Can Osmopriming Induce Cross-Tolerance for Abiotic Stresses in Solanum paniculatum L. Seeds? A Transcriptome Analysis Point of View
Abstract
:1. Introduction
2. Materials and Methods
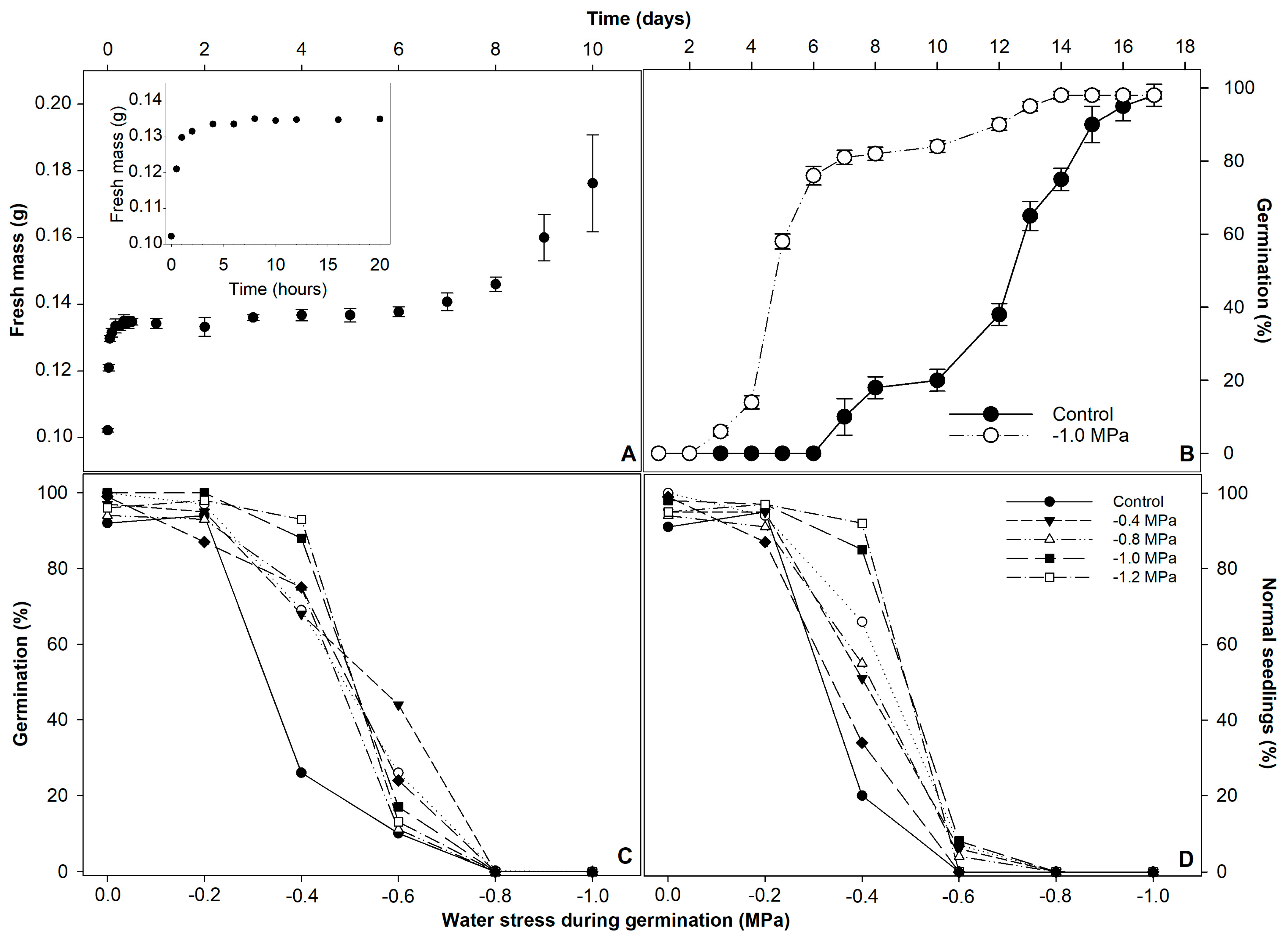
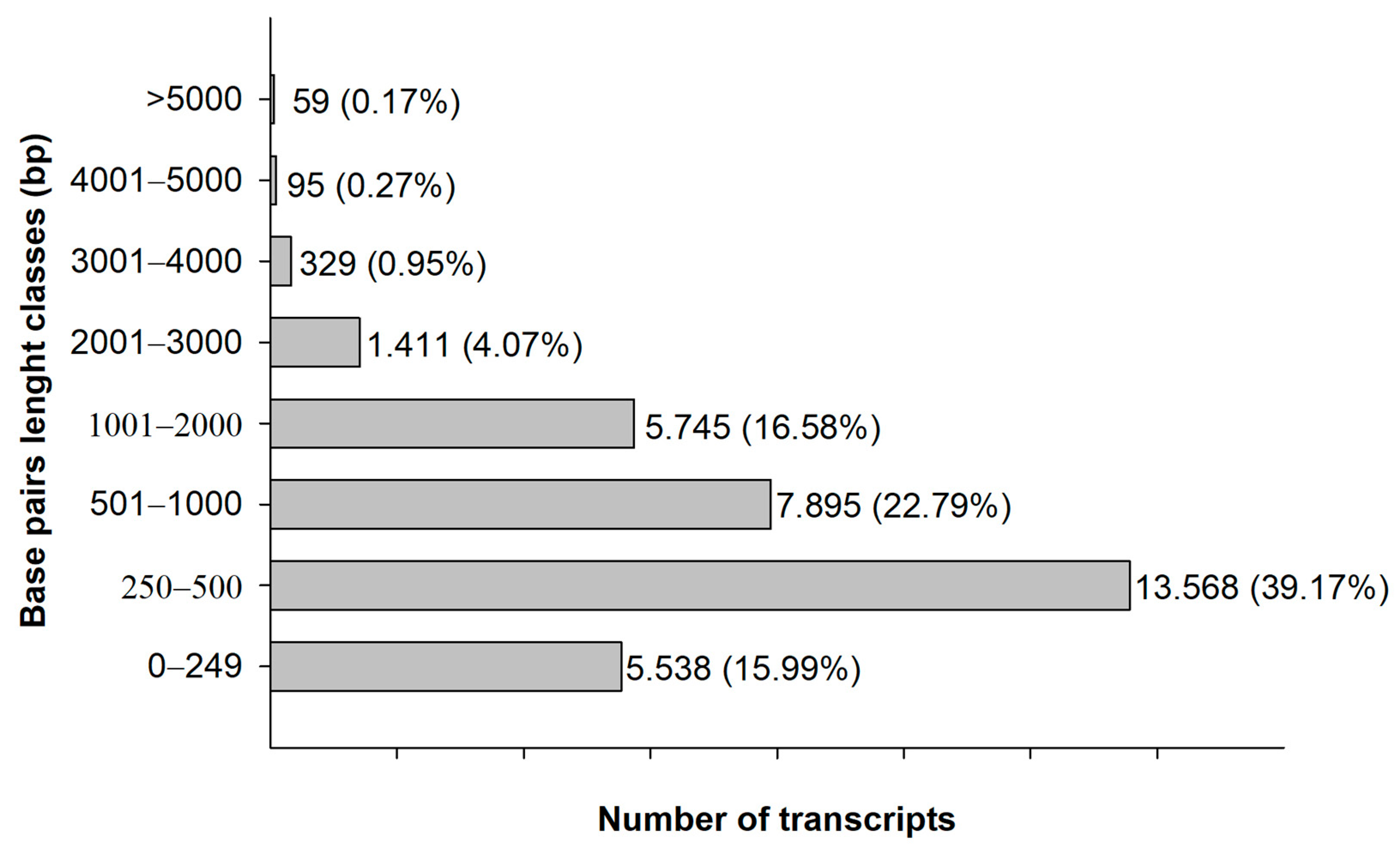
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, A.S.; Men, S.; Wang, L. Chiling and drought stress in crop plants: Implications, cross talk and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef]
- Vidal, M.C.; Stacciarini-Seraphin, E.; Camara, H.H.L.L. Crescimento de plântulas de Solanum lycocarpum St. Hil. (Lobeira) em casa de vegetação. Acta Botânica Brasílica 1999, 13, 271–274. [Google Scholar] [CrossRef]
- Lorenzi, H.; Matos, F.J.A. Plantas Medicinais No Brasil: Nativas E Exóticas, 3rd ed.; Instituto Plantarum: Nova Odessa, Brazil, 2021; p. 576. [Google Scholar]
- Garcia, J.; Jacobson, T.K.B.; Farias, J.G.; Boaventura, R.F. Efectiveness of methods to increase the germination rate of Jurubeba (Solanum paniculatum L.) seeds. Pesqui. Agropecuária Trop. 2008, 38, 223–226. [Google Scholar]
- Martins, A.F. Contribuição Ao Estudo Das Plantas Medicinais: Manual de Preservação Da Natureza; Cabral Editora Universitária: São Paulo, Brazil, 1998. [Google Scholar]
- Cowan, M.F.; Blomstedt, C.K.; Norton, S.L.; Henry, R.J.; Moller, B.L.; Gleadow, R. Crop wild relatives as a genetic resource for generating low-cyanide, drought-tolerant Sorghum. Environ. Exp. Bot. 2020, 169, 103884. [Google Scholar] [CrossRef]
- Coyne, C.J.; Kumar, S.; Von Wettberg, E.J.B.; Marques, E.; Berger, J.D.; Redden, R.J.; Noel Ellis, T.H.; Brus, J.; Zablastzká, L.; Smykal, P. Potentials and limitis of exploitation of crop wild relatives for pea, lentil and chickpea improvement. Legume Sci. 2020, 2, e36. [Google Scholar] [CrossRef]
- Quezada-Martinez, D.; Nyarko, C.P.A.; Schiessi, S.V.; Mason, A.S. Using wild relatives and related species to build climate resilience in Brassica crops. Theor. Appl. Genet. 2021, 134, 1711–1728. [Google Scholar] [CrossRef]
