Harnessing Naturally Occurring Bistable Switches for Their Application in Synthetic Biology
Abstract
:1. Introduction
2. Naturally Occurring Bistable Switches
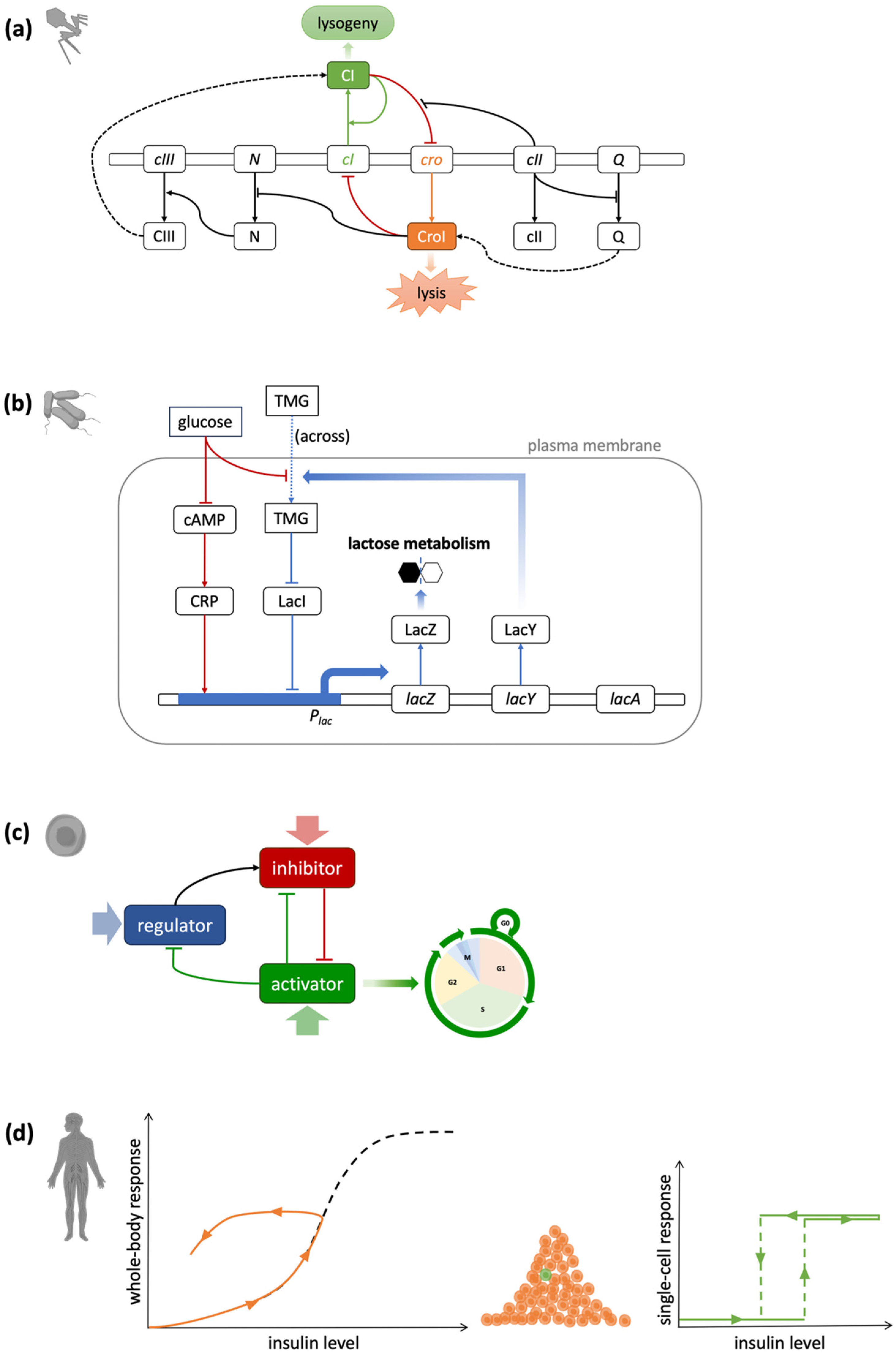
2.1. In Simple Organisms
2.2. In Eukaryotic Cells
2.3. In Multicellular Systems
2.4. Variation and Extension
3. Applications of Bistable Switches in Synthetic Biology
3.1. Design and Basic Applications
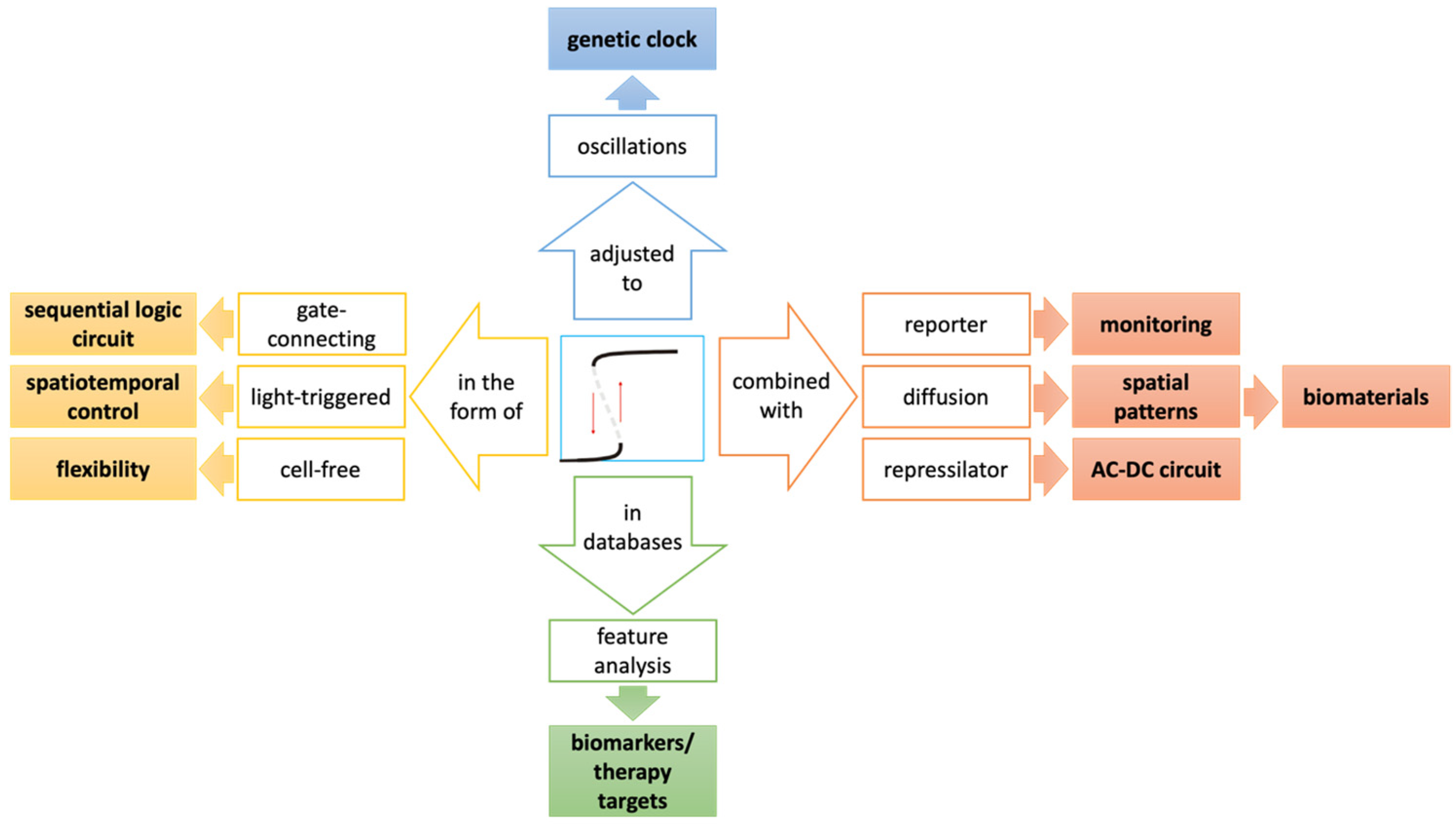
3.2. Expansion of Application
3.3. Improving the Performance of Bistable Switches
4. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Cannon, W.B. Organization for physiological homeostasis. Physiol. Rev. 1929, 9, 399–431. [Google Scholar] [CrossRef]
- Cannon, W.B. The Wisdom of the Body; W W Norton & Co: New York, NY, USA, 1932; p. 312. [Google Scholar]
- Wang, G. Optimal homeostasis necessitates bistable control. J. R. Soc. Interface 2012, 9, 2723–2734. [Google Scholar] [CrossRef] [PubMed]
- Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits; Chapman and Hall/CRC: Boca Raton, FL, USA, 2019. [Google Scholar]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in Bacteriophage Lambda Development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef] [PubMed]
- Cortes, M.G.; Lin, Y.; Zeng, L.; Balázsi, G. From Bench to Keyboard and Back Again: A Brief History of Lambda Phage Modeling. Annu. Rev. Biophys. 2021, 50, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Weitz, J.S.; Mileyko, Y.; Joh, R.I.; Voit, E.O. Collective decision making in bacterial viruses. Biophys. J. 2008, 95, 2673–2680. [Google Scholar] [CrossRef]
- Bednarz, M.; Halliday, J.A.; Herman, C.; Golding, I. Revisiting bistability in the lysis/lysogeny circuit of bacteriophage lambda. PLoS ONE 2014, 9, e100876. [Google Scholar] [CrossRef]
- Jacques, M. Recherches Sur la Croissance des Cultures Bacteriennes; Hermann and Cie: Paris, France, 1942; p. 2. [Google Scholar]
- Santillán, M.; Mackey, M.C. Quantitative approaches to the study of bistability in the lac operon of Escherichia coli. J. R. Soc. Interface 2008, 5 (Suppl. S1), S29–S39. [Google Scholar] [CrossRef]
- Ozbudak, E.M.; Thattai, M.; Lim, H.N.; Shraiman, B.I.; Van Oudenaarden, A. Multistability in the lactose utilization network of Escherichia coli. Nature 2004, 427, 737–740. [Google Scholar] [CrossRef]
- Díaz-Hernández, O.; Santillán, M. Bistable behavior of the lac operon in E. coli when induced with a mixture of lactose and TMG. Front. Physiol. 2010, 1, 22. [Google Scholar] [CrossRef]
- Ferrell, J.E. Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 2002, 14, 140–148. [Google Scholar] [CrossRef]
