Cytoarchitectonic Analysis and 3D Maps of the Mesial Piriform Region in the Human Brain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Postmortem Brains and Histology
2.2. Cytoarchitectonic Mapping
2.3. Texture Analysis
2.4. Computation of Probabilistic Maps and Maximum Probability Map
2.5. Volumetry
2.6. Mapping and Reconstruction of the Piriform Region in the BigBrain
3. Results
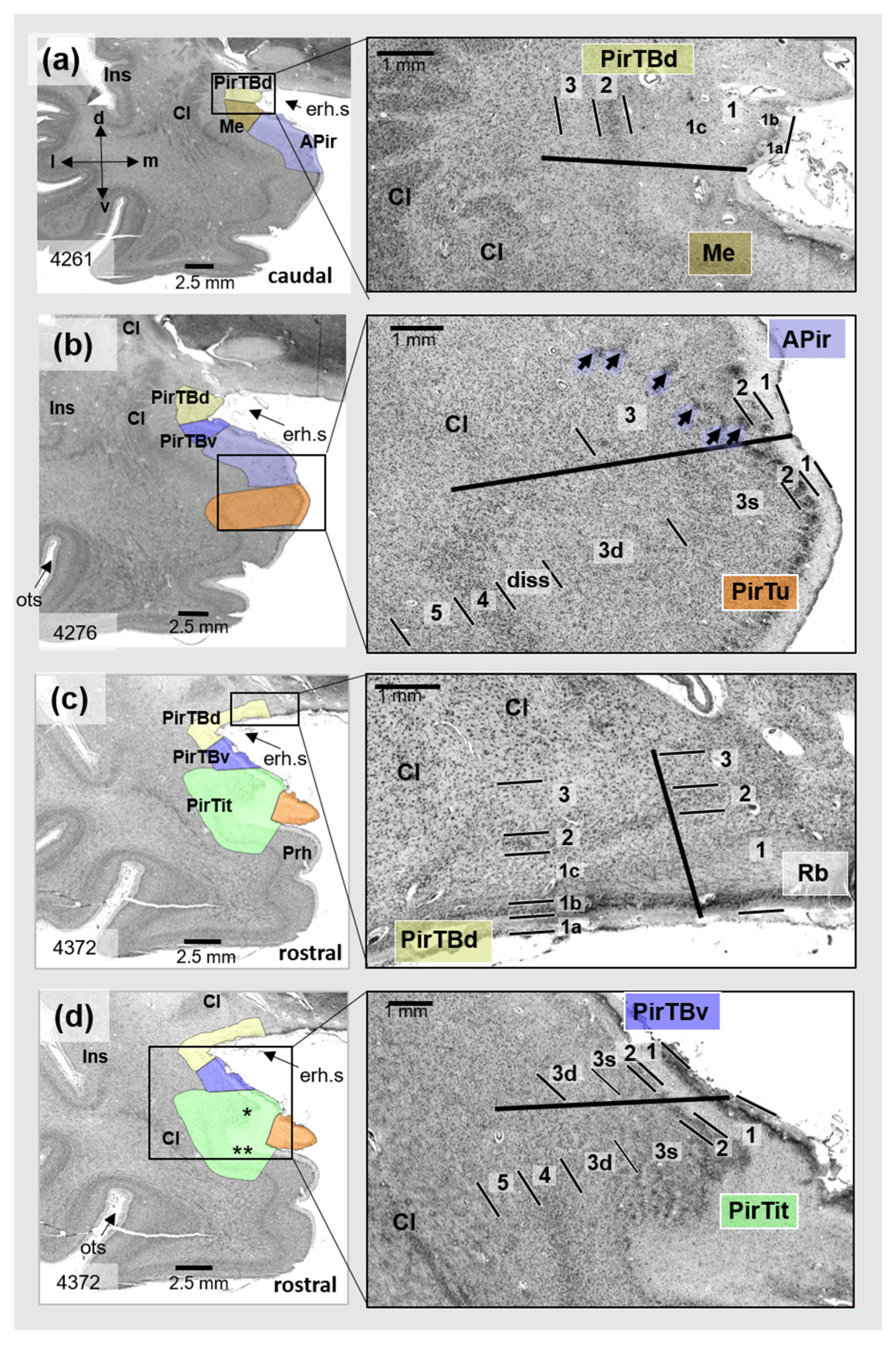
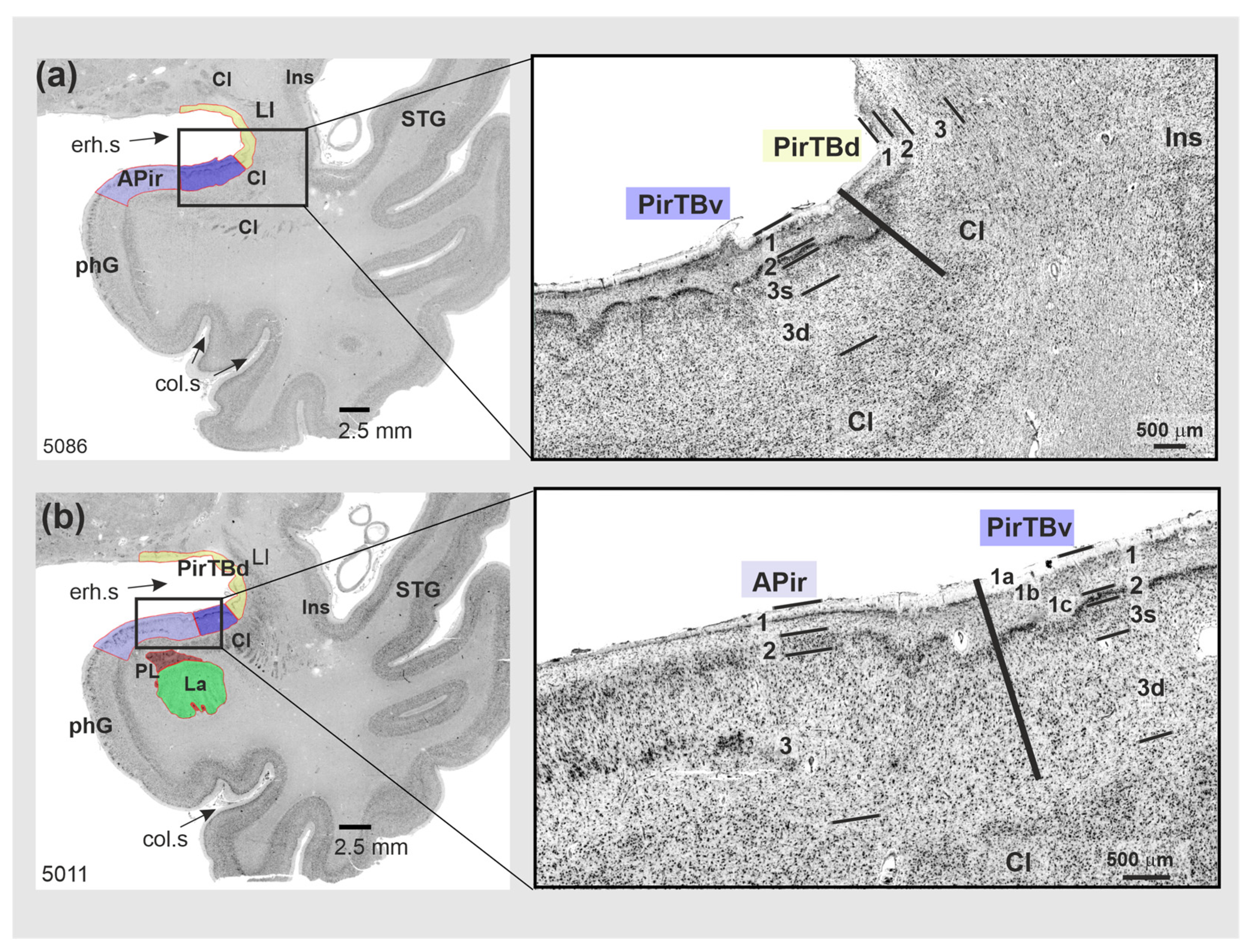
3.1. Cytoarchitecture of Piriform Temporobasal Areas (PirTBd and PirTBv)
3.2. Piriform Temporal Areas (PirTit and PirTu)
3.3. Relation to the Macroanatomical Landmarks
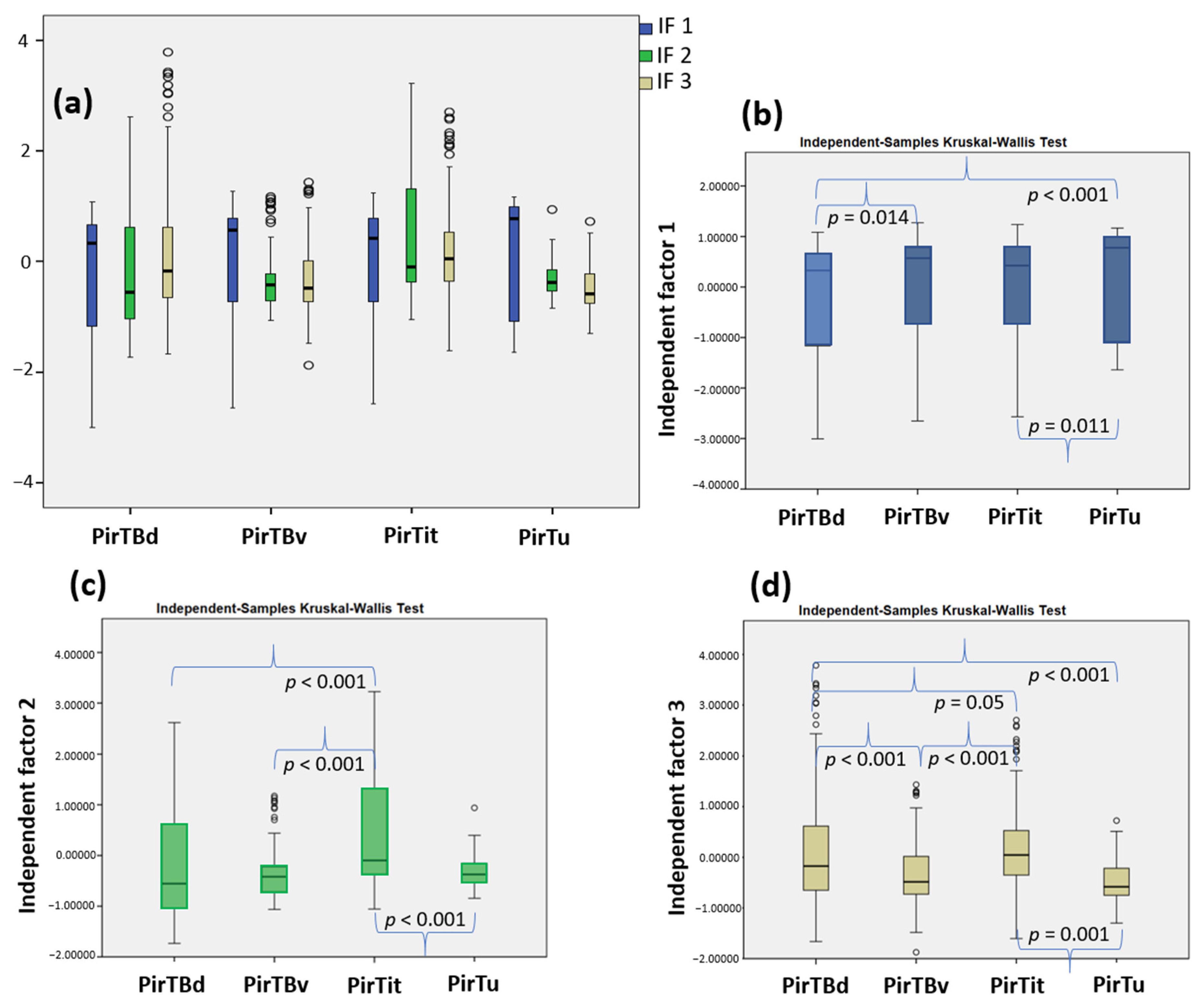
3.4. Texture Analysis
3.5. Interindividual Variability in Volume
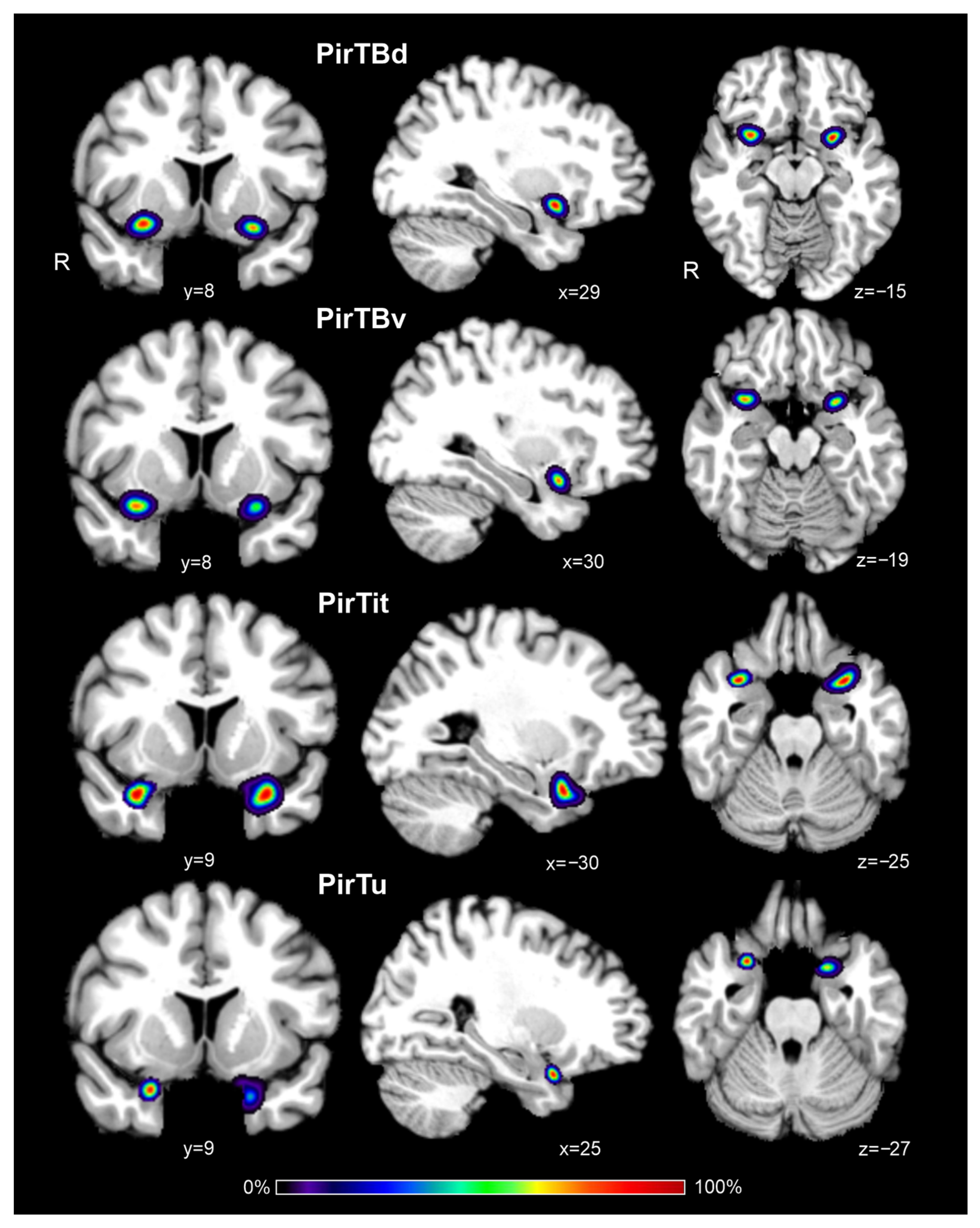
3.6. Interindividual Variability in Extent (Probabilistic Maps and Maximum Probability Maps)
3.7. 3D Reconstruction in the BigBrain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marin, C.; Vilas, D.; Langdon, C.; Alobid, I.; López-Chacón, M.; Haehner, A.; Hummel, T.; Mullol, J. Olfactory Dysfunction in Neurodegenerative Diseases. Curr. Allergy Asthma Rep. 2018, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Ubeda-Bañon, I.; Saiz-Sanchez, D.; Flores-Cuadrado, A.; Rioja-Corroto, E.; Gonzalez-Rodriguez, M.; Villar-Conde, S.; Astillero-Lopez, V.; Cabello-de la Rosa, J.P.; Gallardo-Alcañiz, M.J.; Vaamonde-Gamo, J.; et al. The human olfactory system in two proteinopathies: Alzheimer’s and Parkinson’s diseases. Transl. Neurodegener. 2020, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Leon, M.; Woo, C.C. Olfactory loss is a predisposing factor for depression, while olfactory enrichment is an effective treatment for depression. Front. Neurosci. 2022, 16, 1013363. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, N.; Pearson, J.; Shinoda, K.; Alheid, G.F.; de Olmos, J.S.; Heimer, L. The human basal forebrain. Part I. an overview. In Handbook of Chemical Neuroanatomy; Bloom, F.E., Björklund, A., Hökfelt, T., Eds.; Volume 15: The Primate Nervous System, Part III; Elsevier: Amsterdam, The Netherlands, 1999; pp. 1–56. [Google Scholar]
- Noto, T.; Zhou, G.; Yang, Q.; Lane, G.; Zelano, C. Human Primary Olfactory Amygdala Subregions Form Distinct Functional Networks, Suggesting Distinct Olfactory Functions. Front. Syst. Neurosci. 2021, 15, 752320. [Google Scholar] [CrossRef]
- Gottfried, J.A. Central mechanisms of odour object perception. Nat. Rev. Neurosci. 2010, 11, 628–641. [Google Scholar] [CrossRef] [PubMed]
- Bensafi, M. The Role of the Piriform Cortex in Human Olfactory Perception: Insights from Functional Neuroimaging Studies. Chemosens. Percept. 2012, 5, 4–10. [Google Scholar] [CrossRef]
