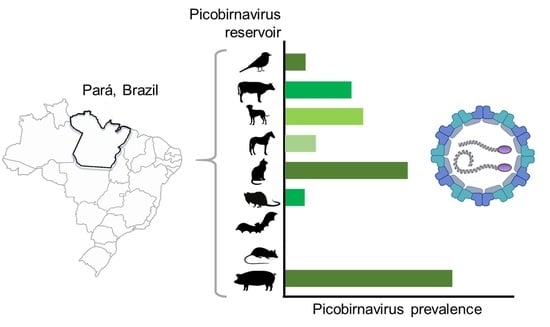
Occurrence of Picobirnavirus in Domestic and Wild Animals from Three Cities of Brazilian Amazon
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Aspects
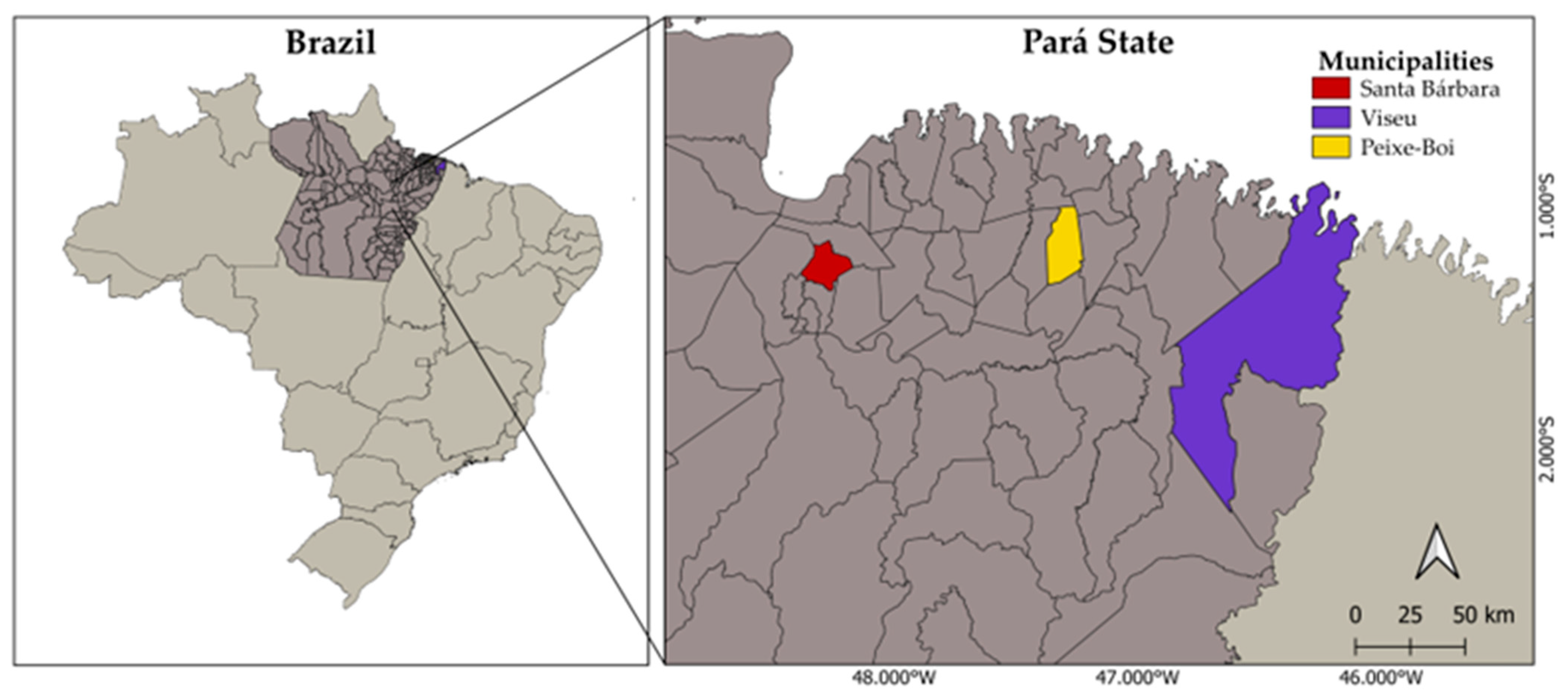
2.2. Study Area
2.3. Sample Collection
2.4. Samples
2.5. Sample Preparation
2.6. Sequence Analysis
2.7. Statistical Analysis
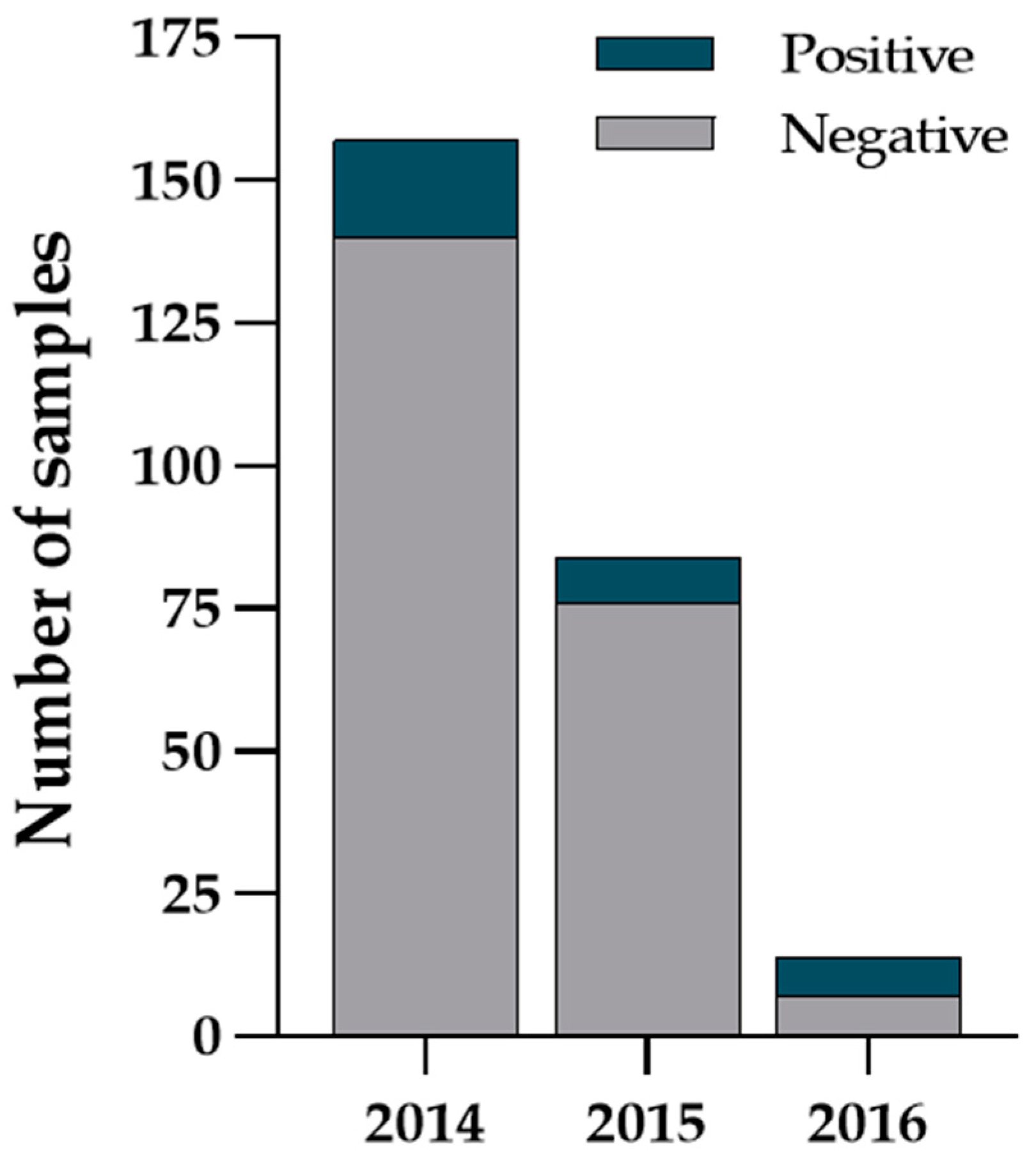
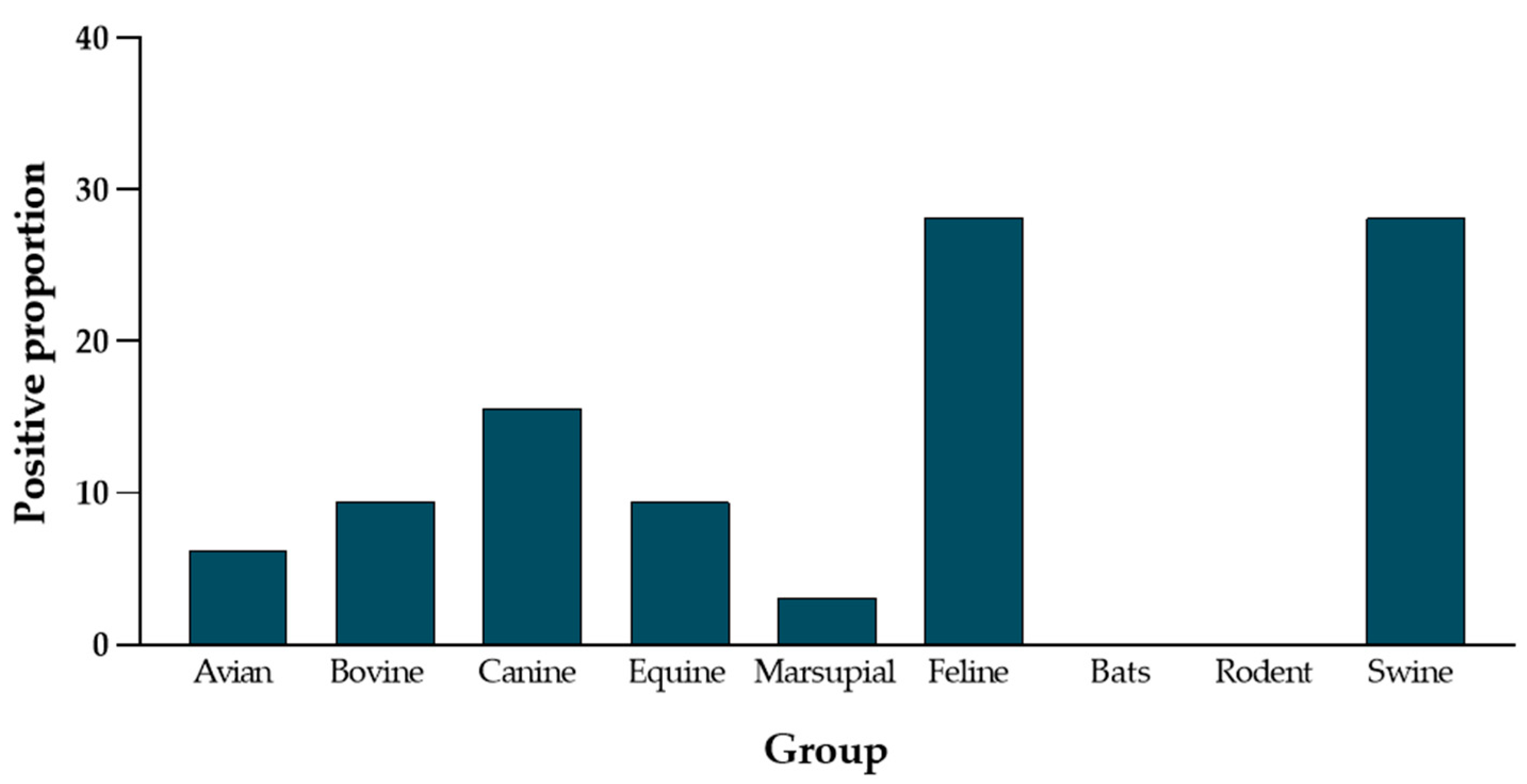
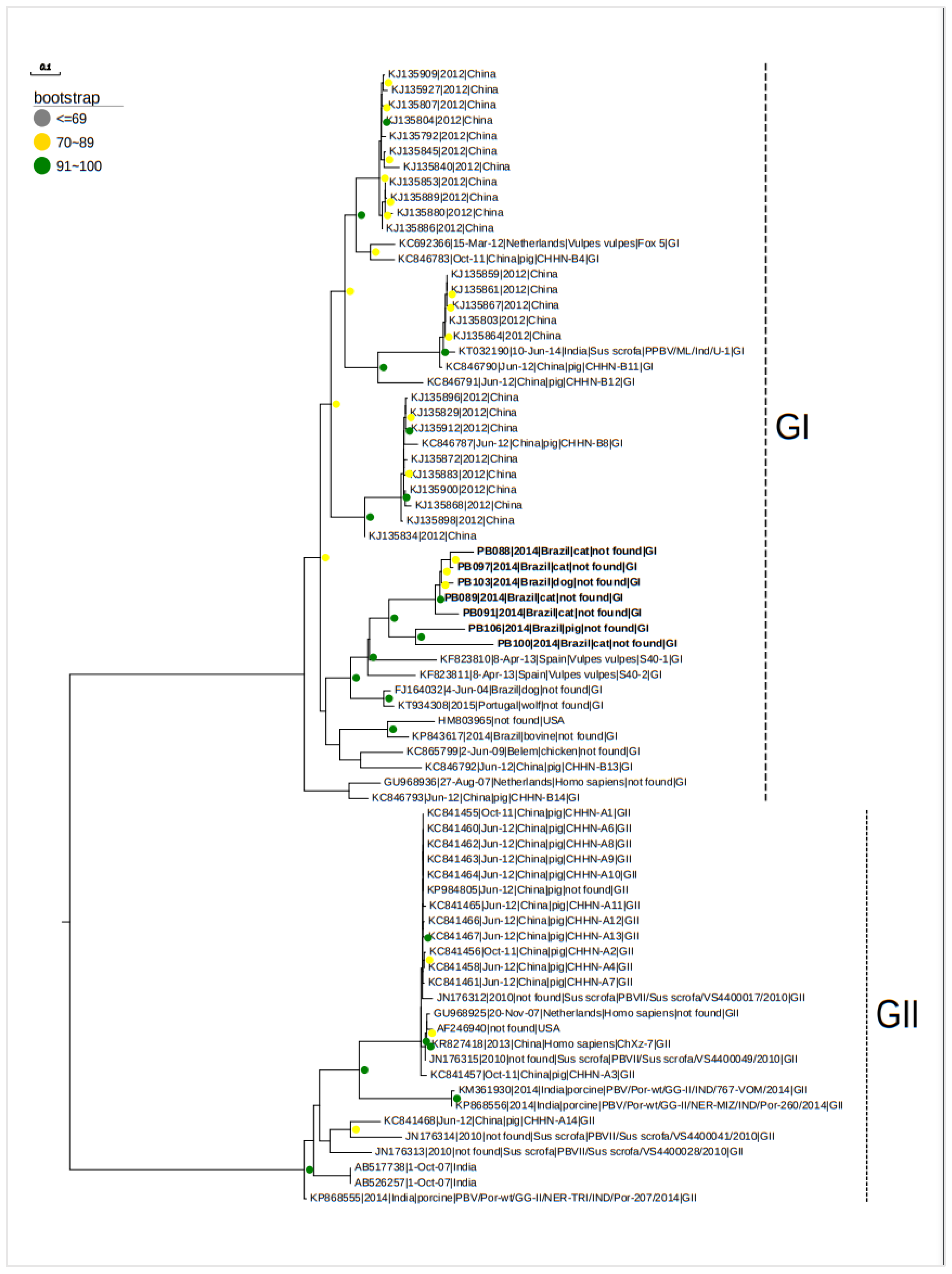
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malik, Y.S.; Ghosh, S. Etymologia: Picobirnavirus. Emerg. Infect. Dis. 2020, 26, 89. [Google Scholar] [CrossRef]
- Collier, A.M.; Lyytinen, O.L.; Guo, Y.R.; Toh, Y.; Poranen, M.M.; Tao, Y.J. Initiation of RNA Polymerization and Polymerase Encapsidation by a Small dsRNA Virus. PLoS Pathog. 2016, 12, e1005523. [Google Scholar] [CrossRef] [PubMed]
- Kashnikov, A.Y.; Epifanova, N.V.; Novikova, N.A. Picobirnaviruses: Prevalence, genetic diversity, detection methods. Vavilov. J. Genet. Breed. 2020, 24, 661. [Google Scholar] [CrossRef] [PubMed]
- Delmas, B.; Attoui, H.; Ghosh, S.; Malik, Y.S.; Mundt, E.; Vakharia, V.N. ICTV virus taxonomy profile: Picobirnaviridae. J. Gen. Virol. 2019, 100, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.J.; Cloherty, G.A.; Berg, M.G. Understanding the genetic diversity of picobirnavirus: A classification update based on phylogenetic and pairwise sequence comparison approaches. Viruses 2021, 13, 1476. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, B.; Masachessi, G.; Mladenova, Z. Animal Picobirnavirus. Virusdisease 2014, 25, 223–238. [Google Scholar] [CrossRef]
- Li, W.; Qiang, X.; Qin, S.; Huang, Y.; Hu, Y.; Bai, B.; Hou, J.; Gao, R.; Zhang, X.; Mi, Z.; et al. Virome diversity analysis reveals novel enteroviruses and a human picobirnavirus in stool samples from African green monkeys with diarrhea. Infect. Genet. Evol. 2020, 82, 104279. [Google Scholar] [CrossRef]
- Malik, Y.S.; Kumar, N.; Sharma, K.; Dhama, K.; Shabbir, M.Z.; Ganesh, B.; Kobayashi, N.; Banyai, K. Epidemiology, Phylogeny, and Evolution of Emerging Enteric Picobirnaviruses of Animal Origin and Their Relationship to Human Strains. Biomed. Res. Int. 2014, 2014, 780752. [Google Scholar] [CrossRef]
- Pereira, H.G.; Fialho, A.M.; Flewett, T.H.; Teixeira, J.M.S.; Andrade, Z.P. Novel viruses in human faeces. Lancet 1988, 2, 103–104. [Google Scholar] [CrossRef]
- Ghosh, S.; Malik, Y.S. The True Host/s of Picobirnaviruses. Front. Vet. Sci. 2021, 7, 615293. [Google Scholar] [CrossRef]
- Yinda, C.K.; Vanhulle, E.; Conceição-Neto, N.; Beller, L.; Deboutte, W.; Shi, C.; Ghogomu, S.M.; Maes, P.; Van Ranst, M.; Matthijnssens, J.; et al. Gut Virome Analysis of Cameroonians Reveals High Diversity of Enteric Viruses, Including Potential Interspecies Transmitted Viruses. mSphere 2019, 4, e00585-18. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.R.; Wang, D. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology 2018, 516, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Neri, U.; Wolf, Y.I.; Roux, S.; Camargo, A.P.; Lee, B.D.; Kazlauskas, D.; Chen, I.M.; Ivanova, N.; Allen, L.Z.; Paez-Espino, D.; et al. A Five-Fold Expansion of the Global RNA Virome Reveals Multiple New Clades of RNA Bacteriophages. SSRN Electron. J. 2022, 185, 4023–4037.e18. [Google Scholar] [CrossRef]
- Zanella, J.R.C. Zoonoses emergentes e reemergentes e sua importância para saúde e produção animal. Pesqui Agropecuária Bras. 2016, 51, 510–519. [Google Scholar] [CrossRef]
- Brown, C. Human-Animal Medicine: Clinical Approaches to Zoonoses, Toxicants and Other Shared Health Risks. Emerg. Infect. Dis. 2010, 16, 1050. [Google Scholar] [CrossRef]
- Monath, T.P. Vaccines against diseases transmitted from animals to humans: A one health paradigm. Vaccine 2013, 31, 5321. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, B.; Banyai, K.; Masachessi, G.; Mladenova, Z.; Nagashima, S.; Ghosh, S.; Nataraju, S.M.; Pativada, M.; Kumar, R.; Kobayashi, N. Genogroup i picobirnavirus in diarrhoeic foals: Can the horse serve as a natural reservoir for human infection? Vet. Res. 2011, 42, 52. [Google Scholar] [CrossRef] [PubMed]