- Kranner, I. What is stress? Concepts, definitions and application in seed science. New Phytol. 2010, 188, 655–673. [Google Scholar] [CrossRef]
- Bowler, C.; Fluhr, R. The role of calcium and activated oxygen as signals for controlling cross-tolerance. Trends Plant Sci. 2000, 5, 241–246. [Google Scholar] [CrossRef]
- Pastori, G.M.; Foyer, C.H. Common components, networks, and pathways of cross-tolerance to stress: The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 2002, 129, 460–468. [Google Scholar] [CrossRef]
- Capiati, D.A.; País, S.M.; Téllez-Iñón, M.T. Wounding increases salt tolerance in tomato plants: Evidence on the participation of calmodulin-like activities in cross-tolerance signaling. J. Exp. Bot. 2006, 57, 2391–2400. [Google Scholar] [CrossRef]
- Bruce, T.J.A.; Matthes, M.C.; Napier, J.A.; Pickett, J.A. Stressful ‘‘memories’’ of plants: Evidence and possible mechanisms. Plant Sci. 2007, 173, 603–608. [Google Scholar] [CrossRef]
- Balbinot, E.; Lopes, H.M. Efeitos do condicionamento fisiológico e da secagem na germinação e no vigor de sementes de cenoura. Rev. Bras. De Sementes 2006, 1, 1–8. [Google Scholar] [CrossRef]
- Patanèa, C.; Cavallaroa, V.; Cosentino, S.L. Germination and radicle growth in unprimed and primed seeds of sweet sorghum as affected by reduced water potential in NaCl at different temperatures. Ind. Crops Prod. 2008, 30, 1–8. [Google Scholar] [CrossRef]
- Hussain, S.; Hussain, S.; Khaliq, A.; Ali, S.; Khan, I. Physiological, biochemical and molecular aspects of seed priming. In Priming and Pre-Treatment of Seeds and Seedlings, 1st ed.; Hasanuzzaman, M., Fotopoulos, V., Eds.; Springer: Singapore, 2019; p. 604. [Google Scholar]
- Heydecker, W.; Higgins, J.; Gulliver, R.L. Accelerated germination by osmotic seed treatment. Nature 1973, 246, 42–44. [Google Scholar] [CrossRef]
- Sher, A.; Sarwar, T.; Nawaz, A.; Ijaz, M.; Sattar, A.; Ahmad, S. Methods of Seed Priming. In Priming and Pre-Treatment of Seeds and Seedlings, 1st ed.; Hasanuzzaman, M., Fotopoulos, V., Eds.; Springer: Singapore, 2019; p. 604. [Google Scholar]