- Sobel, N.; Prabhakaran, V.; Desmond, J.E.; Glover, G.H.; Goode, R.L.; Sullivan, E.V.; Gabrieli, J.D. Sniffing and smelling: Separate subsystems in the human olfactory cortex. Nature 1998, 392, 282–286. [Google Scholar] [CrossRef]
- Royet, J.P.; Plailly, J.; Delon-Martin, C.; Kareken, D.A.; Segebarth, C. fMRI of emotional responses to odors: Influence of hedonic valence and judgment, handedness, and gender. Neuroimage 2003, 20, 713–728. [Google Scholar] [CrossRef]
- Plailly, J.; Bensafi, M.; Pachot-Clouard, M.; Delon-Martin, C.; Kareken, D.A.; Rouby, C.; Segebarth, C.; Royet, J.P. Involvement of right piriform cortex in olfactory familiarity judgments. Neuroimage 2005, 24, 1032–1041. [Google Scholar] [CrossRef]
- Dade, L.A.; Zatorre, R.J.; Jones-Gotman, M. Olfactory learning: Convergent findings from lesion and brain imaging studies in humans. Brain 2002, 125, 86–101. [Google Scholar] [CrossRef]
- Buchanan, T.W.; Tranel, D.; Adolphs, R. A specific role for the human amygdala in olfactory memory. Learn. Mem. 2003, 10, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Hudry, J.; Perrin, F.; Ryvlin, P.; Mauguière, F.; Royet, J.P. Olfactory short-term memory and related amygdala recordings in patients with temporal lobe epilepsy. Brain 2003, 126, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Nigri, A.; Ferraro, S.; D’Incerti, L.; Critchley, H.D.; Bruzzone, M.G.; Minati, L. Connectivity of the amygdala, piriform, and orbitofrontal cortex during olfactory stimulation: A functional MRI study. Neuroreport 2013, 24, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Bzdok, D.; Laird, A.R.; Zilles, K.; Fox, P.T.; Eickhoff, S.B. An investigation of the structural, connectional, and functional subspecialization in the human amygdala. Hum. Brain Mapp. 2013, 34, 3247–3266. [Google Scholar] [CrossRef] [PubMed]
- Hudry, J.; Ryvlin, P.; Royet, J.P.; Mauguière, F. Odorants elicit evoked potentials in the human amygdala. Cereb. Cortex 2001, 11, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Zald, D.H.; Pardo, J.V. Emotion, olfaction, and the human amygdala: Amygdala activation during aversive olfactory stimulation. Proc. Natl. Acad. Sci. USA 1997, 94, 4119–4124. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.K.; Christoff, K.; Stappen, I.; Panitz, D.; Ghahremani, D.G.; Glover, G.; Gabrieli, J.D.; Sobel, N. Dissociated neural representations of intensity and valence in human olfaction. Nat. Neurosci. 2003, 6, 196–202. [Google Scholar] [CrossRef]
- Olofsson, J.K.; Rogalski, E.; Harrison, T.; Mesulam, M.M.; Gottfried, J.A. A cortical pathway to olfactory naming: Evidence from primary progressive aphasia. Brain 2013, 136, 1245–1259. [Google Scholar] [CrossRef]