- Duarte Júnior, J.W.B.; Chagas, E.H.N.; Serra, A.C.S.; Souto, L.C.D.S.; da Penha Júnior, E.T.; Bandeira, R.d.S.; e Guimaraes, R.J.d.P.S.; Oliveira, H.G.d.S.; Sousa, T.K.S.; Lopes, C.T.d.A.; et al. Ocurrence of rotavirus and picobirnavirus in wild and exotic avian from amazon forest. PLoS Negl. Trop. Dis. 2021, 15, e0008792. [Google Scholar] [CrossRef]
- Barros, B.d.C.V.; Chagas, E.N.; Bezerra, L.W.; Ribeiro, L.G.; Duarte Júnior, J.W.B.; Pereira, D.; da Penha Junior, E.T.; Silva, J.R.; Bezerra, D.A.M.; Bandeira, R.S.; et al. Rotavirus A in wild and domestic animals from areas with environmental degradation in the Brazilian Amazon. PLoS ONE 2018, 13, e0209005. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.J.A.; Salimans, M.M.M.; Jansen, C.L.; Wertheim-Van Dillen, P.M.E.; Van Der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Pereira, H.G.; Azeredo, R.S.; Leite, J.P.; Barth, O.M.; Sutmoller, F.; de Farias, V.; Vidal, M.N.P. Comparison of polyacrylamide gel electrophoresis (PAGE), immuno-electron microscopy (IEM) and enzyme immunoassay (EIA) for the rapid diagnosis of rotavirus infection in children. Mem. Inst. Oswaldo Cruz 1983, 78, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.I.; Fang, Z.Y.; Glass, R.I.; Monroe, S.S. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology 2000, 277, 316–329. [Google Scholar] [CrossRef]
