The Crystal Structure of Thermal Green Protein Q66E (TGP-E) and Yellow Thermostable Protein (YTP-E) E148D
Abstract
1. Introduction
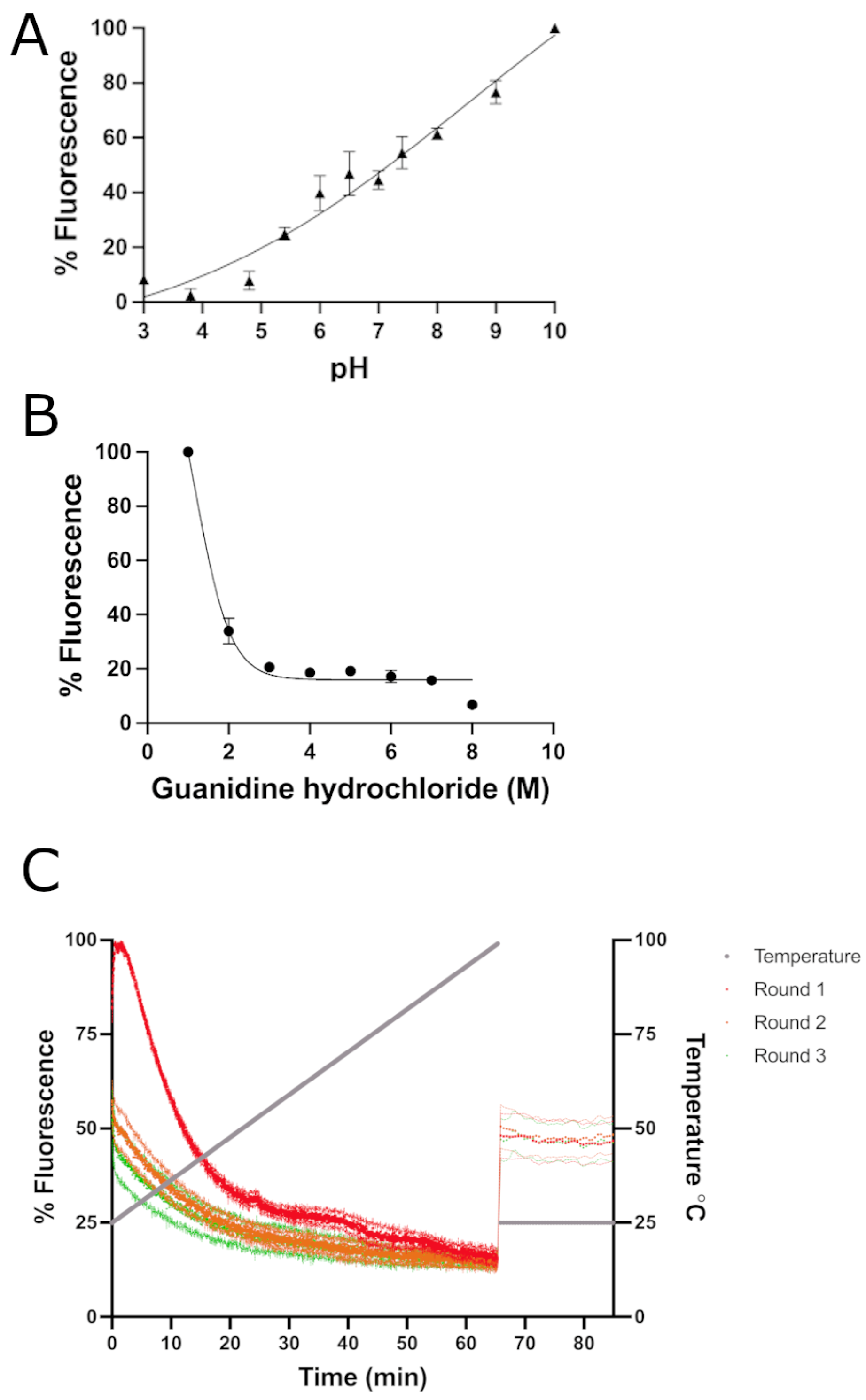
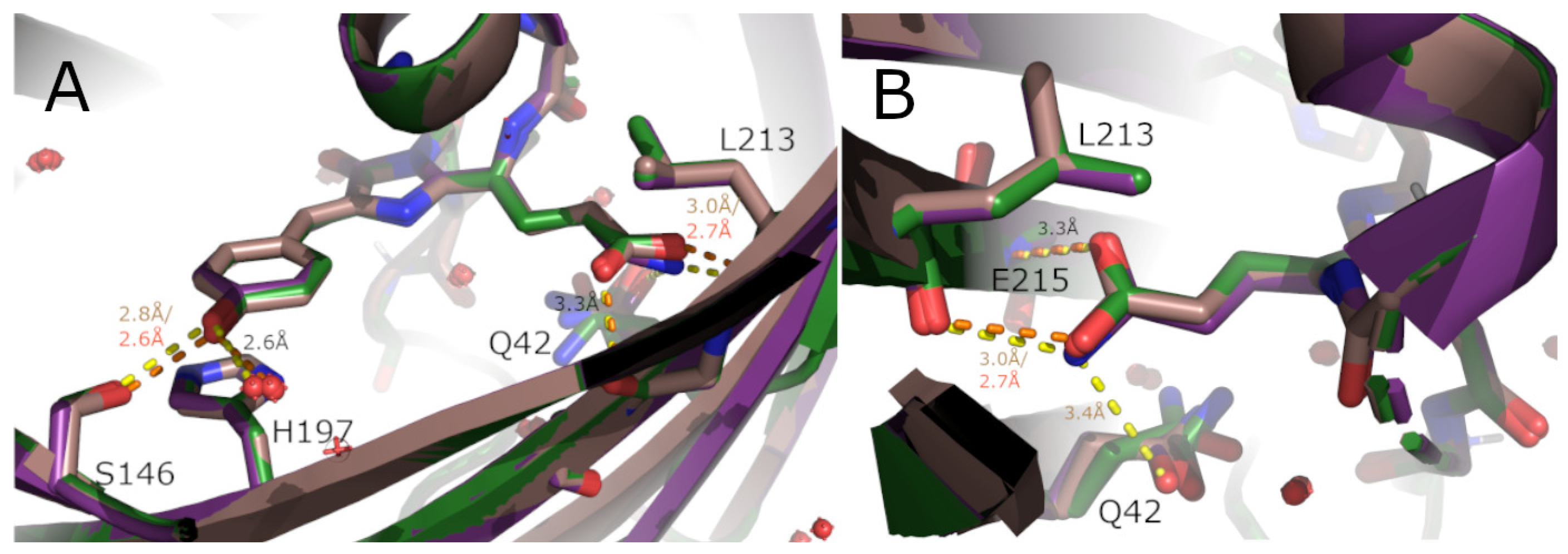
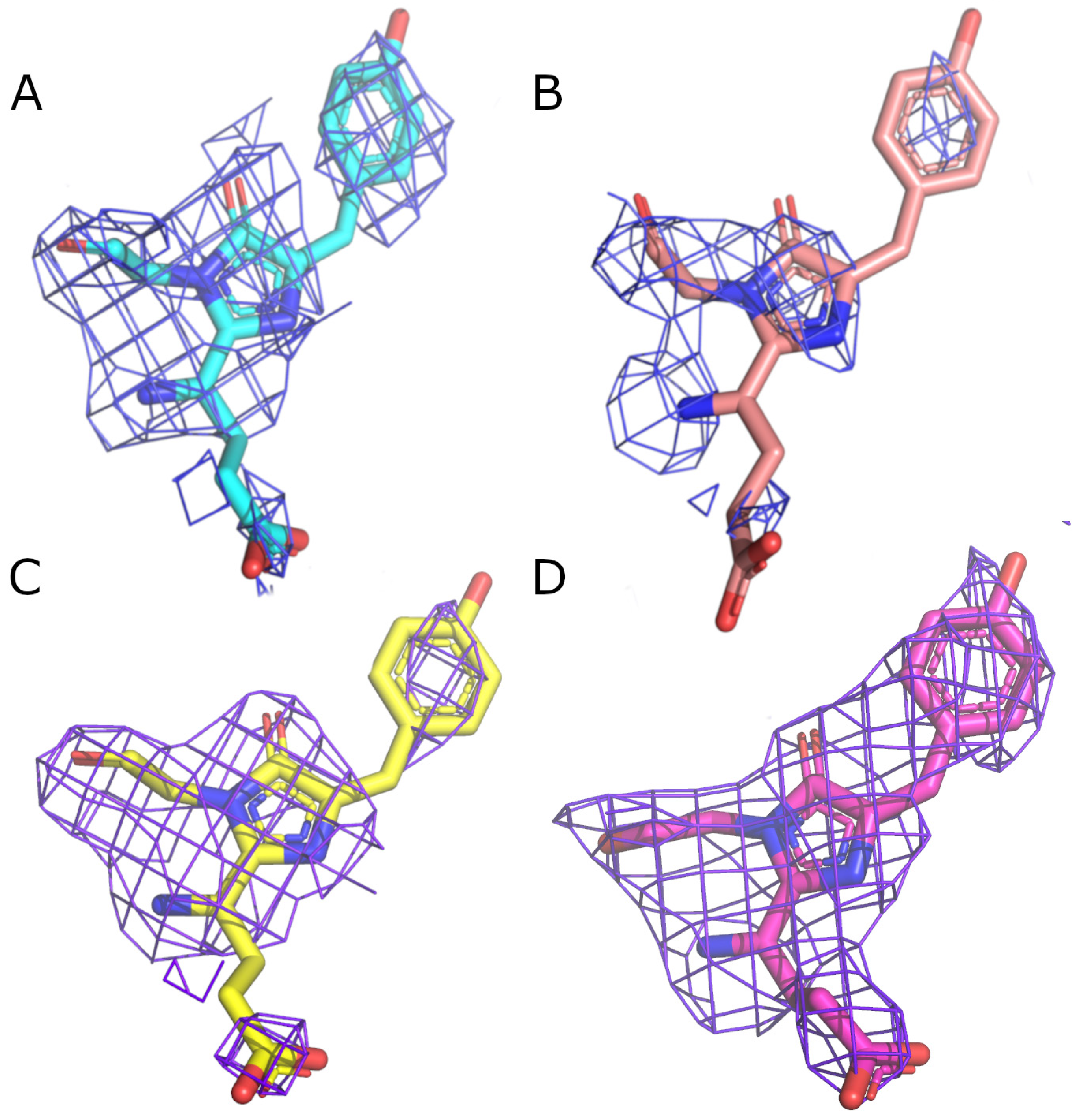
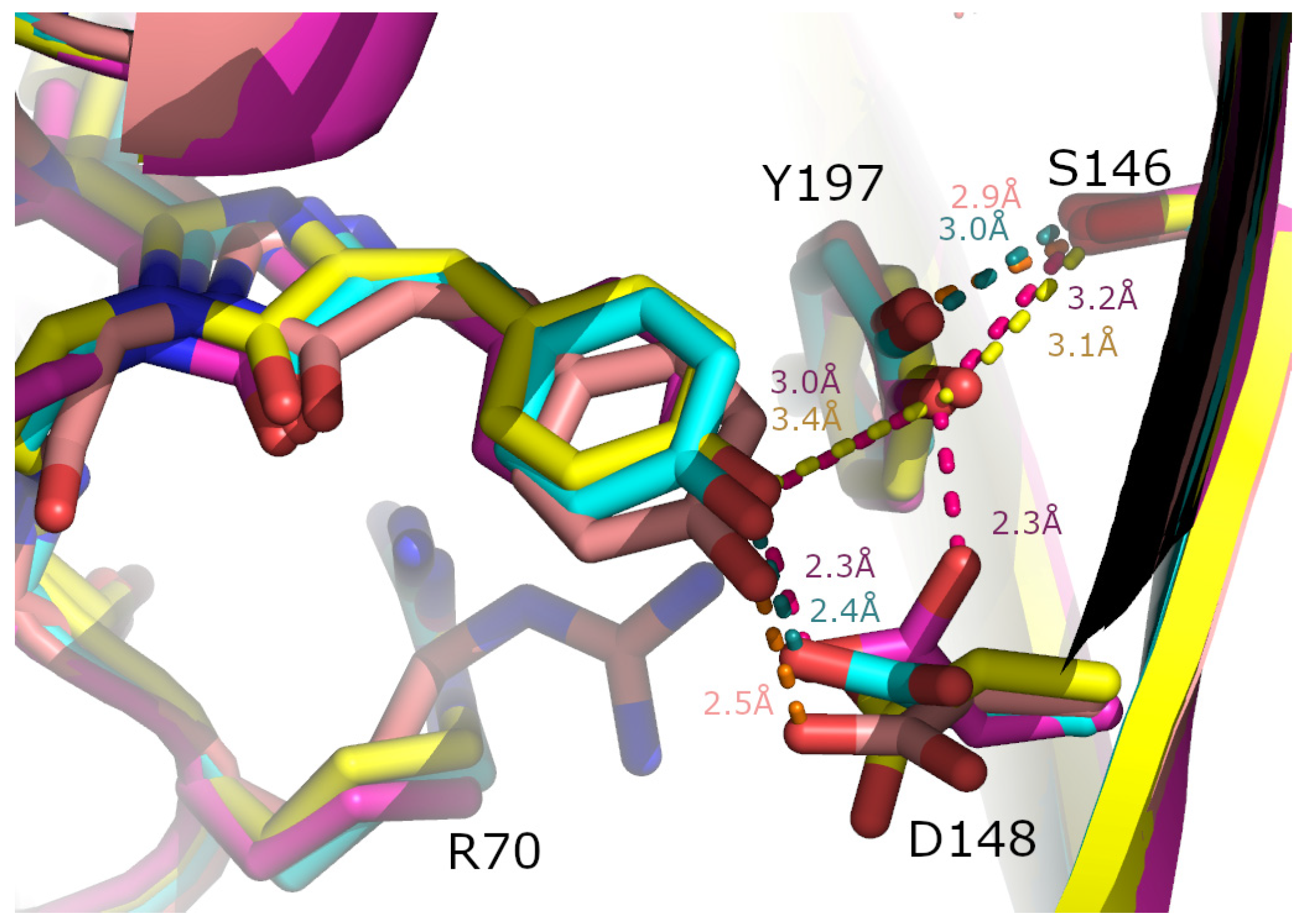
2. Results and Discussion
3. Materials and Methods
3.1. Site-Directed Mutagenesis
3.2. Protein Expression and Purification
3.3. Quantum Yield, pH Stability, Guanidine Hydrochloride Stability, and Thermostability
3.4. Crystallization
3.5. Data Collection and Processing
3.6. Structure Solution and Refinement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aliye, N.; Fabbretti, A.; Lupidi, G.; Tsekoa, T.; Spurio, R. Engineering Color Variants of Green Fluorescent Protein (GFP) for Thermostability, pH-Sensitivity, and Improved Folding Kinetics. Appl. Microbiol. Biotechnol. 2015, 99, 1205–1216. [Google Scholar] [CrossRef]
- Nguyen, A.W.; Daugherty, P.S. Evolutionary Optimization of Fluorescent Proteins for Intracellular FRET. Nat. Biotechnol. 2005, 23, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Holder, A.N.; Ellis, A.L.; Zou, J.; Chen, N.; Yang, J.J. Facilitating Chromophore Formation of Engineered Ca2+ Binding Green Fluorescent Proteins. Arch. Biochem. Biophys. 2009, 486, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. The Green Fluorescent Protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Costantini, L.M.; Baloban, M.; Markwardt, M.L.; Rizzo, M.A.; Guo, F.; Verkhusha, V.V.; Snapp, E.L. A Palette of Fluorescent Proteins Optimized for Diverse Cellular Environments. Nat. Commun. 