- Yao, L.; Pinto, J.M.; Yi, X.; Li, L.; Peng, P.; Wei, Y. Gray matter volume reduction of olfactory cortices in patients with idiopathic olfactory loss. Chem. Senses 2014, 39, 755–760. [Google Scholar] [CrossRef]
- Kedo, O.; Zilles, K.; Palomero-Gallagher, N.; Schleicher, A.; Mohlberg, H.; Bludau, S.; Amunts, K. Receptor-driven, multimodal mapping of the human amygdala. Brain Struct. Funct. 2018, 223, 1637–1666. [Google Scholar] [CrossRef]
- von Economo, C.F.; Koskinas, G.N. Die Cytoarchitektonik der Hirnrinde des Erwachsenen Menschen; Springer: Berlin/Heidelberg, Germany, 1925. [Google Scholar]
- Brockhaus, H. Die Cyto- und Myeloarchitektonik des Cortex claustralis und des Claustrum beim Menschen. J. Psychol. Neurol. 1940, 49, 249–348. [Google Scholar]
- Filimonoff, I.N. Paleocortex, allocortex and cortex intermedius. In Cytoarchitecture of the Human Cortex Cerebri; Sarkiov, S.A., Filimonoff, I.N., Preobrazhenskaya, N.S., Eds.; Medgiz: Moscow, Russia, 1949; pp. 402–433. (In Russian) [Google Scholar]
- Stephan, H. Regio praepiriformis. In Allocortex; Springer: Berlin/Heidelberg, Germany, 1975; pp. 427–464. [Google Scholar]
- Mai, J.K.; Assheuer, J.; Paxinos, G. Atlas of the Human Brain; Thieme: New York, NY, USA, 1997. [Google Scholar]
- Zelano, C.; Bensafi, M.; Porter, J.; Mainland, J.; Johnson, B.; Bremner, E.; Telles, C.; Khan, R.; Sobel, N. Attentional modulation in human primary olfactory cortex. Nat. Neurosci. 2005, 8, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.; Anand, T.; Johnson, B.; Khan, R.M.; Sobel, N. Brain mechanisms for extracting spatial information from smell. Neuron 2005, 47, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Zelano, C.; Montag, J.; Johnson, B.; Khan, R.; Sobel, N. Dissociated representations of irritation and valence in human primary olfactory cortex. J. Neurophysiol. 2007, 97, 1969–1976. [Google Scholar] [CrossRef] [PubMed]
- Zelano, C.; Montag, J.; Khan, R.; Sobel, N. A specialized odor memory buffer in primary olfactory cortex. PLoS ONE 2009, 4, e4965. [Google Scholar] [CrossRef] [PubMed]
- Silveira-Moriyama, L.; Holton, J.L.; Kingsbury, A.; Ayling, H.; Petrie, A.; Sterlacci, W.; Poewe, W.; Maier, H.; Lees, A.J.; Revesz, T. Regional differences in the severity of Lewy body pathology across the olfactory cortex. Neurosci. Lett. 2009, 453, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Gottfried, J.A.; Deichmann, R.; Winston, J.S.; Dolan, R.J. Functional heterogeneity in human olfactory cortex: An event-related functional magnetic resonance imaging study. J. Neurosci. 2002, 22, 10819–10828. [Google Scholar] [CrossRef] [PubMed]
- Gottfried, J.A.; Winston, J.S.; Dolan, R.J. Dissociable codes of odor quality and odorant structure in human piriform cortex. Neuron 2006, 49, 467–479. [Google Scholar] [CrossRef]