- Malik, Y.S.; Sircar, S.; Dhama, K.; Singh, R.; Ghosh, S.; Bányai, K.; Vlasova, A.N.; Nadia, T.; Singh, R.K. Molecular epidemiology and characterization of picobirnaviruses in small ruminant populations in India. Infect. Genet. Evol. 2018, 63, 39–42. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Ruiz-Gonzalez, A.; Schapendonk, C.M.E.; Van Den Brand, J.M.A.; Osterhaus, A.D.M.E.; Smits, S.L. Viral metagenomic analysis of feces of wild small carnivores. Virol. J. 2014, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Silva, R.; Bezerra, D.A.M.; Kaiano, J.H.L.; Oliveira, D.d.S.; Silvestre, R.V.D.; Gabbay, Y.B.; Ganesh, B.; Mascarenhas, J.D.P. Genogroup I avian picobirnavirus detected in Brazilian broiler chickens: A molecular epidemiology study. J. Gen. Virol. 2014, 95, 117–122. [Google Scholar] [CrossRef]
- Ribeiro, A.F.; da Silva, R.R.; Bezerra, D.A.M.; da Silva Bandeira, R.; de Castro, C.M.O.; Mascarenhas, J.D.P. Picobirnavirus genogroup 2 in broiler chickens of the metropolitan Belém mesoregion—PA—Brazil. Braz. J. Anim. Environ. Res. 2019, 2, 2033–2050. [Google Scholar]
- Ng, T.F.F.; Mesquita, J.R.; Nascimento, M.S.J.; Kondov, N.O.; Wong, W.; Reuter, G.; Knowles, N.J.; Vega, E.; Esona, M.D.; Deng, X.; et al. Feline fecal virome reveals novel and prevalent enteric viruses. Vet. Microbiol. 2014, 171, 102–111. [Google Scholar] [CrossRef]
- Kylla, H.; Dutta, T.K.; Roychoudhury, P.; Malik, Y.S.; Mandakini, R.; Subudhi, P.K. Prevalence and molecular characterization of porcine Picobirnavirus in piglets of North East Region of India. Trop. Anim. Health Prod. 2017, 49, 417–422. [Google Scholar] [CrossRef]
