Electrochemical DNA Biosensor for the Detection of Infectious Bronchitis Virus Using a Multi-Walled Carbon Nanotube-Modified Gold Electrode
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of Chitosan-Modified MWCNTs Solution
| Primer | Sequences (5′-3′) | References |
|---|---|---|
| Probe sequence | 5′-NH2-CACCACCAGAACCTGTCACCTC-3′ | [27] |
| Target sequence | 5′-GAGGTGACAGGTTCTGGTGGTG-3′ | (This study) |
| One-base mismatch | 5′-GAGGTGACACGTTCTGGTGGTG-3′ | (This study) |
| Three-base mismatch | 5′-GAGGTCACAGATTCTGGCGGTG-3′ | (This study) |
| Non-complementary | 5′-GCCATGTTGTCACTGTCTATT-3′ | (This study) |
| Target DNA of ND | 5′-GTGCAGGCACCCCRAGTGCT-3′ | [28] |
| Target DNA of MG | 5′-CGCAATTTGGTCCTAATCCCCAACA-3′ | [30] |
| Target DNA of ILT | 5′-CTAACCCGTTCGCCGCACTCG-3′ | [27] |
| Target DNA of AIV | 5′-TCAGGCCCCCTCAAAGCCGA-3′ | [31] |
2.3. Preparation of DNA Oligonucleotides
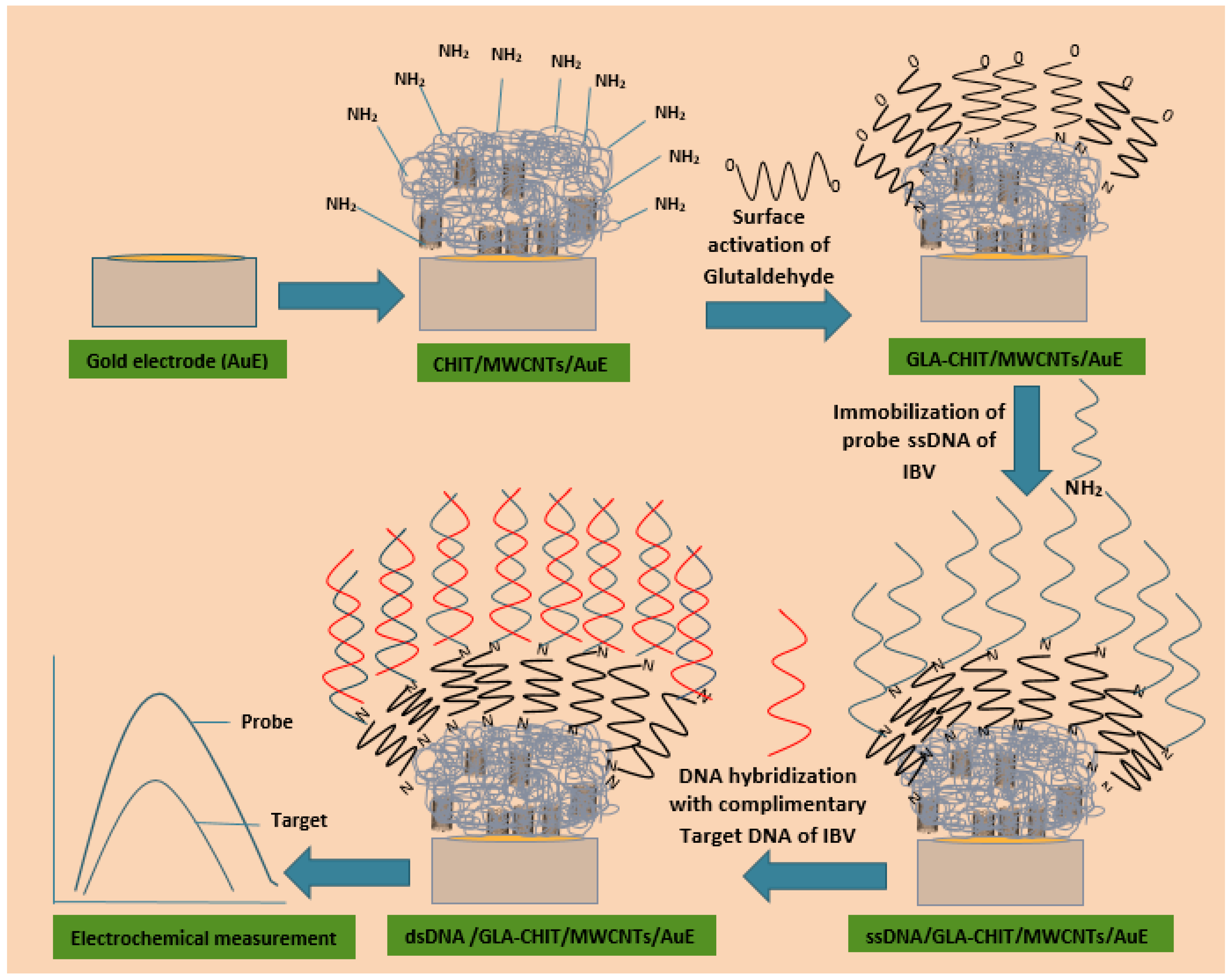
2.4. Construction of DNA Biosensor
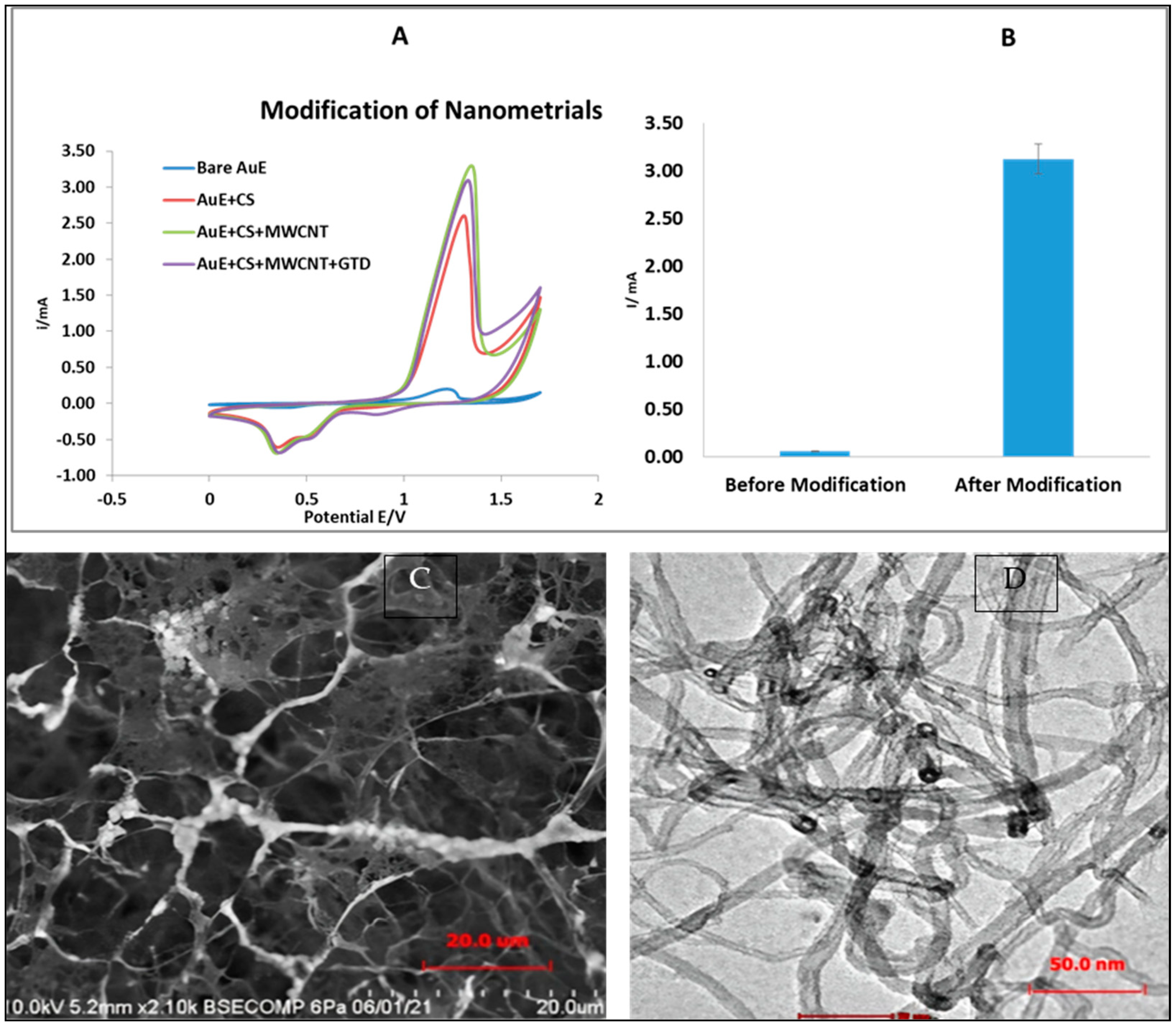
2.5. Physical Characterization
2.6. Optimization of Immobilization and Hybridization
2.7. Electrochemical Analysis
2.8. IBV Screening by RT-PCR
2.9. Hybridization Experiment Using cDNA and PCR Positive Samples
3. Results
3.1. Electrochemical Characteristics of the Modified AuE
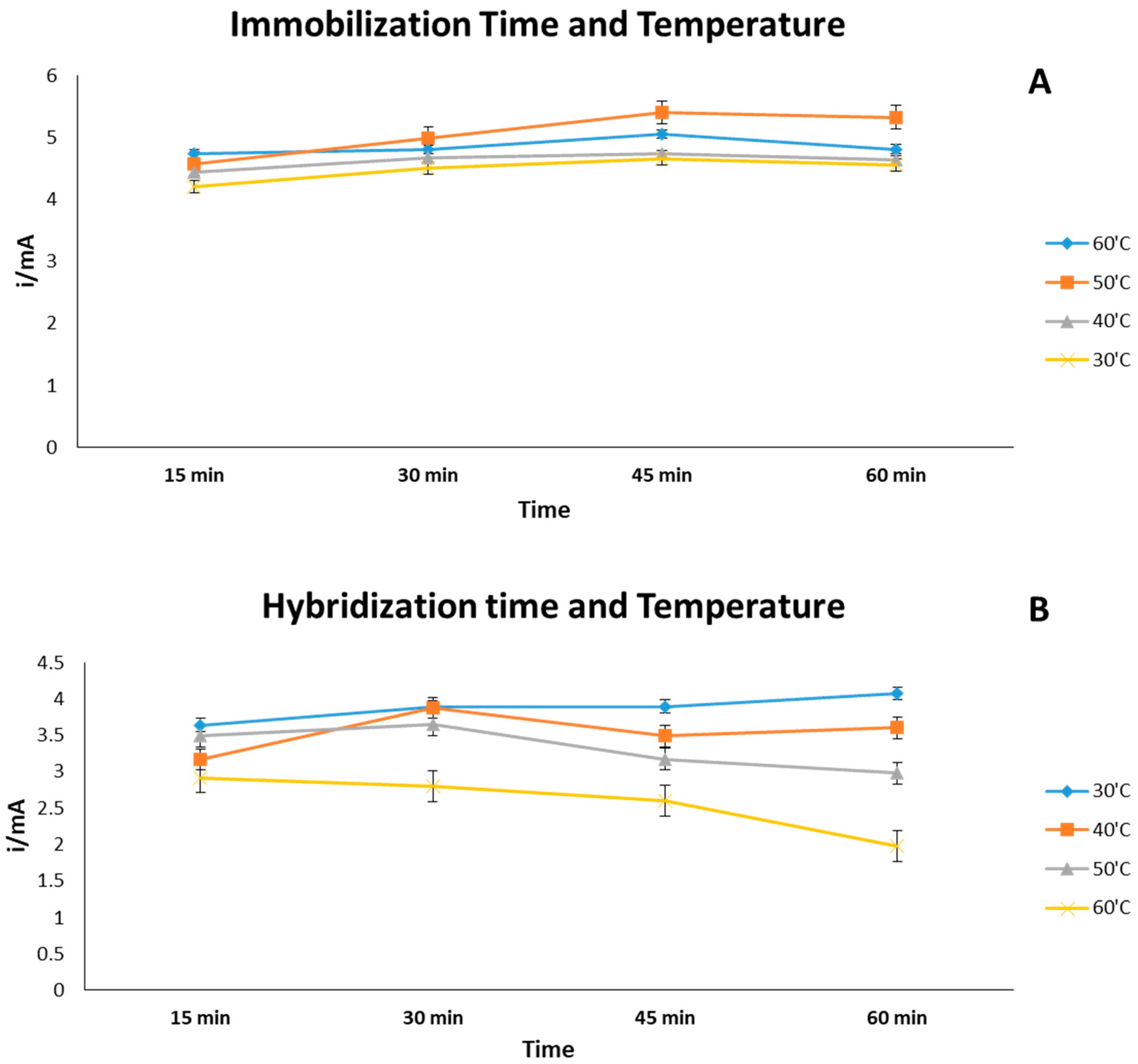
3.2. Effect of Time and Temperature (Immobilization and Hybridization)
3.3. Quantitative Analysis of Target DNA and Sensitivity
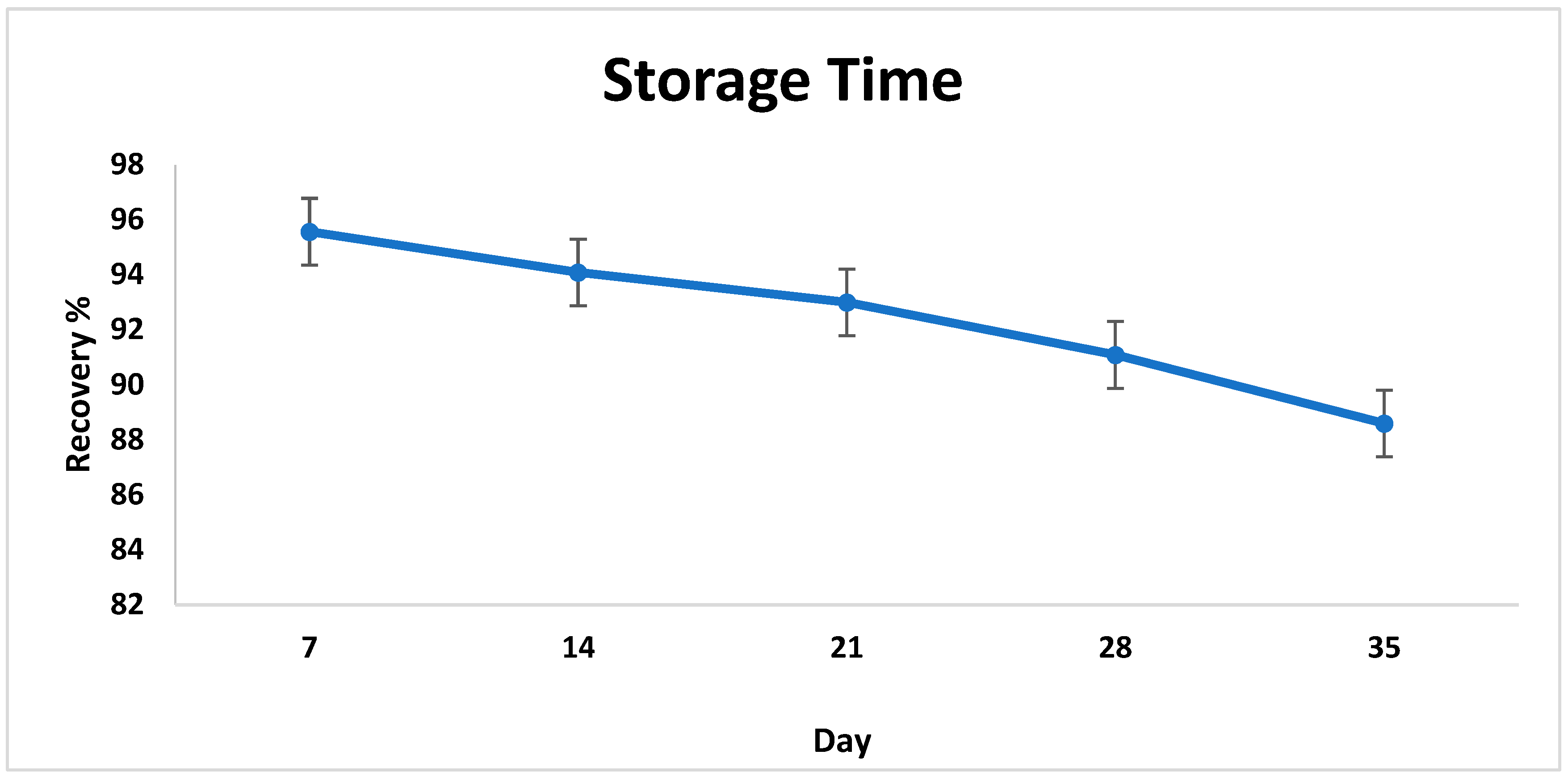
3.4. Reproducibility, Repeatability, and Storage of the Biosensor
3.5. Validation of Developed Biosensor Using Field Samples
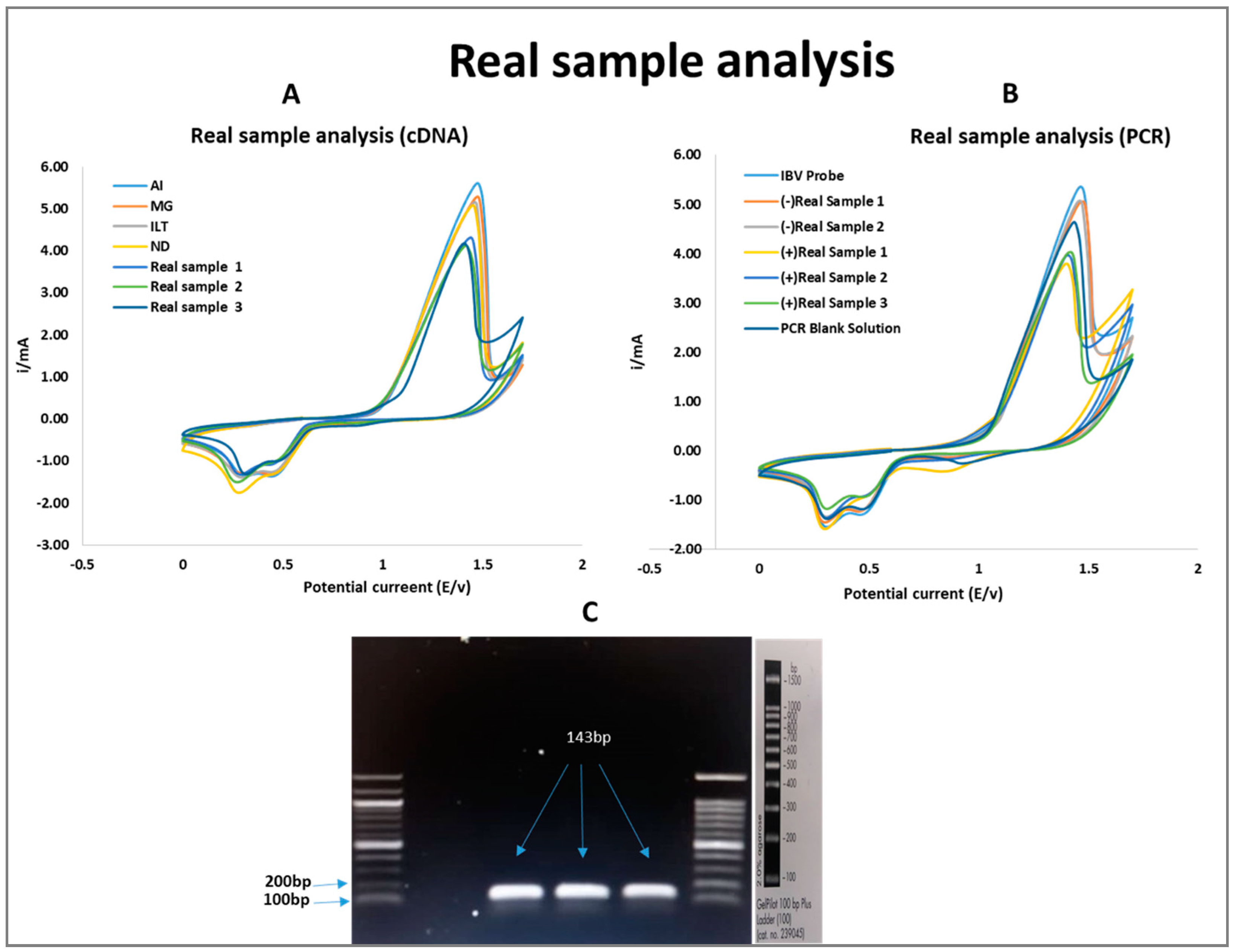
| +Real IBV Sample | cDNA | PCR | |
|---|---|---|---|
| RSD Value | Recovery % | RSD Value | |
| Sample 1 | 4.66 | 95.41 | 4.65 |
| Sample 2 | 1.29 | 99.38 | 3.27 |
| Sample 3 | 2.32 | 99.55 | 7.05 |
| Type of Sensor | Biosensing Concept | Detection Limit | Reference |
|---|---|---|---|
| Electrochemical biosensor (5′-UTR region of the IBV genome) | Modification with nanoparticles (GLU/CS/MWCNTs/AuE) | 2.6 × 10−10 mol L−1 (0.26 nM) | This study |
| Electrochemical biosensor (Spike-S gene) | Modification with nanoparticles and functionalized AuNPs (gold nanoparticles) | 2.96 × 10−16 mol L−1 | [35] |
| Fluorescent immunosensor | Ab (antibody)-functionalized MoS2 (molybdenum disulfide) | 460 EID50/mL | [36] |