- Adnan, M.; Abd-Ur-Rahman, H.; Asif, M.; Hussain, M.; Bilal, H.M.; Adnan, M.; Ur-Rehman, F.; Ahmad, S.; Khalid, M. Seed priming; An effective way to improve plant growth. EC Agric. 2020, 6, 1–5. [Google Scholar]
- Bradford, K.J. Manipulation of seed water relations via osmotic priming to improve germination under stress conditions. HortScience 1986, 21, 1105–1112. [Google Scholar] [CrossRef]
- ISTA. International Rules for Seed Testing; International Seed Testing Association: Zurich, Switzerland, 2004. [Google Scholar]
- Michel, B.E. Evaluation of the Water Potentials of Solutions of Polyethylene Glycol 8000. Plant Physiol. 1983, 72, 66–70. [Google Scholar] [CrossRef]
- Villela, F.A.; Beckert, O.P. Potencial osmótico de soluções aquosas de polietileno glicol 8000. Rev. Bras. De Sementes 2001, 23, 267–275. [Google Scholar] [CrossRef]
- Baggerley, K.; Deng, L.; Morris, J.; Aldaz, C. Differential expression in SAGE: Accounting for normal between-library variation. Bioinformatics 2003, 19, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.O.; Zhao, S.O.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Sehar, S.; Perveen, M.; Gelani, S.; Basra, S.M.A.; Farooq, M. Seed pretreatment with hydrogen peroxide improves heat tolerance in maize at germination and seedling growth stages. Seed Sci. Technol. 2008, 36, 633–645. [Google Scholar] [CrossRef]
- Song, L.; Prince, S.; Valliyodan, B.; Joshi, T.; Maldonado Dos Santos, J.V.; Wang, J.; Lin, L.; Wan, J.; Wang, Y.; Xu, D.; et al. Genome-wide transcriptome analysis of soybean primary root under varying water deficit conditions. BMC Genom. 2016, 17, 57. [Google Scholar] [CrossRef]
- Osthoff, A.; Donà Dalle Rose, P.; Baldauf, J.A.; Piepho, H.P.; Hochholdinger, F. Transcriptomic reprogramming of barley seminal roots by combined water deficit and salt stress. BMC Genom. 2019, 20, 325. [Google Scholar] [CrossRef]
- Gao, G.; Hu, J.; Zhang, X.; Zhang, F.; Li, M.; Wu, X. Transcriptome analysis reveals genes expression pattern of seed response to heat stress in Brassica napus L. Oil Crop. Sci. 2021, 6, 87–96. [Google Scholar] [CrossRef]