- Malik, Y.S.; Chandrashekar, K.M.; Sharma, K.; Haq, A.A.; Vaid, N.; Chakravarti, S.; Batra, M.; Singh, R.; Pandey, A.B. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop. Anim. Health Prod. 2011, 43, 1475–1478. [Google Scholar] [CrossRef]
- Fregolente, M.C.D.; de Castro-Dias, E.; Martins, S.S.; Spilki, F.R.; Allegretti, S.M.; Gatti, M.S.V. Molecular characterization of picobirnaviruses from new hosts. Virus Res. 2009, 143, 134–136. [Google Scholar] [CrossRef]
- Takiuchi, E.; Macedo, R.; Kunz, A.F.; Gallego, J.C.; de Mello, J.L.; Otonel, R.A.A.; Alfieri, A.A. Electrophoretic RNA genomic profiles of Brazilian Picobirnavirus (PBV) strains and molecular characterization of a PBV isolated from diarrheic calf. Virus Res. 2016, 211, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.P.; Cubel Garcia, R.C.N.; Labarthe, N.V.; Leite, J.P.G. Detection of double-stranded RNA viruses in fecal samples of dogs with gastroenteritis in Rio de Janeiro, Brazil. Arq. Bras. Med. Vet. E Zootec. 2004, 56, 554–557. [Google Scholar] [CrossRef]
- Assunção, J.; Lipscomb, M.; Mobarak, A.M.; Szerman, D. Agricultural Productivity and Deforestation in Brazil. 2016. Available online: https://climatepolicyinitiative.org/wp-content/uploads/2017/06/Agricultural-Productivity-and-Deforestation-in-Brazil-CPI.pdf (accessed on 15 March 2018).
