Tropical Fruit Virus Resistance in the Era of Next-Generation Plant Breeding
Abstract
:1. Introduction
2. Viral Infections in Tropical Fruit Crops
2.1. Banana
2.2. Citrus
2.3. Pineapple
2.4. Papaya
2.5. Melon
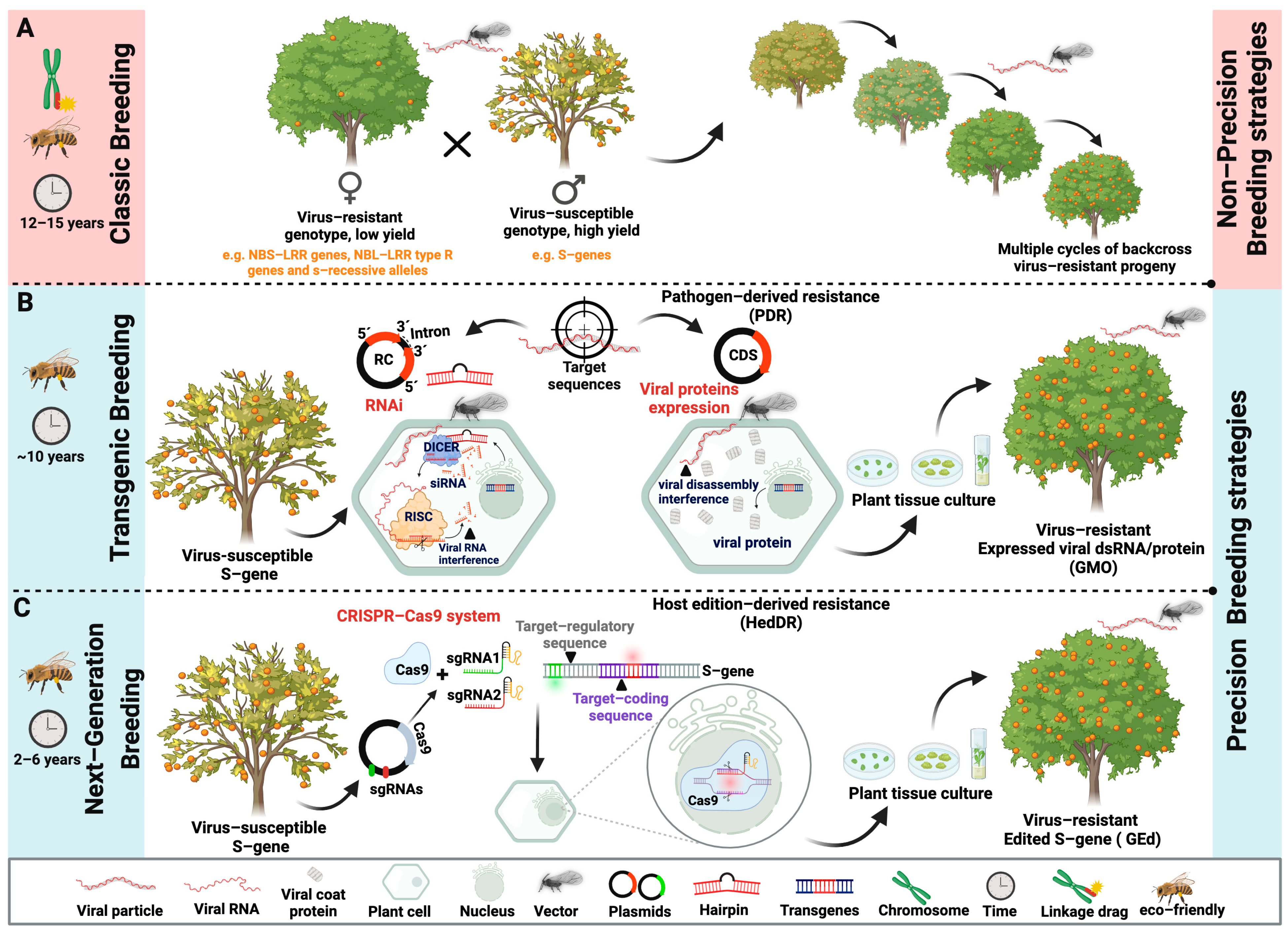
3. Management Strategies
4. Disease-Resistant Crops: Research Trends in Tropical Fruit Virology
4.1. Pathogen-Derived Resistance by RNA Silencing
4.2. CRISPR/Cas
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Major Tropical Fruits Market Review—Preliminary Results 2023; FAO: Rome, Italy, 2024. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Fruit and Vegetables—Your Dietary Essentials. The International Year of Fruits and Vegetables; FAO: Rome, Italy, 2021. [Google Scholar]
- Associação Brasileira dos Produtores Exportadores de Frutas e Derivados. A Revolução da Fruticultura Sustentável: Tendências e Inovações; Food Safety Brazil: Campinas, Brazil, 2023. [Google Scholar]
- Food and Agriculture Organization of the United Nations. World Food and Agriculture—Statistical Yearbook 2022; FAO: Rome, Italy, 2022. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Agricultural Production Statistics 2020–2022; FAOSTAT Analytical Brief 79; Food and Agriculture Organization of the United Nations: Rome, Italy, 2022; ISSN 2709-0078. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/dc1b16d6-44af-4d50-9544-10fcb8a3779f/content (accessed on 13 May 2024).