- Song, J.; Keppler, B.D.; Wise, R.R.; Bent, A.F. PARP2 Is the Predominant Poly (ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. PLoS Genet. 2015, 11, e1005200. [Google Scholar] [CrossRef] [PubMed]
- Ventura, L.; Dona, M.; Macovei, A.; Carbonera, D.; Buttafava, A.; Mondoni, A.; Rossi, G.; Balestrazzi, A. Understanding the molecular pathways associated with seed vigor. Plant Physiol. Biochem. 2012, 60, 196–206. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, Y.; Pu, Z.; Wang, J.; Zheng, Y.; Li, Y.; Wei, Y. Regulation, evolution, and functionality of flavonoids in cereal crops. Biotechnol. Lett. 2013, 35, 1765–1780. [Google Scholar] [CrossRef]
- Peterbauer, T.; Lahuta, L.B.; Blochl, A.; Mucha, J.; Jones, D.A.; Hedley, C.L.; Gorecki, R.J.; Richter, A. Analysis of the raffinose family oligosaccharide pathway in pea seeds with contrasting carbohydrate composition. Plant Physiol. 2001, 127, 1764–1772. [Google Scholar] [CrossRef] [PubMed]
- Philippe, R.N.; Ralph, S.G.; Bohlmann, S.D.; Mansfield, J. Transcriptome profiles of hybrid poplar (Populus trichocarpa deltoides) reveal rapid changes in undamaged, systemic sink leaves after simulated feeding by forest tent caterpillar (Malacosoma disstria). New Phytol. 2010, 188, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef]
- Su, P.H.; Li, H.M. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 2010, 22, 1516–1531. [Google Scholar] [CrossRef]
- Prieto-Dapena, P.; Castaño, R.; Almoguera, C.; Jordano, J. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol. 2006, 142, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.M.; Nonogaki, H. Seeds: Physiology of Development, Germination and Dormancy; Springer: New York, NY, USA, 2013; 392p. [Google Scholar]
- Zhao, Z.; Zhang, W.; Yan, J.; Zhang, J.; Liu, Z.L.X.; Yi, Y. Over-expression of Arabidopsis DnaJ (Hsp40) contributes to NaCl stress tolerance. Afr. J. Biotechnol. 2010, 9, 972–978. [Google Scholar] [CrossRef]