- Verdugo, A.; Vinod, P.; Tyson, J.J.; Novak, B. Molecular mechanisms creating bistable switches at cell cycle transitions. Open Biol. 2013, 3, 120179. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, M.; Tyson, J.J.; Novák, B. Cell-cycle transitions: A common role for stoichiometric inhibitors. Mol. Biol. Cell 2017, 28, 3437–3446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tyson, J.J.; Novák, B. Role for regulated phosphatase activity in generating mitotic oscillations in Xenopus cell-free extracts. Proc. Natl. Acad. Sci. USA 2013, 110, 20539–20544. [Google Scholar] [CrossRef] [PubMed]
- Dragoi, C.-M.; Kaur, E.; Barr, A.R.; Tyson, J.J.; Novák, B. The oscillation of mitotic kinase governs cell cycle latches in mammalian cells. J. Cell Sci. 2024, 137, 2805–2816. [Google Scholar] [CrossRef] [PubMed]
- Stallaert, W.; Kedziora, K.M.; Chao, H.X.; Purvis, J.E. Bistable switches as integrators and actuators during cell cycle progression. FEBS Lett. 2019, 593, 2805–2816. [Google Scholar] [CrossRef]
- Rombouts, J.; Gelens, L. Dynamic bistable switches enhance robustness and accuracy of cell cycle transitions. PLoS Comput. Biol. 2021, 17, e1008231. [Google Scholar] [CrossRef]
- Xiong, W.; Ferrell, J.E. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 2003, 426, 460–465. [Google Scholar] [CrossRef]
- Kueh, H.Y.; Champhekar, A.; Nutt, S.L.; Elowitz, M.B.; Rothenberg, E.V. Positive feedback between PU. 1 and the cell cycle controls myeloid differentiation. Science 2013, 341, 670–673. [Google Scholar] [CrossRef]
- Fregoso Lomas, M.; De Vito, S.; Boisclair Lachance, J.-F.; Houde, J.; Nilson, L.A. Determination of EGFR Signaling Output by Opposing Gradients of BMP and JAK/STAT Activity. Curr. Biol. CB 2016, 26, 2572–2582. [Google Scholar] [CrossRef]
- Cantoria, M.J.; Alizadeh, E.; Ravi, J.; Varghese, R.P.; Bunnag, N.; Pond, K.W.; Kettenbach, A.N.; Ahmed, Y.; Paek, A.L.; Tyson, J.J. Feedback in the β-catenin destruction complex imparts bistability and cellular memory. Proc. Natl. Acad. Sci. USA 2023, 120, e2208787120. [Google Scholar] [CrossRef]
- Halder, S.; Ghosh, S.; Chattopadhyay, J.; Chatterjee, S. Bistability in cell signalling and its significance in identifying potential drug-targets. Bioinformatics 2021, 37, 4156–4163. [Google Scholar] [CrossRef] [PubMed]
- Engbers, J.D.T.; Fernandez, F.R.; Turner, R.W. Bistability in Purkinje neurons: Ups and downs in cerebellar research. Neural Netw. 2013, 47, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Buchin, A.; Rieubland, S.; Häusser, M.; Gutkin, B.S.; Roth, A. Inverse Stochastic Resonance in Cerebellar Purkinje Cells. PLoS Comput. Biol. 2016, 12, e1005000. [Google Scholar] [CrossRef] [PubMed]
- Hahn, G.; Kumar, A.; Schmidt, H.; Knösche, T.R.; Deco, G. Rate and oscillatory switching dynamics of a multilayer visual microcircuit model. eLife 2022, 11, e77594. [Google Scholar] [CrossRef] [PubMed]