2015, 6, 7670. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Shaner, N.C.; Yarbrough, C.A.; Tsien, R.Y.; Remington, S.J. Novel Chromophores and Buried Charges Control Color in mFruits. Biochemistry 2006, 45, 9639–9647. [Google Scholar] [CrossRef]
- Habuchi, S.; Ando, R.; Dedecker, P.; Verheijen, W.; Mizuno, H.; Miyawaki, A.; Hofkens, J. Reversible Single-Molecule Photoswitching in the GFP-like Fluorescent Protein Dronpa. Proc. Natl. Acad. Sci. USA 2005, 102, 9511–9516. [Google Scholar] [CrossRef]
- Jöhr, R.; Bauer, M.S.; Schendel, L.C.; Kluger, C.; Gaub, H.E. Dronpa: A Light-Switchable Fluorescent Protein for Opto-Biomechanics. Nano Lett. 2019, 19, 3176–3181. [Google Scholar] [CrossRef]
- Tubbs, J.L.; Tainer, J.A.; Getzoff, E.D. Crystallographic Structures of Discosoma Red Fluorescent Protein with Immature and Mature Chromophores: Linking Peptide Bond Trans−Cis Isomerization and Acylimine Formation in Chromophore Maturation. Biochemistry 2005, 44, 9833–9840. [Google Scholar] [CrossRef]
- Kiss, C.; Temirov, J.; Chasteen, L.; Waldo, G.S.; Bradbury, A.R.M. Directed Evolution of an Extremely Stable Fluorescent Protein. Protein. Eng. Des. Sel. 2009, 22, 313–323. [Google Scholar] [CrossRef]