| Immunosensor | ICS (Inorganic Carbon Sphere) with Au NP-conjugated antibodies | 104.4 EID50 | [37] |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Serbessa, T.A.; Geleta, Y.G.; Terfa, I.O. Review on diseases and health management of poultry and swine. Int. J. Avian Wildl. Biol. 2023, 7, 27–38. [Google Scholar] [CrossRef]
- Gržinić, G.; Piotrowicz-Cieślak, A.; Klimkowicz-Pawlas, A.; Górny, R.L.; Ławniczek-Wałczyk, A.; Piechowicz, L.; Wolska, L. Intensive poultry farming: A review of the impact on the environment and human health. Sci. Total Environ. 2023, 858, 160014. [Google Scholar] [CrossRef]
- Lu, Y.; Zeng, Y.; Luo, H.; Qiao, B.; Meng, Q.; Dai, Z.; Ping, J. Molecular characteristic, evolution, and pathogenicity analysis of avian infectious bronchitis virus isolates associated with QX type in China. Poult. Sci. 2024, 103, 104256. [Google Scholar] [CrossRef] [PubMed]
- Valastro, V.; Holmes, E.C.; Britton, P.; Fusaro, A.; Jackwood, M.W.; Cattoli, G.; Monne, I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016, 39, 349–364. [Google Scholar] [CrossRef]
- Miranda, C.; Silva, V.; Igrejas, G.; Poeta, P. Genomic evolution of the human and animal coronavirus diseases. Mol. Biol. Rep. 2021, 48, 6645–6653. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.A.; Lee, K.M.; Chiang, H.J.; Huang, S.Y.; Wu, C.J.; Shih, S.R. Molecular virology of SARS-CoV-2 and related coronaviruses. Microbiol. Mol. Biol. Rev. 2022, 86, e0002621. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xie, Z.; Dai, J.; Cao, Y.; Hou, J.; Zheng, Y.; Wei, P. Responses of the Toll-like receptor and melanoma differentiation-associated protein 5 signaling pathways to avian infectious bronchitis virus infection in chicks. Virol. Sin. 2016, 31, 57–68. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.A.; Amin, Z.; Bakar, A.M.S.A.; Saallah, S.; Yusuf, N.H.M.; Shaarani, S.M.; Siddiquee, S. Factor influences for diagnosis and vaccination of avian infectious bronchitis virus (Gammacoronavirus) in chickens. Vet. Sci. 2021, 8, 47. [Google Scholar] [CrossRef]
- Najimudeen, S.M.; Abd-Elsalam, R.M.; Ranaweera, H.A.; Isham, I.M.; Hassan, M.S.; Farooq, M.; Abdul-Careem, M.F. Replication of infectious bronchitis virus (IBV) Delmarva (DMV)/1639 variant in primary and secondary lymphoid organs leads to immunosuppression in chickens. Virology 2023, 587, 109852. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.A.; Sarker, S.; Amin, Z.; Rodrigues, K.F.; Saallah, S.; Shaarani, S.M.; Siddiquee, S. Infectious Bronchitis Virus (Gammacoronavirus) in Poultry, Genomic Architecture, Post-Translational Modifications, and Structural Motifs. Poultry 2023, 2, 363–382. [Google Scholar] [CrossRef]