- Food and Agriculture Organization of the United Nations. Banana Market Review—Preliminary Results 2023; FAO: Rome, Italy, 2023. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Citrus Fruit Statistical Compendium 2020; FAO: Rome, Italy, 2021. [Google Scholar]
- Nicaise, V. Crop Immunity against Viruses: Outcomes and Future Challenges. Front. Plant Sci. 2014, 5, 118474. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Liu, J.; You, H.; Xu, C.; Dong, K.; Luo, N.; Kong, L.; Li, X.; Hong, N.; Wang, G.; et al. Genomic, Morphological and Biological Traits of the Viruses Infecting Major Fruit Trees. Viruses 2019, 11, 515. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H.; Galtier, N. Determinants of Genetic Diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, P.M.B.; Favaratto, L.; Fernandes, A.A.R.; Vicien, C.; Capalbo, D.M.F.; Zerbini, F.M. To Become More Sustainable Organic Agriculture Needs Genome Editing Technology. Front. Bioeng. Biotechnol. 2022, 10, 912793. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.C.; Naidu, R.A. Global Dimensions of Plant Virus Diseases: Current Status and Future Perspectives. Annu. Rev. Virol. 2019, 6, 387–409. [Google Scholar] [CrossRef]
- He, S.; Creasey Krainer, K.M. Pandemics of People and Plants: Which Is the Greater Threat to Food Security? Mol. Plant 2020, 13, 933–934. [Google Scholar] [CrossRef]
- Jin, X.; Cao, X.; Wang, X.; Jiang, J.; Wan, J.; Laliberté, J.F.; Zhang, Y. Three-Dimensional Architecture and Biogenesis of Membrane Structures Associated with Plant Virus Replication. Front. Plant Sci. 2018, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Underwood, W. The Plant Cell Wall: A Dynamic Barrier against Pathogen Invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef]
- Sett, S.; Prasad, A.; Prasad, M. Resistance Genes on the Verge of Plant–Virus Interaction. Trends Plant Sci. 2022, 27, 1242–1252. [Google Scholar] [CrossRef]
- Kozieł, E.; Otulak-Kozieł, K.; Bujarski, J.J. Plant Cell Wall as a Key Player During Resistant and Susceptible Plant-Virus Interactions. Front. Microbiol. 2021, 12, 656809. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, C.; Selvarajan, R.; Jebasingh, T.; Sankara Naynar, P. Evidence of viral genome linked protein of banana bract mosaic virus interaction with translational eukaryotic initiation factor 4E of plantain cv. Nendran based on yeast two hybrid system study. Virus Dis. 2021, 32, 23–130. [Google Scholar] [CrossRef] [PubMed]
- Watt, L.G.; Crawshaw, S.; Rhee, S.-J.; Murphy, A.M.; Canto, T.; Carr, J.P. The cucumber mosaic virus 1a protein regulates interactions between the 2b protein and ARGONAUTE 1 while maintaining the silencing suppressor activity of the 2b protein. PLoS Pathog. 2020, 16, e1009125. [Google Scholar] [CrossRef]
- Leastro, M.O.; Pallás, V.; Sánchez-Navarro, J.A. The capsid protein of citrus leprosis virus C shows a nuclear distribution and interacts with the nucleolar fibrillarin protein. Virus Res. 2024, 340, 199297. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Y.; Wang, G.; Wen, S.; Wang, Y.; Li, L.; Xiao, F.; Hong, N. The p23 of citrus tristeza virus interacts with host FKBP-type peptidyl-prolylcis-trans isomerase 17-2 and is involved in the intracellular movement of the viral coat protein. Cells 2021, 10, 934. [Google Scholar] [CrossRef]
- Reyes, C.A.; Ocolotobiche, E.E.; Marmisollé, F.E.; Robles Luna, G.; Borniego, M.B.; Bazzini, A.A.; Asurmendi, S.; García, M.L. Citrus Psorosis Virus 24K Protein Interacts with Citrus MiRNA Precursors, Affects Their Processing and Subsequent MiRNA Accumulation and Target Expression. Mol. Plant Pathol. 2016, 17, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.K.; Borth, W.B.; Melzer, M.J.; Wang, M.-L.; Hu, J.S. Analysis of pineapple mealybug wilt associated virus-1 and-2 for potential RNA silencing suppressors and pathogenicity factors. Viruses 2015, 7, 969–995. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Shen, W.; Yan, P.; Tuo, D.; Li, X.; Zhou, P. NIa-pro of Papaya ringspot virus interacts with papaya methionine sulfoxide reductase B1. Virology 2012, 434, 78–87. [Google Scholar] [CrossRef]
- Sahana, N.; Kaur, H.; Basavaraj; Tena, F.; Jain, R.K.; Palukaitis, P.; Canto, T.; Praveen, S. Inhibition of the host proteasome facilitates papaya ringspot virus accumulation and proteosomal catalytic activity is modulated by viral factor HcPro. PLoS ONE 2012, 7, e52546. [Google Scholar] [CrossRef]
- Maurastoni, M.; Antunes, T.F.S.; Abreu, E.F.M.; Ribeiro, S.G.; Mehta, A.; Sanches, M.M.; Fontes, W.; Kitajima, E.W.; Cruz, F.T.; Santos, A.M.C.; et al. A Capsid Protein Fragment of a Fusagra-like Virus Found in Carica papaya Latex Interacts with the 50S Ribosomal Protein L17. Viruses 2023, 15, 541. [Google Scholar] [CrossRef]