- Dai, M.; Fisher, H.E.; Temirov, J.; Kiss, C.; Phipps, M.E.; Pavlik, P.; Werner, J.H.; Bradbury, A.R.M. The Creation of a Novel Fluorescent Protein by Guided Consensus Engineering. Protein. Eng. Des. Sel. 2007, 20, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Close, D.W.; Paul, C.D.; Langan, P.S.; Wilce, M.C.J.; Traore, D.A.K.; Halfmann, R.; Rocha, R.C.; Waldo, G.S.; Payne, R.J.; Rucker, J.B.; et al. Thermal Green Protein, an Extremely Stable, Nonaggregating Fluorescent Protein Created by Structure-Guided Surface Engineering. Proteins 2015, 83, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Ebisawa, T.; Yamamura, A.; Kameda, Y.; Hayakawa, K.; Nagata, K.; Tanokura, M. The Structure of mAG, a Monomeric Mutant of the Green Fluorescent Protein Azami-Green, Reveals the Structural Basis of Its Stable Green Emission. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 485–489. [Google Scholar] [CrossRef]
- Cai, H.; Yao, H.; Li, T.; Hutter, C.A.J.; Li, Y.; Tang, Y.; Seeger, M.A.; Li, D. An Improved Fluorescent Tag and Its Nanobodies for Membrane Protein Expression, Stability Assay, and Purification. Commun. Biol. 2020, 3, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Velappan, N.; Close, D.; Hung, L.-W.; Naranjo, L.; Hemez, C.; DeVore, N.; McCullough, D.K.; Lillo, A.M.; Waldo, G.S.; Bradbury, A.R.M. Construction, Characterization and Crystal Structure of a Fluorescent Single-Chain Fv Chimera. Protein Eng. Des. Sel. 2021, 34, gzaa029. [Google Scholar] [CrossRef]
- Anderson, M.R.; Padgett, C.M.; Dargatz, C.J.; Nichols, C.R.; Vittalam, K.R.; DeVore, N.M. Engineering a Yellow Thermostable Fluorescent Protein by Rational Design. ACS Omega 2023, 8, 436–443. [Google Scholar] [CrossRef]