- Ron Mizrachi, B.; Tendler, A.; Karin, O.; Milo, T.; Haran, D.; Mayo, A.; Alon, U. Major depressive disorder and bistability in an HPA-CNS toggle switch. PLoS Comput. Biol. 2023, 19, e1011645. [Google Scholar] [CrossRef]
- Wang, G.; Krueger, G.R. Computational analysis of mTOR signaling pathway: Bifurcation, carcinogenesis, and drug discovery. Anticancer Res. 2010, 30, 2683–2688. [Google Scholar]
- Wang, G. Singularity analysis of the AKT signaling pathway reveals connections between cancer and metabolic diseases. Phys. Biol. 2010, 7, 046015. [Google Scholar] [CrossRef]
- Wang, G. Raison d’être of insulin resistance: The adjustable threshold hypothesis. J. R. Soc. Interface 2014, 11, 20140892. [Google Scholar] [CrossRef]
- Akhtar, J.; Han, Y.; Han, S.; Lin, W.; Cao, C.; Ge, R.; Babarinde, I.A.; Jia, Q.; Yuan, Y.; Chen, G.; et al. Bistable insulin response: The win-win solution for glycemic control. IScience 2022, 25, 105561. [Google Scholar] [CrossRef]
- Kaimachnikov, N.P.; Kholodenko, B.N. Toggle switches, pulses and oscillations are intrinsic properties of the Src activation/deactivation cycle. FEBS J. 2009, 276, 4102–4118. [Google Scholar] [CrossRef]
- Ehrmann, A.; Nguyen, B.; Seifert, U. Interlinked GTPase cascades provide a motif for both robust switches and oscillators. J. R. Soc. Interface 2019, 16, 20190198. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Walker, B.L.; Iannaccone, S.; Bhatt, D.; Kennedy, P.J.; Tse, W.T. Bistable switches control memory and plasticity in cellular differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 6638–6643. [Google Scholar] [CrossRef] [PubMed]
- Doncic, A.; Atay, O.; Valk, E.; Grande, A.; Bush, A.; Vasen, G.; Colman-Lerner, A.; Loog, M.; Skotheim, J.M. Compartmentalization of a bistable switch enables memory to cross a feedback-driven transition. Cell 2015, 160, 1182–1195. [Google Scholar] [CrossRef]
- Zaytsev, A.V.; Segura-Pena, D.; Godzi, M.; Calderon, A.; Ballister, E.R.; Stamatov, R.; Mayo, A.M.; Peterson, L.; Black, B.E.; Ataullakhanov, F.I. Bistability of a coupled Aurora B kinase-phosphatase system in cell division. eLife 2016, 5, e10644. [Google Scholar] [CrossRef] [PubMed]
- Heldt, F.S.; Tyson, J.J.; Cross, F.R.; Novák, B. A Single Light-Responsive Sizer Can Control Multiple-Fission Cycles in Chlamydomonas. Curr. Biol. CB 2020, 30, 634–644.e7. [Google Scholar] [CrossRef] [PubMed]
- Frolov, V.A.; Lizunov, V.A.; Dunina-Barkovskaya, A.Y.; Samsonov, A.V.; Zimmerberg, J. Shape bistability of a membrane neck: A toggle switch to control vesicle content release. Proc. Natl. Acad. Sci. USA 2003, 100, 8698–8703. [Google Scholar] [CrossRef]