- Sun, Z.; Wu, Y.-X.; Liu, L.-Z.; Tian, Y.-P.; Li, X.-D.; Geng, C. P3N-PIPO but not P3 is the avirulence determinant in melon carrying the Wmr resistance against watermelon mosaic virus, although they contain a common genetic determinant. J. Virol. 2024, 98, e00507-24. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Yeh, S.; Chen, T. Leucine 127 of Cucurbit Chlorotic Yellows Virus P22 Is Crucial for Its RNA Silencing Suppression Activity and Pathogenicity. Phytopathology 2024, 114, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sun, X.; Shi, Y.; Wei, Y.; Han, X.; Li, H.; Chen, L.; Sun, B.; Sun, H.; Shi, Y. Cucurbit chlorotic yellows virus p22 protein interacts with cucumber SKP1LB1 and its F-box-like motif is crucial for silencing suppressor activity. Viruses 2019, 11, 818. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wei, Y.; Shi, Y.; Han, X.; Chen, S.; Yang, L.; Li, H.; Sun, B.; Shi, Y. Cucumber ribosomal protein CsRPS21 interacts with P22 protein of Cucurbit chlorotic yellows virus. Front. Microbiol. 2021, 12, 654697. [Google Scholar] [CrossRef] [PubMed]
- Sinijadas, K.; Paul, A.; Radhika, N.S.; Johnson, J.M.; Manju, R.V.; Anuradha, T. Piriformospora indica Suppresses the Symptoms Produced by Banana Bract Mosaic Virus by Inhibiting Its Replication and Manipulating Chlorophyll and Carotenoid Biosynthesis and Degradation in Banana. 3 Biotech 2024, 14, 141. [Google Scholar] [CrossRef]
- Sankaranarayanan, R.; Palani, S.N.; Kumar, A.; Punitha Selvakumar, A.S.; Tennyson, J. Prediction and Experimental Confirmation of Banana Bract Mosaic Virus Encoding miRNAs and Their Targets. ExRNA 2020, 2, 5. [Google Scholar] [CrossRef]
- Chabi, M.; Dassou, A.G.; Adoukonou-Sagbadja, H.A.; Thomas, J.; Omondi, H.A. Variation in Symptom Development and Infectivity of Banana Bunchy Top Disease among Four Cultivars of Musa sp. Crops 2023, 3, 158–169. [Google Scholar] [CrossRef]
- Qazi, J. Banana Bunchy Top Virus and the Bunchy Top Disease. J. Gen. Plant Pathol. 2016, 82, 2–11. [Google Scholar] [CrossRef]
- Ji, X.; Yu, N.; Qu, L.; Li, B.; Liu, Z. Banana Bunchy Top Virus (BBTV) Nuclear Shuttle Protein Interacts and Re-Distributes BBTV Coat Protein in Nicotiana benthamiana. 3 Biotech 2019, 9, 121. [Google Scholar] [CrossRef]
- Lantican, D.V.; Nocum, J.D.L.; Manohar, A.N.C.; Mendoza, J.V.S.; Gardoce, R.R.; Lachica, G.C.; Gueco, L.S.; Dela Cueva, F.M. Comparative RNA-Seq Analysis of Resistant and Susceptible Banana Genotypes Reveals Molecular Mechanisms in Response to Banana Bunchy Top Virus (BBTV) Infection. Sci. Rep. 2023, 13, 18719. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N.; Kumar, P.L. Application of CRISPR/Cas for Diagnosis and Management of Viral Diseases of Banana. Front. Microbiol. 2021, 11, 609784. [Google Scholar] [CrossRef] [PubMed]
- Nasim, N.; Dey, N. Pararetroviruses: Plant Infecting DsDNA Viruses. Plant Mol. Biol. Rep. 2022, 40, 106–118. [Google Scholar] [CrossRef]
- Rajeswaran, R.; Seguin, J.; Chabannes, M.; Duroy, P.-O.; Laboureau, N.; Farinelli, L.; Iskra-Caruana, M.-L.; Pooggin, M.M. Evasion of Short Interfering RNA-Directed Antiviral Silencing in Musa Acuminata Persistently Infected with Six Distinct Banana Streak Pararetroviruses. J. Virol. 2014, 88, 19. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Ohki, S.T. Cucumber Mosaic Virus: Viral Genes as Virulence Determinants. Mol. Plant Pathol. 2012, 13, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Tabara, M.; Miyashita, S.; Ando, S.; Kawano, S.; Kanayama, Y.; Fukuhara, T.; Kormelink, R. Cucumber Mosaic Virus Infection in Arabidopsis: A Conditional Mutualistic Symbiont? Front. Microbiol. 2022, 12, 770925. [Google Scholar] [CrossRef] [PubMed]
- Tzean, Y.; Lee, M.C.; Jan, H.H.; Chiu, Y.S.; Tu, T.C.; Hou, B.H.; Chen, H.M.; Chou, C.N.; Yeh, H.H. Cucumber Mosaic Virus-Induced Gene Silencing in Banana. Sci. Rep. 2019, 9, 11553. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.J.; Zhou, Y.H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Interplay between Reactive Oxygen Species and Hormones in the Control of Plant Development and Stress Tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed]
- Arena, G.D.; Ramos-González, P.L.; Falk, B.W.; Casteel, C.L.; Freitas-Astúa, J.; Machado, M.A. Plant Immune System Activation Upon Citrus Leprosis Virus C Infection Is Mimicked by the Ectopic Expression of the P61 Viral Protein. Front. Plant Sci. 2020, 11, 1188. [Google Scholar] [CrossRef]
- Folimonova, S.Y.; Sun, Y.-D. Annual Review of Virology Citrus Tristeza Virus: From Pathogen to Panacea. Annu. Rev. Virol. 2022, 9, 417–435. [Google Scholar] [CrossRef]
- Folimonova, S.Y. Citrus Tristeza Virus: A Large RNA Virus with Complex Biology Turned into a Valuable Tool for Crop Protection. PLoS Pathog. 2020, 16, e1008416. [Google Scholar] [CrossRef]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.X.; Falk, B.W.; Dawson, W.O.; Ding, S.W. Three Distinct Suppressors of RNA Silencing Encoded by a 20-Kb Viral RNA Genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef]
- Kang, S.H.; Sun, Y.D.; Atallah, O.O.; Huguet-Tapia, J.C.; Noble, J.D.; Folimonova, S.Y. A Long Non-Coding RNA of Citrus Tristeza Virus: Role in the Virus Interplay with the Host Immunity. Viruses 2019, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Xu, J.; Jiang, Q.; Zhang, Q.; Wang, C.; Bin, Y.; Song, Z. Construction of Full-Length Infectious CDNA Clones of Citrus Mosaic Virus RNA1 and RNA2 and Infection of Citrus Seedlings by Agrobacterium-Mediated Vacuum-Infiltration. Phytopathology 2023, 113, 6–10. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Family: Secoviridae. ICTV 9th Report. 2011. Available online: https://ictv.global/report_9th/RNApos/Picornavirales/Secoviridae (accessed on 9 May 2024).