- Li, W.; Luxenberg, E.; Parrish, T.; Gottfried, J.A. Learning to smell the roses: Experience-dependent neural plasticity in human piriform and orbitofrontal cortices. Neuron 2006, 52, 1097–1108. [Google Scholar] [CrossRef]
- Kjelvik, G.; Evensmoen, H.R.; Hummel, T.; Engedal, K.; Selbæk, G.; Saltvedt, I.; Håberg, A.K. The Human Brain Representation of Odor Identification in Amnestic Mild Cognitive Impairment and Alzheimer’s Dementia of Mild Degree. Front. Neurol. 2020, 11, 607566. [Google Scholar] [CrossRef]
- Li, W.; Howard, J.D.; Gottfried, J.A. Disruption of odour quality coding in piriform cortex mediates olfactory deficits in Alzheimer’s disease. Brain 2010, 133, 2714–2726. [Google Scholar] [CrossRef] [PubMed]
- Löscher, W.; Ebert, U. The role of the piriform cortex in kindling. Prog. Neurobiol. 1996, 50, 427–481. [Google Scholar] [CrossRef] [PubMed]
- Haberly, L.B. Parallel-distributed processing in olfactory cortex: New insights from morphological and physiological analysis of neuronal circuitry. Chem. Senses 2001, 26, 551–576. [Google Scholar] [CrossRef] [PubMed]
- Litaudon, P.; Amat, C.; Bertrand, B.; Vigouroux, M.; Buonviso, N. Piriform cortex functional heterogeneity revealed by cellular responses to odours. Eur. J. Neurosci. 2003, 17, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, S.T.; Clugnet, M.C.; Price, J.L. Central olfactory connections in the macaque monkey. J. Comp. Neurol. 1994, 346, 403–434. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, D.N.; Jackson, G.D. The piriform cortex and human focal epilepsy. Front. Neurol. 2014, 5, 259. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.K.; Majtanik, M.; Paxinos, G. Atlas of the Human Brain, 4th ed.; Academic Press: Cantabrigian, MA, USA, 2016. [Google Scholar]
- Ding, S.L.; Royall, J.J.; Sunkin, S.M.; Ng, L.; Facer, B.A.; Lesnar, P.; Guillozet-Bongaarts, A.; McMurray, B.; Szafer, A.; Dolbeare, T.A.; et al. Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol. 2016, 524, 3127–3481. [Google Scholar] [CrossRef]
- Ding, S.L.; Van Hoesen, G.W.; Cassell, M.D.; Poremba, A. Parcellation of human temporal polar cortex: A combined analysis of multiple cytoarchitectonic, chemoarchitectonic, and pathological markers. J. Comp. Neurol. 2009, 514, 595–623. [Google Scholar] [CrossRef] [PubMed]
- Amunts, K.; Lepage, C.; Borgeat, L.; Mohlberg, H.; Dickscheid, T.; Rousseau, M.É.; Bludau, S.; Bazin, P.L.; Lewis, L.B.; Oros-Peusquens, A.M.; et al. BigBrain: An ultrahigh-resolution 3D human brain model. Science 2013, 340, 1472–1475. [Google Scholar] [CrossRef]
- Merker, B. Silver staining of cell bodies by means of physical development. J. Neurosci. Methods 1983, 9, 235–241. [Google Scholar] [CrossRef]