- Vidic, J.; Vizzini, P.; Manzano, M.; Kavanaugh, D.; Ramarao, N.; Zivkovic, M.; Radonic, V.; Knezevic, N.; Giouroudi, I.; Gadjanski, I. Point-of-Need DNA Testing for Detection of Foodborne Pathogenic Bacteria. Sensors 2019, 19, 1100. [Google Scholar] [CrossRef] [PubMed]
- Abu-Salah, K.M.; Zourob, M.M.; Mouffouk, F.; Alrokayan, S.A.; Alaamery, M.A.; Ansari, A.A. DNA-based nanobiosensors as an emerging platform for detection of disease. Sensors 2015, 15, 14539–14568. [Google Scholar] [CrossRef]
- Ye, Q.; Lu, D.; Zhang, T.; Mao, J.; Shang, S. Recent advances and clinical application in point-of-care testing of SARS-CoV-2. J. Med. Virol. 2022, 94, 1866–1875. [Google Scholar] [CrossRef]
- Arega, S.M.; Conraths, F.J.; Ameni, G. Prevalence of tuberculosis in pigs slaughtered at two abattoirs in Ethiopia and molecular characterization of Mycobacterium tuberculosisisolated from tuberculous-like lesions in pigs. BMC Vet. Res. 2013, 9, 97. [Google Scholar] [CrossRef]
- Grabowska, I.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Olejniczak, A.B.; Leśnikowski, Z.J.; Radecka, H. DNA probe modified with 3-iron bis (dicarbollide) for electrochemical determination of DNA sequence of Avian Influenza Virus H5N1. Biosens. Bioelectron. 2014, 51, 170–176. [Google Scholar] [CrossRef]
- Awad, F.; Forrester, A.; Baylis, M.; Lemiere, S.; Jones, R.; Ganapathy, K. Immune responses and interactions following simultaneous application of live Newcastle disease, infectious bronchitis and avian metapneumovirus vaccines in specific-pathogen-free chicks. Res. J. Vet. Sci. 2015, 98, 127–133. [Google Scholar] [CrossRef]
- Ahmed, W.; Simpson, S.L.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Blackall, L.L.; Shanks, O.C. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022, 805, 149877. [Google Scholar] [CrossRef] [PubMed]
- Robinson-McCarthy, L.R.; Mijalis, A.J.; Filsinger, G.T.; De Puig, H.; Donghia, N.M.; Schaus, T.E.; Church, G.M. Laboratory-generated DNA can cause anomalous pathogen diagnostic test results. Microbiol. Spectr. 2021, 9, e00313-21. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; CHamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens. Bioelectron. 2016, 80, 194–200. [Google Scholar] [CrossRef]