- Hyun, J.W.; Hwang, R.Y.; Choi, C.W.; Jung, K.E.; Han, S.G. Symptomatology of Citrus Mosaic Sadwavirus (CiMV) in Some Citrus Cultivars and Effect of CiMV Infection on Citrus Fruit Quality. Plant Pathol. J. 2020, 36, 106. [Google Scholar] [CrossRef]
- Reyes, C.A.; De Francesco, A.; Peña, E.J.; Costa, N.; Plata, M.I.; Sendin, L.; Castagnaro, A.P.; García, M.L. Resistance to Citrus Psorosis Virus in Transgenic Sweet Orange Plants Is Triggered by Coat Protein–RNA Silencing. J. Biotechnol. 2011, 151, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; da Graça, J.V.; Freitas-Astúa, J.; Vidalakis, G.; Duran-Vila, N.; Lavagi, I. Citrus Viruses and Viroids. In The Genus Citrus; Woodhead Publishing: Sawston, UK, 2020; pp. 391–410. [Google Scholar]
- Belabess, Z.; Sagouti, T.; Rhallabi, N.; Tahiri, A.; Massart, S.; Tahzima, R.; Lahlali, R.; Haissam Jijakli, M. Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus. Microorganisms 2020, 8, 1197. [Google Scholar] [CrossRef] [PubMed]
- Dey, K.K.; Green, J.C.; Melzer, M.; Borth, W.; Hu, J.S. Mealybug Wilt of Pineapple and Associated Viruses. Horticulturae 2018, 4, 52. [Google Scholar] [CrossRef]
- Green, J.C.; Rwahnih, M.A.; Olmedo-Velarde, A.; Melzer, M.J.; Hamim, I.; Borth, W.B.; Brower, T.M.; Wall, M.; Hu, J.S. Further Genomic Characterization of Pineapple Mealybug Wilt-Associated Viruses Using High-Throughput Sequencing. Trop. Plant Pathol. 2020, 45, 64–72. [Google Scholar] [CrossRef]
- Galeano, E.A.V.; Martins, D.S.; Barros, F.L.S.; Ventura, J.A.; Queiroz, R.B. Cadeia Produtiva do Mamão no Espírito Santo; Incaper: Vitória, Brazil, 2022; pp. 1–172.
- Premchand, U.; Mesta, R.K.; Devappa, V.; Basavarajappa, M.P.; Venkataravanappa, V.; Narasimha Reddy, L.R.C.; Shankarappa, K.S. Survey, Detection, Characterization of Papaya Ringspot Virus from Southern India and Management of Papaya Ringspot Disease. Pathogens 2023, 12, 824. [Google Scholar] [CrossRef]
- Hamim, I.; Borth, W.B.; Marquez, J.; Green, J.C.; Melzer, M.J.; Hu, J.S. Transgene-Mediated Resistance to Papaya Ringspot Virus: Challenges and Solutions. Phytoparasitica 2018, 46, 1–18. [Google Scholar] [CrossRef]
- Vargas-Mejía, P.; Vega-Arreguín, J.; Chávez-Calvillo, G.; Ibarra-Laclette, E.; Silva-Rosales, L. Differential Accumulation of Innate- and Adaptive-Immune-Response-Derived Transcripts during Antagonism between Papaya Ringspot Virus and Papaya Mosaic Virus. Viruses 2020, 12, 230. [Google Scholar] [CrossRef] [PubMed]
- Antunes, T.F.S.; Maurastoni, M.; Madroñero, L.J.; Fuentes, G.; Santamaría, J.M.; Ventura, J.A.; Abreu, E.F.; Fernandes, A.A.R.; Fernandes, P.M.B. Battle of three: The curious case of papaya sticky disease. Plant Dis. 2020, 104, 2754–2763. [Google Scholar] [CrossRef] [PubMed]
- Antunes, T.F.S.; Amaral, R.J.V.; Ventura, J.A.; Godinho, M.T.; Amaral, J.G.; Souza, F.O.; Zerbini, P.A.; Zerbini, F.M.; Fernandes, P.M.B. The DsRNA Virus Papaya Meleira Virus and an SsRNA Virus Are Associated with Papaya Sticky Disease. PLoS ONE 2016, 11, e0155240. [Google Scholar] [CrossRef] [PubMed]
- García-Cámara, I.; Tapia-Tussell, R.; Magaña-álvarez, A.; Velázquez, A.C.; Martín-Mex, R.; Moreno-Valenzuela, O.; Pérez-Brito, D. Empoasca Papayae (Hemiptera: Cicadellidae)-Mediated Transmission of Papaya Meleira Virus-Mexican Variant in Mexico. Plant Dis. 2019, 103, 2015–2023. [Google Scholar] [CrossRef]
- Cornejo-Franco, J.F.; Alvarez-Quinto, R.A.; Quito-Avila, D.F. Transmission of the Umbra-like Papaya Virus Q in Ecuador and Its Association with Meleira-Related Viruses from Brazil. Crop Prot. 2018, 110, 99–102. [Google Scholar] [CrossRef]
- Puthiyaparambil, J.C.; Pagie, M.; Teressita, S.; Jay, P.M.; Bongani, N.; Paul, F.; Candy, M.; Mark, M.; Marion, M.; Ian, M.; et al. Rapid Papaya Crop Improvement through Accelerated in Vitro Breeding and Molecular Diagnostics. Acta Hortic. 2023, 1362, 499–506. [Google Scholar] [CrossRef]
- Soni, S.K.; Kumar Mishra, M.; Mishra, M.; Kumari, S.; Saxena, S.; Shukla, V.; Tiwari, S.; Shirke, P. Papaya Leaf Curl Virus (PaLCuV) Infection on Papaya (Carica papaya L.) Plants Alters Anatomical and Physiological Properties and Reduces Bioactive Components. Plants 2022, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Pandey, V.; Singh, N.; Marwal, A.; Shahid, M.S.; Gaur, R.K. In Silico Identification of Papaya Genome-Encoded MicroRNAs to Target Begomovirus Genes in Papaya Leaf Curl Disease. Front. Microbiol. 2024, 15, 1340275. [Google Scholar] [CrossRef]
- Domingo-Calap, M.L.; Moreno, A.B.; Pendón, J.A.D.; Moreno, A.; Fereres, A.; López-Moya, J.J. Assessing the Impact on Virus Transmission and Insect Vector Behavior of a Viral Mixed Infection in Melon. Phytopathology 2020, 110, 174–186. [Google Scholar] [CrossRef]
- Domingo-Calap, M.L.; Chase, O.; Estapé, M.; Moreno, A.B.; López-Moya, J.J. The P1 Protein of Watermelon Mosaic Virus Compromises the Activity as RNA Silencing Suppressor of the P25 Protein of Cucurbit Yellow Stunting Disorder Virus. Front. Microbiol. 2021, 12, 645530. [Google Scholar] [CrossRef]
- Ouibrahim, L.; Mazier, M.; Estevan, J.; Pagny, G.; Decroocq, V.; Desbiez, C.; Moretti, A.; Gallois, J.L.; Caranta, C. Cloning of the Arabidopsis rwm1 Gene for Resistance to Watermelon Mosaic Virus Points to a New Function for Natural Virus Resistance Genes. Plant J. 2014, 79, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P. Epidemiology of Potyviruses Infecting Crops of Cucurbitaceae. In Plant RNA Viruses Molecular Pathogenesis and Management; Academic Press: Cambridge, MA, USA, 2023; pp. 213–227. [Google Scholar]