- Amunts, K.; Mohlberg, H.; Bludau, S.; Zilles, K. Julich-Brain: A 3D probabilistic atlas of the human brain’s cytoarchitecture. Science 2020, 369, 988–992. [Google Scholar] [CrossRef] [PubMed]
- Stephan, H. Regio peripalaeocorticalis claustralis. In Allocortex; Springer: Berlin/Heidelberg, Germany, 1975; pp. 464–494. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Löfstedt, T.; Brynolfsson, P.; Asklund, T.; Nyholm, T.; Garpebring, A. Gray-level invariant Haralick texture features. PLoS ONE 2019, 14, e0212110. [Google Scholar] [CrossRef]
- Kaiser, H.F. An index of factorial simplicity. Psychometrika 1974, 39, 31–36. [Google Scholar] [CrossRef]
- Mohlberg, H.; Eickhoff, S.B.; Schleicher, A.; Zilles, K.; Amunts, K. A new processing pipeline and release of cytoarchitectonic probabilistic maps. In Proceedings of the 18th Annual Meeting of the Organization for Human Brain Mapping, OHBM 18, Beijing, China, 10 June 2012. [Google Scholar]
- Collins, D.L.; Neelin, P.; Peters, T.M.; Evans, A.C. Automatic 3D Intersubject Registration of MR Volumetric Data in Standardized Talairach Space. J. Comput. Assist. Tomogr. 1994, 18, 192–205. [Google Scholar] [CrossRef]
- Hömke, L. A multigrid method for anisotropic PDE’s in elastic im age registration. Numer. Linear Algebra Appl. 2006, 13, 215–229. [Google Scholar] [CrossRef]
- Eickhoff, S.B.; Stephan, K.E.; Mohlberg, H.; Grefkes, C.; Fink, G.R.; Amunts, K.; Zilles, K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage 2005, 25, 1325–1335. [Google Scholar] [CrossRef]
- Gundersen, H.J.; Bendtsen, T.F.; Korbo, L.; Marcussen, N.; Møller, A.; Nielsen, K.; Nyengaard, J.R.; Pakkenberg, B.; Sørensen, F.B.; Vesterby, A.; et al. Some new, simple and efficient stereological methods and their use in pathological research and diagnosis. Apmis 1988, 96, 379–394. [Google Scholar] [CrossRef]
- Amunts, K.; Kedo, O.; Kindler, M.; Pieperhoff, P.; Mohlberg, H.; Shah, N.J.; Habel, U.; Schneider, F.; Zilles, K. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: Intersubject variability and probability maps. Anat. Embryol. 2005, 210, 343–352. [Google Scholar] [CrossRef]
- Bludau, S.; Eickhoff, S.B.; Mohlberg, H.; Caspers, S.; Laird, A.R.; Fox, P.T.; Schleicher, A.; Zilles, K.; Amunts, K. Cytoarchitecture, probability maps and functions of the human frontal pole. Neuroimage 2014, 93, 260–275. [Google Scholar] [CrossRef]
- Eickhoff, S.B.; Schleicher, A.; Scheperjans, F.; Palomero-Gallagher, N.; Zilles, K. Analysis of neurotransmitter receptor distribution patterns in the cerebral cortex. Neuroimage 2007, 34, 1317–1330. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.; Thörnig, P. JURECA: Modular supercomputer at Jülich Supercomputing Centre. JLSRF 2018, 4, A132. [Google Scholar] [CrossRef]