- Perez-Carrasco, R.; Guerrero, P.; Briscoe, J.; Page, K.M. Intrinsic noise profoundly alters the dynamics and steady state of morphogen-controlled bistable genetic switches. PLoS Comput. Biol. 2016, 12, e1005154. [Google Scholar] [CrossRef]
- Shah, N.A.; Sarkar, C.A. Robust network topologies for generating switch-like cellular responses. PLoS Comput. Biol. 2011, 7, e1002085. [Google Scholar] [CrossRef]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Y.; Cai, Z. Development of a stress-induced mutagenesis module for autonomous adaptive evolution of Escherichia coli to improve its stress tolerance. Biotechnol. Biofuels 2015, 8, 93. [Google Scholar] [CrossRef]
- Grant, P.K.; Szep, G.; Patange, O.; Halatek, J.; Coppard, V.; Csikász-Nagy, A.; Haseloff, J.; Locke, J.C.W.; Dalchau, N.; Phillips, A. Interpretation of morphogen gradients by a synthetic bistable circuit. Nat. Commun. 2020, 11, 5545. [Google Scholar] [CrossRef] [PubMed]
- Kramer, B.P.; Viretta, A.U.; Baba, M.D.-E.; Aubel, D.; Weber, W.; Fussenegger, M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004, 22, 867–870. [Google Scholar] [CrossRef]
- Becskei, A.; Séraphin, B.; Serrano, L. Positive feedback in eukaryotic gene networks: Cell differentiation by graded to binary response conversion. EMBO J. 2001, 20, 2528–2535. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Nemhauser, J.L.; Klavins, E. Synthetic Bistability and Differentiation in Yeast. ACS Synth. Biol. 2019, 8, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Lebar, T.; Bezeljak, U.; Golob, A.; Jerala, M.; Kadunc, L.; Pirš, B.; Stražar, M.; Vučko, D.; Zupančič, U.; Benčina, M. A bistable genetic switch based on designable DNA-binding domains. Nat. Commun. 2014, 5, 5007. [Google Scholar] [CrossRef]
- Oyarzún, D.A.; Chaves, M. Design of a bistable switch to control cellular uptake. J. R. Soc. Interface 2015, 12, 20150618. [Google Scholar] [CrossRef]
- Hsu, C.; Jaquet, V.; Gencoglu, M.; Becskei, A. Protein Dimerization Generates Bistability in Positive Feedback Loops. Cell Rep. 2016, 16, 1204–1210. [Google Scholar] [CrossRef]
- Chen, D.; Arkin, A.P. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 2012, 8, 620. [Google Scholar] [CrossRef]
- Shopera, T.; Henson, W.R.; Ng, A.; Lee, Y.J.; Ng, K.; Moon, T.S. Robust, tunable genetic memory from protein sequestration combined with positive feedback. Nucleic Acids Res. 2015, 43, 9086–9094. [Google Scholar] [CrossRef]
- Mishra, D.; Bepler, T.; Teague, B.; Berger, B.; Broach, J.; Weiss, R. An engineered protein-phosphorylation toggle network with implications for endogenous network discovery. Science 2021, 373, eaav0780. [Google Scholar] [CrossRef]