- Jung, G.; Wiehler, J.; Zumbusch, A. The Photophysics of Green Fluorescent Protein: Influence of the Key Amino Acids at Positions 65, 203, and 222. Biophys. J. 2005, 88, 1932–1947. [Google Scholar] [CrossRef]
- Khrameeva, E.E.; Drutsa, V.L.; Vrzheshch, E.P.; Dmitrienko, D.V.; Vrzheshch, P.V. Mutants of Monomeric Red Fluorescent Protein mRFP1 at Residue 66: Structure Modeling by Molecular Dynamics and Search for Correlations with Spectral Properties. Biochemistry 2008, 73, 1085–1095. [Google Scholar] [CrossRef][Green Version]
- Don Paul, C.; Traore, D.A.K.; Olsen, S.; Devenish, R.J.; Close, D.W.; Bell, T.D.M.; Bradbury, A.; Wilce, M.C.J.; Prescott, M. X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein. PLoS ONE 2015, 10, e0123338. [Google Scholar] [CrossRef]
- Don Paul, C.; Kiss, C.; Traore, D.A.K.; Gong, L.; Wilce, M.C.J.; Devenish, R.J.; Bradbury, A.; Prescott, M. Phanta: A Non-Fluorescent Photochromic Acceptor for pcFRET. PLoS ONE 2013, 8, e75835. [Google Scholar] [CrossRef]
- Liu, Y.; Ke, M.; Gong, H. Protonation of Glu(135) Facilitates the Outward-to-Inward Structural Transition of Fucose Transporter. Biophys. J. 2015, 109, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Arunan, E.; Desiraju, G.R.; Klein, R.A.; Sadlej, J.; Scheiner, S.; Alkorta, I.; Clary, D.C.; Crabtree, R.H.; Dannenberg, J.J.; Hobza, P.; et al. Definition of the hydrogen bond (IUPAC Recommendations 2011). Pure Appl. Chem. 2011, 83, 1637–1641. [Google Scholar] [CrossRef]