- Schiffer, C.; Spitzer, H.; Kiwitz, K.; Unger, N.; Wagstyl, K.; Evans, A.C.; Harmeling, S.; Amunts, K.; Dickscheid, T. Convolutional neural networks for cytoarchitectonic brain mapping at large scale. Neuroimage 2021, 240, 118327. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Omidyeganeh, M.; Lepage, C.; Wagstyl, K.; Spitzer, H.; Dickscheid, T.; Amunts, K.; Evans, A. Non-linear registration of 1 μm histology sections into 3D 20 μm BigBrain space. In Proceedings of the 26th Annual Meeting of the Organization for Human Brain Mapping, Montreal, QC, Canada, 26–30 June 2020. [Google Scholar]
- Stephan, H. Regio retrobulbaris. In Allocortex; Springer: Berlin/Heidelberg, Germany, 1975; pp. 289–309. [Google Scholar]
- Behuet, S.; Bludau, S.; Kedo, O.; Schiffer, C.; Dickscheid, T.; Brandstetter, A.; Massicotte, P.; Omidyeganeh, M.; Evans, A.; Amunts, K. A High-Resolution Model of the Human Entorhinal Cortex in the ‘BigBrain’—Use Case for Machine Learning and 3D Analyses. In Brain-Inspired Computing, Proceedings of the 4th International Workshop, BrainComp 2019, Cetraro, Italy, 15–19 July 2019; Springer: Cham, Switzerland, 2019; pp. 3–21. Available online: https://doi.org/10.1007/978-3-030-82427-3_1 (accessed on 24 March 2024).
- de Olmos, J.S. Amygdala. In The Human Nervous System, 2nd ed.; Paxinos, G., Mai, J.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2004; pp. 739–868. [Google Scholar]
- Catani, M.; Dell’acqua, F.; Thiebaut de Schotten, M. A revised limbic system model for memory, emotion and behaviour. Neurosci. Biobehav. Rev. 2013, 37, 1724–1737. [Google Scholar] [CrossRef]
- Insausti, R.; Juottonen, K.; Soininen, H.; Insausti, A.M.; Partanen, K.; Vainio, P.; Laakso, M.P.; Pitkänen, A. MR volumetric analysis of the human entorhinal, perirhinal, and temporopolar cortices. AJNR Am. J. Neuroradiol. 1998, 19, 659–671. [Google Scholar]
- Gonçalves Pereira, P.M.; Insausti, R.; Artacho-Pérula, E.; Salmenperä, T.; Kälviäinen, R.; Pitkänen, A. MR volumetric analysis of the piriform cortex and cortical amygdala in drug-refractory temporal lobe epilepsy. AJNR Am. J. Neuroradiol. 2005, 26, 319–332. [Google Scholar] [PubMed]
- Palomero-Gallagher, N.; Kedo, O.; Mohlberg, H.; Zilles, K.; Amunts, K. Multimodal mapping and analysis of the cyto- and receptorarchitecture of the human hippocampus. Brain Struct. Funct. 2020, 225, 881–907. [Google Scholar] [CrossRef] [PubMed]
- Brodmann, K. Vergleichende Lokalisationslehre der Großhirnrinde. 1909. Available online: https://wellcomecollection.org/works/vrnkkxtj/items?canvas=2 (accessed on 24 March 2024).
- Heimer, L.; De Olmos, J.S.; Alheid, G.F.; Pearson, J.; Sakamoto, N.; Shinoda, K.; Marksteiner, J.; Switzer Iii, R.C. The human basal forebrain. Part II. In Handbook of Chemical Neuroanatomy; Elsevier: Amsterdam, The Netherlands, 1999; Volume 15, pp. 57–226. [Google Scholar]