- Rashid, J.I.A.; Yusof, N.A.; Abdulla, J.; Shomiad, R.H. Strategies in the optimization of DNA hybridization conditions and its role in electrochemical detection of dengue virus (DENV) using response surface methodology (RSM). RSC Adv. 2023, 13, 18748–18759. [Google Scholar] [CrossRef]
- Kumar, S.; Sidhu, H.K.; Paul, A.K.; Bhardwaj, N.; Thakur, N.S.; Deep, A. Bioengineered multi-walled carbon nanotube (MWCNT) based biosensors and applications thereof. Sens. Diagn. 2023, 2, 1390–1413. [Google Scholar] [CrossRef]
- Speranza, G. The Role of Functionalization in the Applications of Carbon Materials: An Overview. C 2019, 5, 84. [Google Scholar] [CrossRef]
- Moustakim, H.; Mohammadi, H.; Amine, A. Electrochemical DNA Biosensor Based on Immobilization of a Non-Modified ssDNA Using Phosphoramidate-Bonding Strategy and Pencil Graphite Electrode Modified with AuNPs/CB and Self-Assembled Cysteamine Monolayer. Sensors 2022, 22, 9420. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, J.; Xu, Y.; Chu, H.; Shen, H.; Zhang, D.; Zhao, M.; Liu, J.; Li, G. Binding modes and interaction mechanism between different base pairs and methylene blue trihydrate, aquantum mechanics study. Adv. Struct. Bioinform. 2015, 187–203. [Google Scholar]
- Wang, L.; Liao, X.; Ding, Y.; Gao, F.; Wang, Q. DNA biosensor based on a glassy carbon electrode modified with electropolymerized Eriochrome Black T. Microchim. Acta 2014, 181, 155–162. [Google Scholar] [CrossRef]
- Callison, S.A.; Hilt, D.A.; Boynton, T.O.; Sample, B.F.; Robison, R.; Swayne, D.E.; Jackwood, M.W. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods 2006, 138, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Nidzworski, D.; Wasilewska, E.; Smietanka, K.; Szewczyk, B.; Minta, Z. Detection and differentiation of Newcastle disease virus and influenza virus by using duplex real-time PCR. Acta Biochim. Pol. 2013, 60, 475–480. [Google Scholar] [CrossRef]
- Ghica, M.E.; Pauliukaite, R.; Fatibello-Filho, O.; Brett, C.M. Application of functionalised carbon nanotubes immobilised into chitosan films in amperometric enzyme biosensors. Sens. Actuators B Chem. 2009, 142, 308–315. [Google Scholar] [CrossRef]
- Hnatow, L.L.; Keeler, C.L., Jr.; Tessmer, L.L.; Czymmek, K.; Dohms, J.E. Characterization of MGC2, a Mycoplasma gallisepticum cytadhesin with homology to the Mycoplasma pneumoniae 30-kilodalton protein P30 and Mycoplasma genitalium P32. Infect. Immun. 1998, 66, 3436–3442. [Google Scholar] [CrossRef]