- Wachter, R.M.; Elsliger, M.A.; Kallio, K.; Hanson, G.T.; Remington, S.J. Structural Basis of Spectral Shifts in the Yellow-Emission Variants of Green Fluorescent Protein. Structure 1998, 6, 1267–1277. [Google Scholar] [CrossRef]
- Battad, J.M.; Wilmann, P.G.; Olsen, S.; Byres, E.; Smith, S.C.; Dove, S.G.; Turcic, K.N.; Devenish, R.J.; Rossjohn, J.; Prescott, M. A Structural Basis for the pH-Dependent Increase in Fluorescence Efficiency of Chromoproteins. J. Mol. Biol. 2007, 368, 998–1010. [Google Scholar] [CrossRef]
- Andresen, M.; Stiel, A.C.; Trowitzsch, S.; Weber, G.; Eggeling, C.; Wahl, M.C.; Hell, S.W.; Jakobs, S. Structural Basis for Reversible Photoswitching in Dronpa. Proc. Natl. Acad. Sci. USA 2007, 104, 13005–13009. [Google Scholar] [CrossRef]
- Gotthard, G.; von Stetten, D.; Clavel, D.; Noirclerc-Savoye, M.; Royant, A. Chromophore Isomer Stabilization Is Critical to the Efficient Fluorescence of Cyan Fluorescent Proteins. Biochemistry 2017, 56, 6418–6422. [Google Scholar] [CrossRef]
- Agirre, J.; Atanasova, M.; Bagdonas, H.; Ballard, C.B.; Baslé, A.; Beilsten-Edmands, J.; Borges, R.J.; Brown, D.G.; Burgos-Mármol, J.J.; Berrisford, J.M.; et al. The CCP4 Suite: Integrative Software for Macromolecular Crystallography. Acta. Crystallogr. D Struct. Biol. 2023, 79, 449–461. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards Automated Crystallographic Structure Refinement with Phenix.Refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta. Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta. Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]