- Iqbal, S.; Leon-Rojas, J.E.; Galovic, M.; Vos, S.B.; Hammers, A.; de Tisi, J.; Koepp, M.J.; Duncan, J.S. Volumetric analysis of the piriform cortex in temporal lobe epilepsy. Epilepsy Res. 2022, 185, 106971. [Google Scholar] [CrossRef]
- Stephan, H.; Frahm, H.; Baron, G. New and revised data on volumes of brain structures in insectivores and primates. Folia Primatol. 1981, 35, 1–29. [Google Scholar] [CrossRef]
- Koehler, L.; Fournel, A.; Albertowski, K.; Roessner, V.; Gerber, J.; Hummel, C.; Hummel, T.; Bensafi, M. Impaired Odor Perception in Autism Spectrum Disorder Is Associated with Decreased Activity in Olfactory Cortex. Chem. Senses 2018, 43, 627–634. [Google Scholar] [CrossRef]
- Menassa, D.A.; Sloan, C.; Chance, S.A. Primary olfactory cortex in autism and epilepsy: Increased glial cells in autism. Brain Pathol. 2017, 27, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Vasavada, M.M.; Wang, J.; Eslinger, P.J.; Gill, D.J.; Sun, X.; Karunanayaka, P.; Yang, Q.X. Olfactory cortex degeneration in Alzheimer’s disease and mild cognitive impairment. J. Alzheimers Dis. 2015, 45, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Saiz-Sanchez, D.; De la Rosa-Prieto, C.; Ubeda-Banon, I.; Martinez-Marcos, A. Interneurons, tau and amyloid-β in the piriform cortex in Alzheimer’s disease. Brain Struct. Funct. 2015, 220, 2011–2025. [Google Scholar] [CrossRef] [PubMed]
- Wattendorf, E.; Welge-Lüssen, A.; Fiedler, K.; Bilecen, D.; Wolfensberger, M.; Fuhr, P.; Hummel, T.; Westermann, B. Olfactory impairment predicts brain atrophy in Parkinson’s disease. J. Neurosci. 2009, 29, 15410–15413. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, P.S.; Esiri, M.M.; Reading, M.; McShane, R.; Nagy, Z. Alpha-synuclein pathology in the olfactory pathways of dementia patients. J. Anat. 2007, 211, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Baba, T.; Kikuchi, A.; Hasegawa, T.; Sugeno, N.; Konno, M.; Miura, E.; Mori, E. Olfactory dysfunction and dementia in Parkinson’s disease. J. Parkinsons Dis. 2014, 4, 181–187. [Google Scholar] [CrossRef]
- Harding, A.J.; Stimson, E.; Henderson, J.M.; Halliday, G.M. Clinical correlates of selective pathology in the amygdala of patients with Parkinson’s disease. Brain 2002, 125, 2431–2445. [Google Scholar] [CrossRef]
- Cortese, B.M.; McConnell, P.A.; Froeliger, B.; Leslie, K.; Uhde, T.W. Burning odor-elicited anxiety in OEF/OIF combat veterans: Inverse relationship to gray matter volume in olfactory cortex. J. Psychiatr. Res. 2015, 70, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Bitter, T.; Gudziol, H.; Burmeister, H.P.; Mentzel, H.J.; Guntinas-Lichius, O.; Gaser, C. Anosmia leads to a loss of gray matter in cortical brain areas. Chem. Senses 2010, 35, 407–415. [Google Scholar] [CrossRef]