- Kotula, J.W.; Kerns, S.J.; Shaket, L.A.; Siraj, L.; Collins, J.J.; Way, J.C.; Silver, P.A. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. USA 2014, 111, 4838–4843. [Google Scholar] [CrossRef] [PubMed]
- Andrews, L.B.; Nielsen, A.A.; Voigt, C.A. Cellular checkpoint control using programmable sequential logic. Science 2018, 361, eaap8987. [Google Scholar] [CrossRef] [PubMed]
- Perez-Carrasco, R.; Barnes, C.P.; Schaerli, Y.; Isalan, M.; Briscoe, J.; Page, K.M. Combining a Toggle Switch and a Repressilator within the AC-DC Circuit Generates Distinct Dynamical Behaviors. Cell Syst. 2018, 6, 521–530.e3. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.J.; Albert, R.; Goldbeter, A.; Ruoff, P.; Sible, J. Biological switches and clocks. J. R. Soc. Interface 2008, 5 (Suppl. S1), S1–S8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, Y.; Feng, Y.; Klepin, S.; Tsimring, L.S.; Pillus, L.; Hasty, J.; Hao, N. Engineering longevity-design of a synthetic gene oscillator to slow cellular aging. Science 2023, 380, 376–381. [Google Scholar] [CrossRef]
- Gomez, M.M.; Arcak, M. A Tug-of-War Mechanism for Pattern Formation in a Genetic Network. ACS Synth. Biol. 2017, 6, 2056–2066. [Google Scholar] [CrossRef]
- Barbier, I.; Perez-Carrasco, R.; Schaerli, Y. Controlling spatiotemporal pattern formation in a concentration gradient with a synthetic toggle switch. Mol. Syst. Biol. 2020, 16, e9361. [Google Scholar] [CrossRef]
- Müller, K.; Zurbriggen, M.D.; Weber, W. Control of gene expression using a red- and far-red light-responsive bi-stable toggle switch. Nat. Protoc. 2014, 9, 622–632. [Google Scholar] [CrossRef]
- Müller, K.; Engesser, R.; Metzger, S.; Schulz, S.; Kämpf, M.M.; Busacker, M.; Steinberg, T.; Tomakidi, P.; Ehrbar, M.; Nagy, F.; et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013, 41, e77. [Google Scholar] [CrossRef]
- van Sluijs, B.; Maas, R.J.M.; van der Linden, A.J.; de Greef, T.F.A.; Huck, W.T.S. A microfluidic optimal experimental design platform for forward design of cell-free genetic networks. Nat. Commun. 2022, 13, 3626. [Google Scholar] [CrossRef]
- Subsoontorn, P.; Kim, J.; Winfree, E. Ensemble Bayesian analysis of bistability in a synthetic transcriptional switch. ACS Synth. Biol. 2012, 1, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Matsuyama, S.; Kitano, H. Large-scale analysis of network bistability for human cancers. PLoS Comput. Biol. 2010, 6, e1000851. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, L.; Chan, C. Identification of novel targets for breast cancer by exploring gene switches on a genome scale. BMC Genom. 2011, 12, 547. [Google Scholar] [CrossRef] [PubMed]