- Chinnadurai, C.; Kollam, M.; Ramsubhag, A.; Jayaraman, J. Genome Characterization of Zucchini Yellow Mosaic Virus Infecting Cucurbits Reveals the Presence of a New Genotype in Trinidad and Tobago in the Caribbean Region. Arch. Virol. 2021, 166, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Bagewadi, B.; Fauquet, C. Plant Virus Control by Post-Transcriptional Gene Silencing. In Encyclopedia of Agriculture and Food Science; Elsevier: Amsterdam, The Netherlands, 2014; pp. 472–488. [Google Scholar]
- Ali, A. Epidemiology and Evolution of Poytviruses Infecting Cucurbits. In Applied Plant Virology; Academic Press: Cambridge, MA, USA, 2020; pp. 405–417. [Google Scholar]
- Kesh, H.; Kaushik, P. Advances in Melon (Cucumis melo L.) Breeding: An Update. Sci. Hortic. 2021, 282, 110045. [Google Scholar] [CrossRef]
- Normantovich, M.; Amitzur, A.; Offri, S.; Pashkovsky, E.; Shnaider, Y.; Nizan, S.; Yogev, O.; Jacob, A.; Taylor, C.G.; Desbiez, C.; et al. The Melon Fom-1–Prv Resistance Gene Pair: Correlated Spatial Expression and Interaction with a Viral Protein. Plant Direct 2024, 8, e565. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Wintermantel, W.M.; Orfanidou, C.G.; Katisa, N.I. Chapter 12—Criniviruses infecting vegetable crops. In Applied Plant Biotechnology for Improving Resistance to Biotic Stress; Academic Press: Cambridge, MA, USA, 2020; pp. 251–289. [Google Scholar]
- European and Mediterranean Plant Protection Organization (EPPO). EPPO Global Datasheet: Crinivirus Cucurbitae. 2023. Available online: https://gd.eppo.int/taxon/CYSDV0/datasheet (accessed on 18 June 2024).
- Orfanidou, C.; Katsiani, A.; Papayiannis, L.; Katis, N.I.; Maliogka, V.I. Interplay of Cucurbit Yellow Stunting Disorder Virus with Cucurbit Chlorotic Yellows Virus and Transmission Dynamics by Bemisia tabaci MED. Plant Dis. 2021, 105, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Shi, Y.; Han, X.; Chen, S.; Li, H.; Chen, L.; Sun, B.; Shi, Y. Identification of Cucurbit Chlorotic Yellows Virus P4.9 as a Possible Movement Protein. Virol. J. 2019, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; He, H.; Zhang, Z.; Zhang, B.; Zhu, C.; Yan, W.; Zhao, C.; Li, J.; Yan, F. Molecular Basis of Mutual Benefits between Cucurbit Chlorotic Yellows Virus (CCYV) Transmission and Imidacloprid Resistance in Bemisia tabaci. J. Pest Sci. 2023, 96, 489–497. [Google Scholar] [CrossRef]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef]
- Manjunatha, L.; Rajashekara, H.; Uppala, L.S.; Ambika, D.S.; Patil, B.; Shankarappa, K.S.; Nath, V.S.; Kavitha, T.R.; Mishra, A.K. Mechanisms of Microbial Plant Protection and Control of Plant Viruses. Plants 2022, 11, 3449. [Google Scholar] [CrossRef]
- Spadotti, D.M.A.; Favara, G.M.; Novaes, Q.S.; Mello, A.P.O.A.; Freitas, D.M.S.; Edwards Molina, J.P.; Rezende, J.A.M. Long-Lasting Systematic Roguing for Effective Management of CABMV in Passion Flower Orchards through Maintenance of Separated Plants. Plant Pathol. 2019, 68, 1259–1267. [Google Scholar] [CrossRef]
- Rezende, J.A.M.; Kitajima, E.W. Vírus e viroides. In Manual de Fitopatologia—Princípios e Conceitos; Agronômica ceres: São Paulo, Brazil, 2018; Volume 1, pp. 369–376. Available online: https://repositorio.usp.br/item/002905510 (accessed on 9 May 2024).
- Ventura, J.A.; Costa, H.; Tatagiba, J.S.; Andrade, J.S.; Martins, D.S. Meleira do mamoeiro: Etiologia, sintomas e epidemiologia. In Papaya Brasil: Qualidade do Mamão Para o Mercado Interno; Incaper: Vitória, Brazil, 2003; pp. 267–276. [Google Scholar]
- Abiola, A.; Zandjanakou-Tachin, M.; Aoudji, K.N.A.; Avocevou-Ayisso, C.; Kumar, P.L. Adoption of Roguing to Contain Banana Bunchy Top Disease in South-East Bénin: Role of Farmers’ Knowledge and Perception. Int. J. Fruit Sci. 2020, 20, 720–736. [Google Scholar] [CrossRef]
- Krishna, R.; Karkute, S.G.; Ansari, W.A.; Jaiswal, D.K.; Prakash Verma, J.; Singh, M. Transgenic Tomatoes for Abiotic Stress Tolerance: Status and Way Ahead. 3 Biotech 2019, 9, 143. [Google Scholar] [CrossRef]
- Jin, Y.; Goodman, R.E.; Tetteh, A.O.; Lu, M.; Tripathi, L. Bioinformatics Analysis to Assess Potential Risks of Allergenicity and Toxicity of HRAP and PFLP Proteins in Genetically Modified Bananas Resistant to Xanthomonas Wilt Disease. Food Chem. Toxicol. 2017, 109, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Stowe, E.; Dhingra, A. Development of the Arctic® Apple. Plant Breed. Rev. 2020, 273–296. [Google Scholar] [CrossRef]
- Mhatre, M. Agrobacterium-Mediated Genetic Transformation of Pineapple (Ananas comosus L., Merr.). Methods Mol. Biol. 2012, 994, 435–453. [Google Scholar]
- Waltz, E. GABA-Enriched Tomato Is First CRISPR-Edited Food to Enter Market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. CRISPR/Cas9-Based Genome Editing of Banana for Disease Resistance. Curr. Opin. Plant Biol. 2020, 56, 118–126. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Kanchiswamy, C.N. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 230280. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Nian, W. Targeted Genome Editing of Sweet Orange Using Cas9/SgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef]
- Shirazi Parsa, H.; Sabet, M.S.; Moieni, A.; Shojaeiyan, A.; Dogimont, C.; Boualem, A.; Bendahmane, A. CRISPR/Cas9-Mediated Cytosine Base Editing Using an Improved Transformation Procedure in Melon (Cucumis melo L.). Int. J. Mol. Sci. 2023, 24, 11189. [Google Scholar] [CrossRef]
- Ferreira, S.A.; Pitz, K.Y.; Manshardt, R.; Fitch, M.; Gonsalves, D. Virus Coat Protein Transgenic Papaya Provides Practical Control of Papaya Ringspot Virus in Hawaii. Plant Dis. 2007, 86, 101–105. [Google Scholar] [CrossRef]
- Gonsalves, D.; Gonsalves, C.; Ferreira, C.; Fitch, M. Transgenic Virus-Resistant Papaya: From Hope to Reality in Controlling Papaya Ringspot Virus in Hawaii; The American Phytopathological Society (APS): St. Paul, MN, USA, 2004; ISSN 2153-0297. Available online: https://www.apsnet.org/edcenter/apsnetfeatures/Pages/PapayaRingspot.aspx (accessed on 9 May 2024).