- Zhao, Y.; Kong, C.; Cui, X.; Cui, H.; Shi, X.; Zhang, X.; Hu, S.; Hao, L.; Wang, Y. Detection of infectious laryngotracheitis virus by real-time PCR in naturally and experimentally infected chickens. PLoS ONE 2013, 8, e67598. [Google Scholar] [CrossRef]
- Siddiquee, S.; Yusof, N.A.; Salleh, A.B.; Tan, S.G.; Bakar, F.A. Electrochemical DNA biosensor for the detection of Trichoderma harzianum based on a gold electrode modified with a composite membrane made from an ionic liquid, ZnO nanoparticles and chitosan, and by using acridine orange as a redox indicator. Mikrochim. Acta 2011, 172, 357–363. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.A.; Sarker, S.; Amin, Z.; Rodrigues, K.F.; Baka, A.M.S.A.; Saallah, S.; Siddiquee, S. Seroprevalence and molecular characterisation of infectious bronchitis virus (IBV) in broiler farms in Sabah, Malaysia. J. Vet. Med. Sci. 2023, 10, e1153. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.A.; Ringgit, G.; Amin, Z.; Bakar, A.M.S.A.; Saallah, S.; Shaarani, S.M.; Siddiquee, S. Optimizing electrochemical DNA biosensors for the detection of avian infectious bronchitis virus. Malays. J. Microbiol. 2023, 19, 651–663. [Google Scholar]
- Yang, Y.; Yang, D.; Shao, Y.; Li, Y.; Chen, X.; Xu, Y.; Miao, J. A label-free electrochemical assay for coronavirus IBV H120 strain quantification based on equivalent substitution effect and AuNPs-assisted signal amplification. Mikrochim. Acta 2020, 187, 624. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Neethirajan, S. Immunosensor based on antibody-functionalized MoS 2 for rapid detection of avian coronavirus on cotton thread. IEEE Sens. J. 2018, 18, 4358–4363. [Google Scholar] [CrossRef]
- Liu, I.L.; Lin, Y.C.; Lin, Y.C.; Jian, C.Z.; Cheng, I.C.; Chen, H.W. A novel immune-chromatographic strip for antigen detection of avian infectious bronchitis virus. Int. J. Mol. Sci. 2019, 20, 2216. [Google Scholar] [CrossRef]
- Nordin, N.; Yusof, N.A.; Radu, S.; Hushiarian, R. Development of an Electrochemical DNA Biosensor to Detect a Foodborne Pathogen. J. Vis. Exp. 2018, 136, e56585. [Google Scholar]
- Wang, Q.; Zhang, B.; Lin, X.; Weng, W. Hybridization biosensor based on the covalent immobilization of probe DNA on chitosan–mutiwalled carbon nanotubes nanocomposite by using glutaraldehyde as an arm linker. Sens. Actuators B Chem. 2011, 156, 599–605. [Google Scholar] [CrossRef]
- Zhao, W.R.; Kang, T.F.; Lu, L.P.; Cheng, S.Y. Electrochemical magnetic imprinted sensor based on MWCNTs@CS/CTABr surfactant composites for sensitive sensing of diethylstilbestrol. J. Electroanal. Chem. 2018, 818, 181–190. [Google Scholar] [CrossRef]