- Crespo, I.; Del Sol, A. A general strategy for cellular reprogramming: The importance of transcription factor cross-repression. Stem Cells 2013, 31, 2127–2135. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, H.; Shi, H.; Ji, W.; Feng, J.; Gong, Y.; Yang, Z.; Ouyang, Q. Automated design of genetic toggle switches with predetermined bistability. ACS Synth. Biol. 2012, 1, 284–290. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, Q.; Wang, X. Design of Adjacent Transcriptional Regions to Tune Gene Expression and Facilitate Circuit Construction. Cell Syst. 2018, 6, 206–215.e6. [Google Scholar] [CrossRef]
- Gyorgy, A. Context-Dependent Stability and Robustness of Genetic Toggle Switches with Leaky Promoters. Life 2021, 11, 1150. [Google Scholar] [CrossRef]
- Yong, C.; Gyorgy, A. Stability and Robustness of Unbalanced Genetic Toggle Switches in the Presence of Scarce Resources. Life 2021, 11, 271. [Google Scholar] [CrossRef]
- Melendez-Alvarez, J.R.; Tian, X.-J. Emergence of qualitative states in synthetic circuits driven by ultrasensitive growth feedback. PLoS Comput. Biol. 2022, 18, e1010518. [Google Scholar] [CrossRef]
- Zhang, R.; Li, J.; Melendez-Alvarez, J.; Chen, X.; Sochor, P.; Goetz, H.; Zhang, Q.; Ding, T.; Wang, X.; Tian, X.-J. Topology-dependent interference of synthetic gene circuit function by growth feedback. Nat. Chem. Biol. 2020, 16, 695–701. [Google Scholar] [CrossRef]
- Lyons, S.M.; Xu, W.; Medford, J.; Prasad, A. Loads bias genetic and signaling switches in synthetic and natural systems. PLoS Comput. Biol. 2014, 10, e1003533. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, M. Tunable ultrasensitivity: Functional decoupling and biological insights. Sci. Rep. 2016, 6, 20345. [Google Scholar] [CrossRef] [PubMed]
- Santos-Moreno, J.; Tasiudi, E.; Stelling, J.; Schaerli, Y. Multistable and dynamic CRISPRi-based synthetic circuits. Nat. Commun. 2020, 11, 2746. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; del Rio-Salgado, J.M.; Garcia-Ojalvo, J.; Elowitz, M.B. Synthetic multistability in mammalian cells. Science 2022, 375, eabg9765. [Google Scholar] [CrossRef] [PubMed]
- Mileyko, Y.; Joh, R.I.; Weitz, J.S. Small-scale copy number variation and large-scale changes in gene expression. Proc. Natl. Acad. Sci. USA 2008, 105, 16659–16664. [Google Scholar] [CrossRef]
- Lee, J.W.; Gyorgy, A.; Cameron, D.E.; Pyenson, N.; Choi, K.R.; Way, J.C.; Silver, P.A.; Del Vecchio, D.; Collins, J.J. Creating Single-Copy Genetic Circuits. Mol. Cell 2016, 63, 329–336. [Google Scholar] [CrossRef]
- Salzano, D.; Fiore, D.; di Bernardo, M. Ratiometric control of cell phenotypes in monostrain microbial consortia. J. R. Soc. Interface 2022, 19, 20220335. [Google Scholar] [CrossRef]