| TGP-E | YTP-E E148D | |
|---|---|---|
| Diffraction source | Rigaku XtaLab Synergy-S | Rigaku XtaLab Synergy-S |
| Wavelength (Å) | 1.54 | 1.54 |
| Temperature (K) | 100 | 100 |
| Detector | HyPix-6000HE | HyPix-6000HE |
| Crystal-to-detector distance (mm) | 36.34 | 36.34 |
| Rotation range per image (°) | 0.5 | 0.15 |
| Exposure time per image (s) | 45 | 21.29 |
| Space group | P1 | P 1 21 1 |
| a, b, c (Å) | 38.74, 49.06, 59.11 | 42.78, 137.78, 74.56 |
| α, β, γ (°) | 78.55, 72.08, 71.41 | 90.00, 97.71, 90.00 |
| Mosaicity (°) | 1.37 | 1.23 |
| Resolution range (Å) | 20.42–2.00 | 20.32–3.00 |
| Total no. of reflections | 149,158 (10,544) | 79,636 (15,409) |
| No. of unique reflections | 26,322 (1957) | 16,717 (2738) |
| Completeness (%) | 99.9 (100.0) | 98.6 (97.0) |
| Redundancy | 5.7 (5.4) | 4.8 (5.6) |
| <I/σ(I)> from merged data | 7.6 (2.1) | 11.5 (5.7) |
| CC1/2 | 0.990 (0.776) | 0.986 (0.970) |
| Rr.i.m. or Rmeas | 0.125 (0.552) | 0.051 (0.099) |
| Overall B factor from Wilson plot (Ã2) | 12.68 | 30.84 |
| TGP-E | YTP-E E148D | |
|---|---|---|
| Resolution range (Å) | 20.42–2.00 | 20.32–3.00 |
| Completeness (%) | 99.75 (100.0) | 97.37 (99.71) |
| No. of reflections, working set | 26,282 (2606) | 16,706 (1702) |
| No. of reflections, test set | 1248 (124) | 1671 (170) |
| Final Rcryst | 0.1692 (0.2078) | 0.2207 (0.3394) |
| Final Rfree | 0.2291 (0.2821) | 0.2807 (0.4392) |
| No. of non-H atoms | 3706 | 126 |
| Protein/nucleic acid | 3429 | 6787 |
| Ions | 0 | 0 |
| Ligands | 48 | 96 |