- Laurance, W.F.; Vasconcelos, H.L. Consequências ecológicas da fragmentação florestal na amazônia. Oecologia Bras. 2009, 13, 434–451. [Google Scholar] [CrossRef]
- Suzán, G.; Marcé, E.; Giermakowski, J.T.; Armién, B.; Pascale, J.; Mills, J.; Ceballos, G.; Gomez, A.; Aguirre, A.A.; Salazar-Bravo, J.; et al. The Effect of Habitat Fragmentation and Species Diversity Loss on Hantavirus Prevalence in Panama. Ann. N. Y. Acad. Sci. 2008, 1149, 80–83. [Google Scholar] [CrossRef]
- Morand, S.; Lajaunie, C. Outbreaks of Vector-Borne and Zoonotic Diseases Are Associated with Changes in Forest Cover and Oil Palm Expansion at Global Scale. Front. Vet. Sci. 2021, 8, 230. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Kulmann-Leal, B.; Kaminski, V.L.; Valverde-Villegas, J.M.; Da Veiga, A.B.G.; Spilki, F.R.; Fearnside, P.M.; Caesar, L.; Giatti, L.L.; Wallau, G.L.; et al. Beyond diversity loss and climate change: Impacts of Amazon deforestation on infectious diseases and public health. An. Acad. Bras. Cienc. 2020, 92, e20191375. [Google Scholar] [CrossRef]
- Masachessi, G.; Ganesh, B.; Martinez, L.C.; Giordano, M.O.; Barril, P.A.; Isa, M.B.; Pavan, G.V.; Mateos, C.A.; Nates, S.V. Maintenance of picobirnavirus (PBV) infection in an adult orangutan (Pongo pygmaeus) and genetic diversity of excreted viral strains during a three-year period. Infect. Genet. Evol. 2015, 29, 196–202. [Google Scholar] [CrossRef]
- Lessa, L.G.; Geise, L. Hábitos alimentares de masupiais didelfídeos brasileiros: Análise do estado de conhecimento atual. Oecologia Aust. 2010, 14, 901–910. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chagas, E.H.N.; da Silva, J.R.; de Barros, B.d.C.V.; Duarte Júnior, J.W.B.; dos Santos, F.d.S.; Sousa Júnior, E.C.; Bezerra, D.A.M.; dos Santos, M.I.; Pinheiro, H.H.C.; Malik, Y.S.; et al. Occurrence of Picobirnavirus in Domestic and Wild Animals from Three Cities of Brazilian Amazon. Zoonotic Dis. 2024, 4, 74-85. https://doi.org/10.3390/zoonoticdis4010008
Chagas EHN, da Silva JR, de Barros BdCV, Duarte Júnior JWB, dos Santos FdS, Sousa Júnior EC, Bezerra DAM, dos Santos MI, Pinheiro HHC, Malik YS, et al. Occurrence of Picobirnavirus in Domestic and Wild Animals from Three Cities of Brazilian Amazon. Zoonotic Diseases. 2024; 4(1):74-85. https://doi.org/10.3390/zoonoticdis4010008
Chicago/Turabian StyleChagas, Elaine Hellen Nunes, Julia Rezende da Silva, Bruno de Cássio Veloso de Barros, José Wandilson Barbosa Duarte Júnior, Fabiolla da Silva dos Santos, Edivaldo Costa Sousa Júnior, Delana Andreza Melo Bezerra, Maria Inês dos Santos, Helder Henrique Costa Pinheiro, Yashpal Singh Malik, and et al. 2024. "Occurrence of Picobirnavirus in Domestic and Wild Animals from Three Cities of Brazilian Amazon" Zoonotic Diseases 4, no. 1: 74-85. https://doi.org/10.3390/zoonoticdis4010008