- Gonsalves, D. Hawaii’s Transgenic Papaya Story 1978–2012: A Personal Account. In Genetics and Genomics of Papaya. Plant Genetics and Genomics: Crops and Models; Springer: New York, NY, USA, 2014; pp. 115–142. [Google Scholar]
- Wu, Z.; Mo, C.; Zhang, S.; Li, H. Characterization of Papaya Ringspot Virus Isolates Infecting Transgenic Papaya ‘Huanong No.1’ in South China. Sci. Rep. 2018, 8, 8206. [Google Scholar] [CrossRef]
- Christou, P. Plant genetic engineering and agricultural biotechnology 1983–2013. Trends Biotechnol. 2013, 31, 125–127. [Google Scholar] [CrossRef]
- Meissner Filho, P.E.; Vilarinhos, A.D.; Oliveira, V.J.D.S.D.; Silva, D.D.C.S.D.; Santos, V.D.S.; Dantas, J.L.L. Resistance of transgenic papaya trees to papaya ringspot in Brazil. Pesqui. Agropecuária Bras. 2021, 56, e01954. [Google Scholar] [CrossRef]
- Ye, C.; Li, H. 20 Years of transgenic research in China for resistance to Papaya ringspot virus. Transgenic Plant J. 2010, 4, 58–63. [Google Scholar]
- Gao, L.; Luo, J.; Ding, X.; Wang, T.; Hu, T.; Song, P.; Zhai, R.; Zhang, H.; Zhang, K.; Li, K.; et al. Soybean RNA Interference Lines Silenced for EIF4E Show Broad Potyvirus Resistance. Mol. Plant Pathol. 2020, 21, 303–317. [Google Scholar] [CrossRef]
- Atarashi, H.; Jayasinghe, W.H.; Kwon, J.; Kim, H.; Taninaka, Y.; Igarashi, M.; Ito, K.; Yamada, T.; Masuta, C.; Nakahara, K.S. Artificially Edited Alleles of the Eukaryotic Translation Initiation Factor 4E1 Gene Differentially Reduce Susceptibility to Cucumber Mosaic Virus and Potato Virus Y in Tomato. Front. Microbiol. 2020, 11, 564310. [Google Scholar] [CrossRef]
- Lucioli, A.; Tavazza, R.; Baima, S.; Fatyol, K.; Burgyan, J.; Tavazza, M. CRISPR-Cas9 Targeting of the EIF4E1 Gene Extends the Potato Virus Y Resistance Spectrum of the Solanum tuberosum L. Cv. Desirée. Front. Microbiol. 2022, 13, 873930. [Google Scholar] [CrossRef]
- Zulfiqar, S.; Farooq, M.A.; Zhao, T.; Wang, P.; Tabusam, J.; Wang, Y.; Zuan, S.; Zhao, J.; Chen, X.; Shen, S.; et al. Virus-induced gene silencing (VIGS): A powerful tool for crop improvement and its advancement towards epigenetics. Int. J. Mol. Sci. 2023, 24, 5608. [Google Scholar] [CrossRef]
- Sharma, N.; Prasad, M. Silencing AC1 of tomato leaf curl virus using artificial microRNA confers resistance to leaf curl disease in transgenic tomato. Plant Cell Rep. 2020, 39, 1565–1579. [Google Scholar] [CrossRef]
- Kim, J.; Lee, S.; Baek, K.; Jin, E. Site-specific gene knock-out and on-site heterologous gene overexpression in Chlamydomonas reinhardtii via a CRISPR-Cas9-mediated knock-in method. Front. Plant Sci. 2020, 11, 306. [Google Scholar] [CrossRef]
- Lee, J.E.; Neumann, M.; Duro, D.I.; Schmid, M. CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS ONE 2019, 14, e0222778. [Google Scholar] [CrossRef]
- Khalid, A.; Zhang, Q.; Yasir, M.; Li, F. Small RNA based genetic engineering for plant viral resistance: Application in crop protection. Front. Microbiol. 2017, 8, 43. [Google Scholar] [CrossRef]
- Van Esse, H.P.; Reuber, T.L.; Van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, X.; Zhou, G.; Zhang, T. Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020, 18, 328–336. [Google Scholar] [CrossRef]
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vasquez, V.; Blevins, B.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef]
- Yu, H.; Wang, Y.; Fu, F.; Li, W. Transgenic improvement for biotic resistance of crops. Int. J. Mol. Sci. 2022, 23, 14370. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, U.K.; Ganapathi, T.R.; Hadapad, A.B. Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high-level resistance to banana bunchy top virus infection. J. Gen. Virol. 2012, 93, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.J.; Yu, T.A.; Huang, C.H.; Wang, H.C.; Wang, S.L.; Yeh, S.D. Generation of hermaphrodite transgenic papaya lines with virus resistance via transformation of somatic embryos derived from adventitious roots of in vitro shoots. Transgenic Res. 2010, 19, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Sarrocco, S. Transgenic approaches for plant disease control: Status and prospects 2021. Plant Pathol. 2022, 71, 207–225. [Google Scholar] [CrossRef]
- Niehl, A.; Soininen, M.; Poranen, M.M.; Heinlein, M. Synthetic biology approach for plant protection using ds RNA. Plant Biotechnol. J. 2018, 16, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Samarskaya, V.; Zavriev, S.K.; Fesenko, I.; Kalinina, N.O.; Love, A.J. RNA-based technologies for engineering plant virus resistance. Plants 2021, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, M.; Sakina, A.; Wani, S.H.; Shikari, A.B.; Tripathi, P.; Zaid, A.; Gala, A.; Abdelrahman, M.; Sharma, M.; Singh, A.K.; et al. Harnessing genome editing techniques to engineer disease resistance in plants. Front. Plant Sci. 2019, 10, 550. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops—Bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [PubMed]