| Waters | 422 | 30 |
| Total | 3706 | 6913 |
| R.m.s. deviations from ideality | Molprobity validation | Molprobity validation |
| Bonds (Å) | 0.036 | 0.003 |
| Angles (°) | 2.1 | 0.67 |
| Average B factors (Å2) | 16.36 | 31.08 |
| Protein/nucleic acid | 16.05 | 31.04 |
| Ions | N/A | N/A |
| Ligands | 9.03 | 37.65 |
| Waters | 22.56 | 19.74 |
| Ramachandran plot | Molprobity validation | Molprobity validation |
| Favoured regions (%) | 99.52 | 96.26 |
| Outliers (%) | 0 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, M.R.; Padgett, C.M.; Ogbeifun, V.O.; DeVore, N.M. The Crystal Structure of Thermal Green Protein Q66E (TGP-E) and Yellow Thermostable Protein (YTP-E) E148D. SynBio 2024, 2, 298-310. https://doi.org/10.3390/synbio2030018
Anderson MR, Padgett CM, Ogbeifun VO, DeVore NM. The Crystal Structure of Thermal Green Protein Q66E (TGP-E) and Yellow Thermostable Protein (YTP-E) E148D. SynBio. 2024; 2(3):298-310. https://doi.org/10.3390/synbio2030018
Chicago/Turabian StyleAnderson, Matthew R., Caitlin M. Padgett, Victoria O. Ogbeifun, and Natasha M. DeVore. 2024. "The Crystal Structure of Thermal Green Protein Q66E (TGP-E) and Yellow Thermostable Protein (YTP-E) E148D" SynBio 2, no. 3: 298-310. https://doi.org/10.3390/synbio2030018
APA StyleAnderson, M. R., Padgett, C. M., Ogbeifun, V. O., & DeVore, N. M. (2024). The Crystal Structure of Thermal Green Protein Q66E (TGP-E) and Yellow Thermostable Protein (YTP-E) E148D. SynBio, 2(3), 298-310. https://doi.org/10.3390/synbio2030018