- Shalauddin, M.; Akhter, S.; Bagheri, S.; Karim, M.M.A.; Kadri, N.A.; Basirun, W. Immobilized copper ions on MWCNTs-Chitosan thin film, Enhanced amperometric sensor for electrochemical determination of diclofenac sodium in aqueous solution. Int. J. Hydrogen Energy 2017, 42, 19951–19960. [Google Scholar] [CrossRef]
- Markegard, C.B.; Gallivan, C.P.; Cheng, D.D.; Nguyen, H.D. Effects of concentration and temperature on DNA hybridization by two closely related sequences via large-scale coarse-grained simulations. J. Phys. Chem. B 2016, 120, 7795–7806. [Google Scholar] [CrossRef] [PubMed]
- Nimse, S.B.; Song, K.; Sonawane, M.D.; Sayyed, D.R.; Kim, T. Immobilization techniques for microarray, challenges and applications. Sensors 2014, 14, 22208–22229. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Rajabzadeh, N.; Mazloum-Ardakani, M.; Heidari, M.M. Comparison of impedimetric detection of DNA hybridization on chemically and electrochemically functionalized multi-wall carbon nanotubes modified electrode. Sens. Actuators B Chem. 2015, 207, 673–682. [Google Scholar] [CrossRef]
- Hassan, R.A.; Heng, L.Y.; Tan, L.L. Novel DNA biosensor for direct determination of carrageenan. Sci. Rep. 2019, 9, 6379. [Google Scholar] [CrossRef]
- Kusnin, N.; Yusof, N.A.; Abdullah, J.; Sabri, S.; Mohammad, F.; Mustafa, S.; Al-Lohedan, H.A. Electrochemical sensory detection of Sus scrofa mtDNA for food adulteration using hybrid ferrocenylnaphthalene diimide intercalator as a hybridization indicator. RSC Adv. 2020, 10, 27336–27345. [Google Scholar] [CrossRef]
- Ali, M.R.; Bacchu, M.S.; Setu, M.A.A.; Akter, S.; Hasan, M.N.; Chowdhury, F.T.; Khan, M.Z.H. Development of an advanced DNA biosensor for pathogenic Vibrio cholerae detection in real sample. Biosens. Bioelectron. 2021, 188, 113338. [Google Scholar] [CrossRef]
- Moarefdoust, M.M.; Jahani, S.; Moradalizadeh, M.; Motaghi, M.M.; Foroughi, M.M.A. DNA Biosensor Based on a Raspberry-like Hierarchical Nanostructure for the Determination of the Anticancer Drug Nilotinib. ChemistryOpen 2022, 11, e202100261. [Google Scholar] [CrossRef]
- Nordin, N.; Yusof, N.A.; Abdullah, J.; Radu, S.; Hushiarian, R. A simple, portable, electrochemical biosensor to screen shellfish for Vibrio parahaemolyticus. AMB Express 2017, 7, 41. [Google Scholar] [CrossRef]