- Sung, D.Y.; Guy, C.L. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol. 2003, 132, 979–987. [Google Scholar] [CrossRef]
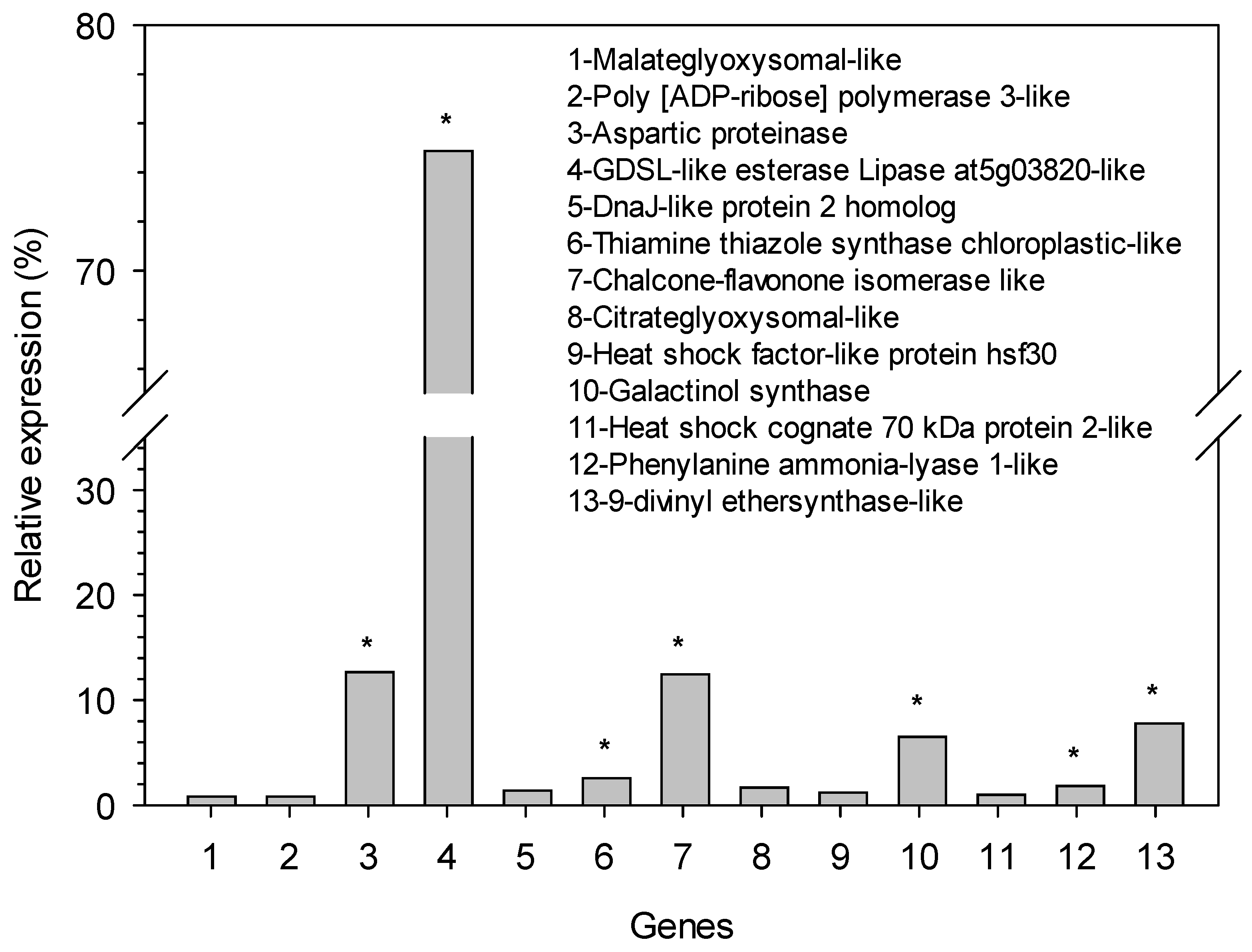
| Contig | Functional Description of Transcript | Size (bp) | Primers Forward (5′-3′) | Primers Reverse (5′-3′) |
|---|---|---|---|---|
| 339 | MALATEGLYOXYSOMAL-LIKE | 1168 | TCCACAACTATGCCAACTTCC | TTTCTCTGCCCTCTCAAACAC |
| 1056 | POLY [ADP-RIBOSE] POLYMERASE 3-LIKE | 2445 | TCCACAACTATGCCAACTTCC | TTTCTCTGCCCTCTCAAACAC |
| 2688 | ASPARTIC PROTEINASE | 1599 | TCAACCGAAACACAAAGGAAG | TTTACCACCGATCAGAACATCA |
| 3417 | GDSL-LIKE ESTERASE LIPASE AT5G03820-LIKE | 1199 | ATGCCTCAACATTGAAGCCT | AGAGCCTTCCCAACAAGATG |
| 3929 | DNAJ-LIKE PROTEIN 2 HOMOLOG | 1168 | ATATTTGTTCCGAGTGCCGA | GTAACATCCCTTTCTCAACTTTCA |
| 7098 | THIAMINE THIAZOLE SYNTHASE CHLOROPLASTIC-LIKE | 1094 | AACCCGTTAAATCAACTCACCA | CGTCATTTCCCTAGCAACAATC |
| 10,620 | CHALCONE-FLAVONONE ISOMERASE LIKE | 545 | AAGAATGAAGTGATGGTGGATGA | CTATGTCTGTTATTCCATGTCCCA |
| 9460 | CITRATEGLYOXYSOMAL-LIKE | 389 | CCAGAGTTTATTGAGGGCGT | CTTCTTCAGCAAGCTTCTTAATCA |
| 6576 | HEAT SHOCK FACTOR-LIKE PROTEIN HSF30 | 595 | AGAAAGCAGTATCCACAGCAA | TTAGCCTCAGTATTTCCATCCTC |
| 14,206 | GALACTINOL SYNTHASE | 637 | TCAACTACTCAAAGCTTCGCAT | TATCGCATACACAATCCGCC |
| 8235 | HEAT SHOCK COGNATE 70 KDA PROTEIN 2-LIKE | 1152 | TTCAACTTTCCTCCCAACAG | CAATATCACAGAAATTCGCAGG |
| 6440 | PHENYLANINE AMMONIA-LYASE 1-LIKE | 1488 | GTACAATGCTGTGAAATTCCCT | GAATGGTCAATCATGCTGTCA |
| 21,837 | 9-DIVINYL ETHERSYNTHASE-LIKE | 1115 | GGTTACACGACAAATTCATCCC | AGAACACTTTCATGCCTCCAT |
| * 34 | CYTOCHROME P450 87A3-LIKE | 1676 | TGTATTCTCAAGCTGTCCACT | TTATACCACCTCCAAATGCCA |
| * 327 | HEAT SHOCK PROTEIN 70 | 989 | AGATTACCATCACCAACGACA | GCATAGTTCTCCAAAGCATTCT |
| * 416 | SUBTILISIN-LIKE PROTEASE-LIKE | 2737 | TGGTGTTGGAGTCGTTGTAG | TGGTGTTGGAGTCGTTGTAG |
| DET (Contig) | Fold-Change (Primed vs. Unprimed) | Functional Description | Gene Ontology Classification | ||
|---|---|---|---|---|---|
| 5548 | −53.08 | benzoquinone reductase | response to abiotic stimulus | response to chemical | response to stress |