- Prado, G.S.; Rocha, D.C.; Santos, L.N.D.; Contiliani, D.F.; Nobile, P.M.; Martinati-Schenk, J.C.; Padilha, L.; Maluf, M.P.; Lubini, G.; Pereira, T.C.; et al. CRISPR technology towards genome editing of the perennial and semi-perennial crops citrus, coffee and sugarcane. Front. Plant Sci. 2024, 14, 1331258. [Google Scholar] [CrossRef]
- Schiml, S.; Puchta, H. Revolutionizing plant biology: Multiple ways of genome engineering by CRISPR/Cas. Plant Methods 2016, 12, 8. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Khan, Z.A.; Kumar, R.; Dasgupta, I. CRISPR/Cas-mediated resistance against viruses in plants. Int. J. Mol. Sci. 2023, 23, 2303. [Google Scholar] [CrossRef]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004. [Google Scholar] [CrossRef]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.E.A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 80. [Google Scholar] [CrossRef]
- Ntui, V.O.; Tripathi, J.N.; Tripathi, L. Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 2020, 21, 100–128. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. Control of bacterial diseases of banana using CRISPR/Cas-based gene editing. Int. J. Mol. Sci. 2022, 23, 3619. [Google Scholar] [CrossRef]
- Jia, H.; Orbović, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019, 17, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Omar, A.A.; Orbović, V.; Wang, N. Biallelic editing of the LOB1 promoter via CRISPR/Cas9 creates canker-resistant ‘Duncan’ grapefruit. Phytopathology 2022, 112, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Hooghvorst, I.; López-Cristoffanini, C.; Nogués, S. Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 2019, 9, 170–177. [Google Scholar] [CrossRef]
- Nizan, S.; Amitzur, A.; Dahan-Meir, T.; Benichou, J.I.; Bar-Ziv, A.; Perl-Treves, R. Mutagenesis of the melon Prv gene by CRISPR/Cas9 breaks papaya ringspot virus resistance and generates an autoimmune allele with constitutive defense responses. J. Exp. Bot. 2023, 74, 4579–4596. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Toshiya, Y.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef]
- Osakabe, Y.; Liang, Z.; Ren, C.; Nishitani, C.; Osakabe, K.; Wada, M.; Komori, S.; Malnoy, M.; Velasco, R.; Poli, M.; et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018, 13, 2844–2863. [Google Scholar] [CrossRef]
- Brewer, S.E.; Chambers, A.H. CRISPR/Cas9-mediated genome editing of phytoene desaturase in Carica papaya L. J. Hortic. Sci. Biotechnol. 2022, 97, 580–592. [Google Scholar] [CrossRef]
- Jagram, N.; Dasgupta, I. Principles and practice of virus induced gene silencing for functional genomics in plants. Virus Genes 2023, 59, 173–187. [Google Scholar] [CrossRef]
- Tuo, D.; Ma, C.; Yan, P.; Kong, H.; Zhou, P.; Guo, A.; Zhao, H.; Shen, W. Genetic transformation and gene delivery strategies in Carica papaya L. Trop. Plants 2023, 2, 5. [Google Scholar] [CrossRef]
- Chen, T.Y.; Pai, H.; Hou, L.Y.; Lee, S.C.; Lin, T.T.; Chang, C.H.; Hsu, F.C.; Hsu, Y.H.; Lin, N.S. Dual resistance of transgenic plants against Cymbidium mosaic virus and Odontoglossum ringspot virus. Sci. Rep. 2019, 9, 102–130. [Google Scholar] [CrossRef]
- Cheng, H.W.; Lin, T.T.; Huang, C.H.; Raja, J.A.; Yeh, S.D. Modification of papaya ringspot virus HC-Pro to generate effective attenuated mutants for overcoming the problem of strain-specific cross protection. Plant Dis. 2023, 107, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- LaTourrette, K.; Garcia-Ruiz, H. Determinants of Virus Variation, Evolution, and Host Adaptation. Pathogens 2022, 11, 1039. [Google Scholar] [CrossRef] [PubMed]
- Desbiez, C.; Domingo-Calap, M.L.; Pitrat, M.; Wipf-Scheibel, C.; Girardot, G.; Ferriol, I.; Lopez-Moya, J.J.; Lecoq, H. Specificity of Resistance and Tolerance to Cucumber Vein Yellowing Virus in Melon Accessions and Resistance Breaking with a Single Mutation in VPg. Phytopathology 2022, 112, 1185–1191. [Google Scholar] [CrossRef]
| Fruit Crop | Symptoms | Disease | Virus(es) | Family (Genome Type) | Geographic Distribution |
|---|---|---|---|---|---|
| Banana | Mosaic and streak on the inflorescence bracts and petioles | Bract mosaic | banana bract mosaic virus (BBrMV) | Potyviridae (+ssRNA) | Asia, Africa, South America and South Pacific Islands |
| Dwarfism, dark green streaks/marks | Bunchy top | banana bunchy top virus (BBTV) | Nanoviridae (ssDNA) | Asia, Africa, Oceania, South Pacific Islands | |
| Discontinuous chlorotic streak, necrosis | Streak disease | banana streak virus (BSV) | Caulimoviridae (dsDNA) | Asia, Africa, Oceania and South Pacific Islands | |
| Mosaic in leaves (dark/light green and yellowish areas) | Mosaic disease | cucumber mosaic virus (CMV) | Bromoviridae (+ssRNA) | Tropical and subtropical countries | |
| Citrus | Green or yellow, smooth and circular lesions | Leprosis | citrus leprosis virus (CiLV) | Kitaviridae (+ssRNA) | South and Central America |
| Dwarfism, intense yellowing | Tristeza | citrus tristeza virus (CTV) | Closteroviridae (+ssRNA) | Mediterranean region, North and South America | |