- López-Vidriero, I.; Godoy, M.; Grau, J.; Peñuelas, M.; Solano, R.; Franco-Zorrilla, J.M. DNA features beyond the transcription factor binding site specify target recognition by plant MYC2-related bHLH proteins. Plant Commun. 2021, 2, 100232. [Google Scholar] [CrossRef]
- Iyuke, S.; Ntombenhle-Hlongwane, G.; Dodoo-Arhin, D.; Wamwangi, D.; Daramola, M.O.; Moothi, K. DNA hybridisation sensors for product authentication and tracing, State of the art and challenges. S. Afr. J. Chem. Eng. 2019, 27, 16–34. [Google Scholar]
- Doðru, E.; Erhan, E.; Arikan, O.A. Investigation of Dilution Agent Effect onto Interactions Between Methylene Blue and DNA using Carbon Fiber Based DNA Biosensor. J. New Mater. Electrochem. Syst. 2017, 20, 65. [Google Scholar]
- Campos-Ferreira, D.S.; Nascimento, G.A.; Souza, E.V.; Souto-Maior, M.A.; Arruda, M.S.; Zanforlin, D.M.; Lima-Filho, J.L. Electrochemical DNA biosensor for human papillomavirus 16 detection in real samples. Anal. Chim. Acta 2013, 804, 258–263. [Google Scholar] [CrossRef]
- Ortega-Zamora, C.; Jiménez-Skrzypek, G.; González-Sálamo, J.; Mazzapioda, L.; Navarra, M.A.; Gentili, A.; Hernández-Borges, J. Extraction of Emerging Contaminants from Environmental Waters and Urine by Dispersive Liquid–Liquid Microextraction with Solidification of the Floating Organic Droplet Using Fenchol, Acetic Acid Deep Eutectic Mixtures. ACS Sustain. Chem. Eng. 2022, 10, 15714–15725. [Google Scholar] [CrossRef]
- Jamaluddin, R.Z.A.R.; Tan, L.L.; Chong, K.F.; Heng, L.Y. An electrochemical DNA biosensor fabricated from graphene decorated with graphitic nanospheres. Nanotechnology 2020, 31, 485501. [Google Scholar] [CrossRef]
- Bohari, N.A.; Siddiquee, S.; Saallah, S.; Misson, M.; Arshad, S.E. Optimization and analytical behavior of electrochemical sensors based on the modification of indium tin oxide (ITO) using PANI/MWCNTs/AuNPs for mercury detection. Sensors 2020, 20, 6502. [Google Scholar] [CrossRef]
- Ahuja, S.; Kumar, M.S.; Nandeshwar, R.; Kondabagil, K.; Tallur, S. Longer amplicons provide better sensitivity for electrochemical sensing of viral nucleic acid in water samples using PCB electrodes. Sci. Rep. 2022, 12, 8814. [Google Scholar] [CrossRef]
- Smith, M.; Withnall, R.; Anastasova, S.; Gil-Rosa, B.; Blackadder-Coward, J.; Taylor, N. Developing a multimodal biosensor for remote physiological monitoring. BMJ Mil. Health 2023, 169, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, J. Biosensors and sensors for dopamine detection. View 2021, 2, 20200102. [Google Scholar] [CrossRef]
