occurring Green Tea Polyphenols as Anti-Mycobacterial Agents. Medical Sciences Forum. 2021; 7(1):5. https://doi.org/10.3390/ECMS2021-10844
Chicago/Turabian StyleMali, Suraj N., and Anima Pandey. 2021. "Naturally Occurring Green Tea Polyphenols as Anti-Mycobacterial Agents" Medical Sciences Forum 7, no. 1: 5. https://doi.org/10.3390/ECMS2021-10844