- Sadeghpour, M.; Veliz-Cuba, A.; Orosz, G.; Josić, K.; Bennett, M.R. Bistability and oscillations in co-repressive synthetic microbial consortia. Quant. Biol. 2017, 5, 55–66. [Google Scholar] [CrossRef]
- Ishimatsu, K.; Hata, T.; Mochizuki, A.; Sekine, R.; Yamamura, M.; Kiga, D. General applicability of synthetic gene-overexpression for cell-type ratio control via reprogramming. ACS Synth. Biol. 2014, 3, 638–644. [Google Scholar] [CrossRef]
- Sekine, R.; Yamamura, M.; Ayukawa, S.; Ishimatsu, K.; Akama, S.; Takinoue, M.; Hagiya, M.; Kiga, D. Tunable synthetic phenotypic diversification on Waddington’s landscape through autonomous signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 17969–17973. [Google Scholar] [CrossRef]
- Lugagne, J.-B.; Sosa Carrillo, S.; Kirch, M.; Köhler, A.; Batt, G.; Hersen, P. Balancing a genetic toggle switch by real-time feedback control and periodic forcing. Nat. Commun. 2017, 8, 1671. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.K.; Narang, A. Inducer exclusion, by itself, cannot account for the glucose-mediated lac repression of Escherichia coli. Biophys. J. 2022, 121, 820–829. [Google Scholar] [CrossRef] [PubMed]

| Natural Bistable Switches | Synthetic Bistable Switches | Main Differences | |
|---|---|---|---|
| Structural functional basis |
|
| In natural life, bistable switches based on protein–protein interactions are very common; in synthetic biology, most bistable switches are currently implemented through genetic regulation. |
| Topology complexities |
|
| The complexity of natural bistable switches is much higher than that of synthetic bistable switches. |
| Adjustment levels |
|
| The regulation scope of natural bistable switches is much wider than that of current synthetic bistable switches. |
| Functions |
|
| The application of synthetic bistable switches focuses on further regulating and changing the functional state of organisms, or giving organisms additional new functions. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huan, M.; Wang, G. Harnessing Naturally Occurring Bistable Switches for Their Application in Synthetic Biology. SynBio 2024, 2, 363-377. https://doi.org/10.3390/synbio2040023
Huan M, Wang G. Harnessing Naturally Occurring Bistable Switches for Their Application in Synthetic Biology. SynBio. 2024; 2(4):363-377. https://doi.org/10.3390/synbio2040023
Chicago/Turabian StyleHuan, Ma, and Guanyu Wang. 2024. "Harnessing Naturally Occurring Bistable Switches for Their Application in Synthetic Biology" SynBio 2, no. 4: 363-377. https://doi.org/10.3390/synbio2040023
APA StyleHuan, M., & Wang, G. (2024). Harnessing Naturally Occurring Bistable Switches for Their Application in Synthetic Biology. SynBio, 2(4), 363-377. https://doi.org/10.3390/synbio2040023