- Li, H.; Wan, C.; Wang, Z.; Tan, J.; Tan, M.; Zeng, Y.; Guo, X. Rapid diagnosis of duck Tembusu virus and goose astrovirus with TaqMan-based duplex real-time PCR. Front. Microbiol. 2023, 14, 1146241. [Google Scholar] [CrossRef]
- Patel, S.A.; Thompson, N.G.; Nguyen, T.T. Electrochemical DNA biosensors for viral diagnostics. ACS Omega 2021, 6, 7123–7132. [Google Scholar]

| Parameters | Variation |
|---|---|
| Buffer solution | Acetate, Phosphate, Tris-HCl, Ammonium, Citrate |
| Redox indicator | MB, PB, and K3 [Fe(CN)6] |
| Scan rate/mVs−1 | 50, 100, 150, 200, 250, 300 |
| pH | 6.0, 6.5, 7.0, 7.5, 80, 8.5 |
| Accumulation time/sec | 5, 10, 15, 20, 25 |
| Probe volume/µL | 5, 10, 15, 20, 25 |
| Target volume/µL | 5, 10, 15, 20, 25 |
| Immobilization temperature/°C | 30, 40, 50, 60 |
| Immobilization time/min | 15, 30, 45, 60 |
| hybridization temperature/°C | 30, 40, 50, 60 |
| hybridization time/min | 15, 30, 45, 60 |
| Hybridization specificity (comparative study) | ND, MG, AIV, ILT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhuiyan, M.S.A.; Ringgit, G.; Sarker, S.; Abu Bakar, A.M.S.; Saallah, S.; Amin, Z.; Shaarani, S.M.; Siddiquee, S. Electrochemical DNA Biosensor for the Detection of Infectious Bronchitis Virus Using a Multi-Walled Carbon Nanotube-Modified Gold Electrode. Poultry 2025, 4, 12. https://doi.org/10.3390/poultry4010012
Bhuiyan MSA, Ringgit G, Sarker S, Abu Bakar AMS, Saallah S, Amin Z, Shaarani SM, Siddiquee S. Electrochemical DNA Biosensor for the Detection of Infectious Bronchitis Virus Using a Multi-Walled Carbon Nanotube-Modified Gold Electrode. Poultry. 2025; 4(1):12. https://doi.org/10.3390/poultry4010012
Chicago/Turabian StyleBhuiyan, Md Safiul Alam, Gilbert Ringgit, Subir Sarker, Ag Muhammad Sagaf Abu Bakar, Suryani Saallah, Zarina Amin, Sharifudin Md. Shaarani, and Shafiquzzaman Siddiquee. 2025. "Electrochemical DNA Biosensor for the Detection of Infectious Bronchitis Virus Using a Multi-Walled Carbon Nanotube-Modified Gold Electrode" Poultry 4, no. 1: 12. https://doi.org/10.3390/poultry4010012
APA StyleBhuiyan, M. S. A., Ringgit, G., Sarker, S., Abu Bakar, A. M. S., Saallah, S., Amin, Z., Shaarani, S. M., & Siddiquee, S. (2025). Electrochemical DNA Biosensor for the Detection of Infectious Bronchitis Virus Using a Multi-Walled Carbon Nanotube-Modified Gold Electrode. Poultry, 4(1), 12. https://doi.org/10.3390/poultry4010012