| 488 | −26.63 | small heat shock protein chloroplastic-like | response to stress | ||
| 2004 | 1.52 | elongation factor 1-alpha | response to chemical | ||
| 1923 | 1.76 | late embryogenesis abundant protein Lea5 | response to abiotic stimulus | response to chemical | response to stress |
| 334 | 1.76 | aspartic proteinase-like | response to abiotic stimulus | response to chemical | response to stress |
| 145 | 1.84 | polyadenylate-binding protein 8-like | response to chemical | ||
| 194 | 1.93 | dnaJ protein homolog | response to abiotic stimulus | response to stress | |
| 9183 | 2.03 | elongation factor 1-alpha | response to chemical | ||
| 3485 | 2.17 | peroxidase 12-like | response to stress | ||
| 4082 | 2.26 | cold shock protein cs66-like | response to abiotic stimulus | response to chemical | response to stress |
| 1334 | 2.38 | cation transport regulator-like protein | response to chemical | ||
| 8235 | 2.48 | heat shock cognate 70 kda protein 2-like | response to stress | ||
| 3929 | 2.60 | dnaj protein homolog 2-like | response to abiotic stimulus | response to stress | |
| 16,971 | 2.92 | dehydrogenase/reductase SDR family protein 7-like | response to chemical | ||
| 23,208 | 3.04 | heat shock cognate 70 kDa protein 2-like | response to stress | ||
| 10,417 | 3.44 | heat shock cognate 70 kda protein 2-like | response to stress | ||
| 2828 | 3.50 | em protein H5-like | response to chemical | response to stress | |
| 1719 | 3.52 | non-specific lipid-transfer protein 2-like | response to stress | ||
| 3594 | 3.98 | 17.9 kDa class I heat shock protein-like | response to abiotic stimulus | response to chemical | response to stress |
| 9460 | 4.32 | citrate glyoxysomal-like | response to chemical | response to stress | |
| 2973 | 5.26 | 11S globulin precursor | response to chemical | ||
| 7098 | 5.32 | thiamine thiazole synthase chloroplastic-like | response to abiotic stimulus | response to stress | |
| 3565 | 5.46 | tubulin beta-1 chain | response to abiotic stimulus | ||