| Leaves show yellowing and mottling; coloring ringspot in fruit | Mosaic | citrus mosaic virus (CiMV) | Secoviridae (+ssRNA) | Mediterranean region and Asia | |
| Bark scaling in both the trunk and branches | Citrus Psorosis | citrus psorosis virus (CPsV) | Aspiviridae (-ssRNA) | North and South America | |
| Pineapple | Red–bronze or yellow coloring on central leaves; margins tend to curve down | Mealybug wilt | pineapple mealybug wilt-associated virus complex (PMWaV-1, 2, 3) | Closteoviridae (+ssRNA) | South and Central America, Australia and West Africa |
| Papaya | Aqueous latex exudation | Sticky disease | papaya meleira virus complex (PMeV-1, 2) | PMeV Fusagraviridae (dsRNA) PMeV2 Tombusviridae (+ssRNA) | Central and South America |
| Mosaic, ringspot on the fruit | Ringspot (mosaic) | papaya ringspot virus (PRSV) | Potyviridae (+ssRNA) | Tropical and subtropical countries | |
| Curling and distortion of the leaves | Leaf curl disease | papaya leaf curl virus (PaLCuV) | Geminiviridae (ssDNA) | Indian Subcontinent | |
| Melon | Leaves with yellow spots; the same can occur to fruits | Watermelon mosaic | watermelon mosaic virus (WMV) | Potyviridae (+ssRNA) | Mediterranean region, Eastern Asia, North and South America |
| Yellow mosaic, leaf distortion and blistering | Zucchini yellow mosaic | zucchini yellow mosaic virus (ZYMV) | Potyviridae (+ssRNA) | Tropical and subtropical countries | |
| Mosaic with puckering and blistering on leaves | Mosaic | papaya ringspot virus—watermelon strain (PRSV-W) | Potyviridae (+ssRNA) | Tropical and subtropical countries | |
| Interveinal chlorosis, leaves become thickened | Yellow stunting | cucurbit yellow stunting disorder virus (CYSDV) | Closteroviridae (+ssRNA) | Mediterranean region, North America, Africa and Asia | |
| Chlorotic mottle in leaves | Cucurbit yellows disease | cucurbit chlorotic yellows virus (CCYV) | Closteroviridae (+ssRNA) | Middle East, Mediterranean region and North America |
| Host | Virus(es) | Viral Protein | Host Factors | Function | Reference |
|---|---|---|---|---|---|
| Banana | BBrMV | eIF4E | VPg | Genome viral translation | [18] |
| CMV | CMV 2b | AGO1 | Viral suppression of RNA silencing | [19] | |
| CMV | CMV 1a | AGO1 | Regulation of CMV 2b-AGO1 interaction, and inhibition of another layer of AGO2-mediated antiviral silencing | ||
| Citrus | CiLV | p29 | Fib2 | Possible promotion of viral movement and/or suppression of the plant’s antiviral defense RNA silencing mechanism | [20] |
| CTV | p23 and CP | CaFKBP17-2 | Intracellular trafficking movement of the p23-CP-CaFKBP17-2 complex | [21] | |
| CPsV | 24K | miR156/miR171 | Affects the normal processing of miRNA biogenesis and reduces mature miRNAs in citrus | [22] | |
| Pineapple | PMWaV-1 | p61 | RNAi system | Systemic RNA silencing suppressive activity | [23] |
| PMWaV-2 | p20/CP | Local and systemic RNA silencing suppressive activity | |||
| Papaya | PRSV-P | NIa-Po | PaMsrB1 | Interferes with the elimination of reactive oxygen species (ROS) by PaMsrB1 in chloroplasts due to viral infection | [24] |
| MG132 | proteasome 20s | Increased viral accumulation due to inhibition of protease and RNAse functions of the 20s proteasome protein | [25] | ||
| HcPro | proteasome 20s PAA domain | ||||
| PMeV-1 | CP2 and CP4 | 50S ribosomal protein L17 (RPL17) | Downregulation of RPL17 delays papaya sticky disease symptoms | [26] | |
| Melon | WMV | P3N-PIPO | Wmr | Disruption of viral virulence of the wmr gene resulting in cell death phenotype | [27] |
| CCYV | P22 (L127 residue) | RNAi system | Maintains the stability of the RNA silencing suppression system and is essential for increasing virulence | [28] | |
| P22(F-Box motif) | CsSKP1LB1 | Inhibition of the suppressive activity of RNA silencing mediated by the viral protein P22 | [29] | ||
| P22 | CsRPS21 | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vieira, M.S.; Cabral, R.L.R.; Favaratto, L.; Maciel, L.S.; Xavier, A.d.S.; Zerbini, F.M.; Fernandes, P.M.B. Tropical Fruit Virus Resistance in the Era of Next-Generation Plant Breeding. SynBio 2024, 2, 267-284. https://doi.org/10.3390/synbio2030016
Vieira MS, Cabral RLR, Favaratto L, Maciel LS, Xavier AdS, Zerbini FM, Fernandes PMB. Tropical Fruit Virus Resistance in the Era of Next-Generation Plant Breeding. SynBio. 2024; 2(3):267-284. https://doi.org/10.3390/synbio2030016
Chicago/Turabian StyleVieira, Marcella Silva, Rafael Lara Rezende Cabral, Luíza Favaratto, Laiane Silva Maciel, André da Silva Xavier, Francisco Murilo Zerbini, and Patricia M. B. Fernandes. 2024. "Tropical Fruit Virus Resistance in the Era of Next-Generation Plant Breeding" SynBio 2, no. 3: 267-284. https://doi.org/10.3390/synbio2030016
APA StyleVieira, M. S., Cabral, R. L. R., Favaratto, L., Maciel, L. S., Xavier, A. d. S., Zerbini, F. M., & Fernandes, P. M. B. (2024). Tropical Fruit Virus Resistance in the Era of Next-Generation Plant Breeding. SynBio, 2(3), 267-284. https://doi.org/10.3390/synbio2030016