| 7666 | 5.98 | Low-temperature-induced 66 | response to abiotic stimulus | response to chemical | response to stress |
| 1638 | 6.22 | 11s seed storage globulin | response to chemical | ||
| 5501 | 6.53 | RING/U-box domain-containing protein | response to abiotic stimulus | response to chemical | response to stress |
| 6576 | 7.24 | heat shock factor protein hsf30-like | response to abiotic stimulus | response to stress | |
| 14,206 | 7.42 | galactinol synthase | response to abiotic stimulus | response to chemical | response to stress |
| 22,593 | 8.09 | chalcone isomerase | response to abiotic stimulus | response to chemical | |
| 10,620 | 10.51 | chalcone isomerase | response to abiotic stimulus | response to chemical | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, P.B.; Vaz, T.A.A.; Acencio, M.L.; Bovolenta, L.A.; Hilhorst, H.W.M.; da Silva, E.A.A. Can Osmopriming Induce Cross-Tolerance for Abiotic Stresses in Solanum paniculatum L. Seeds? A Transcriptome Analysis Point of View. Seeds 2023, 2, 382-393. https://doi.org/10.3390/seeds2040029
da Silva PB, Vaz TAA, Acencio ML, Bovolenta LA, Hilhorst HWM, da Silva EAA. Can Osmopriming Induce Cross-Tolerance for Abiotic Stresses in Solanum paniculatum L. Seeds? A Transcriptome Analysis Point of View. Seeds. 2023; 2(4):382-393. https://doi.org/10.3390/seeds2040029
Chicago/Turabian Styleda Silva, Pedro Bento, Tatiana Arantes Afonso Vaz, Marcio Luis Acencio, Luiz Augusto Bovolenta, Henk W. M. Hilhorst, and Edvaldo A. Amaral da Silva. 2023. "Can Osmopriming Induce Cross-Tolerance for Abiotic Stresses in Solanum paniculatum L. Seeds? A Transcriptome Analysis Point of View" Seeds 2, no. 4: 382-393. https://doi.org/10.3390/seeds2040029
APA Styleda Silva, P. B., Vaz, T. A. A., Acencio, M. L., Bovolenta, L. A., Hilhorst, H. W. M., & da Silva, E. A. A. (2023). Can Osmopriming Induce Cross-Tolerance for Abiotic Stresses in Solanum paniculatum L. Seeds? A Transcriptome Analysis Point of View. Seeds, 2(4), 382-393. https://doi.org/10.3390/seeds2040029