- Bitter, T.; Brüderle, J.; Gudziol, H.; Burmeister, H.P.; Gaser, C.; Guntinas-Lichius, O. Gray and white matter reduction in hyposmic subjects—A voxel-based morphometry study. Brain Res. 2010, 1347, 42–47. [Google Scholar] [CrossRef]
- Esposito, F.; Cirillo, M.; De Micco, R.; Caiazzo, G.; Siciliano, M.; Russo, A.G.; Monari, C.; Coppola, N.; Tedeschi, G.; Tessitore, A. Olfactory loss and brain connectivity after COVID-19. Hum. Brain Mapp. 2022, 43, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Aroniadou-Anderjaska, V.; Fritsch, B.; Qashu, F.; Braga, M.F. Pathology and pathophysiology of the amygdala in epileptogenesis and epilepsy. Epilepsy Res. 2008, 78, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Chee, K.; Razmara, A.; Geller, A.S.; Harris, W.B.; Restrepo, D.; Thompson, J.A.; Kramer, D.R. The role of the piriform cortex in temporal lobe epilepsy: A current literature review. Front. Neurol. 2022, 13, 1042887. [Google Scholar] [CrossRef] [PubMed]
- Makhalova, J.; Le Troter, A.; Aubert-Conil, S.; Giusiano, B.; McGonigal, A.; Trebuchon, A.; Carron, R.; Medina Villalon, S.; Bénar, C.G.; Ranjeva, J.P.; et al. Epileptogenic networks in drug-resistant epilepsy with amygdala enlargement: Assessment with stereo-EEG and 7 T MRI. Clin. Neurophysiol. 2022, 133, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Ciumas, C.; Lindström, P.; Aoun, B.; Savic, I. Imaging of odor perception delineates functional disintegration of the limbic circuits in mesial temporal lobe epilepsy. Neuroimage 2008, 39, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Galovic, M.; Baudracco, I.; Wright-Goff, E.; Pillajo, G.; Nachev, P.; Wandschneider, B.; Woermann, F.; Thompson, P.; Baxendale, S.; McEvoy, A.W.; et al. Association of Piriform Cortex Resection With Surgical Outcomes in Patients With Temporal Lobe Epilepsy. JAMA Neurol. 2019, 76, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Gogshelidze, D.; Chervakov, P.; Amunts, K.; Dickscheid, T. Siibra-Explorer—Interactive Web Viewer for Multilevel Brain Atlases (v2.10.1); Zenodo: Geneva, Switzerland, 2023. [Google Scholar] [CrossRef]
- Young, J.C.; Vaughan, D.N.; Paolini, A.G.; Jackson, G.D. Electrical stimulation of the piriform cortex for the treatment of epilepsy: A review of the supporting evidence. Epilepsy Behav. 2018, 88, 152–161. [Google Scholar] [CrossRef]
- Cota, V.R.; Drabowski, B.M.; de Oliveira, J.C.; Moraes, M.F. The epileptic amygdala: Toward the development of a neural prosthesis by temporally coded electrical stimulation. J. Neurosci. Res. 2016, 94, 463–485. [Google Scholar] [CrossRef]



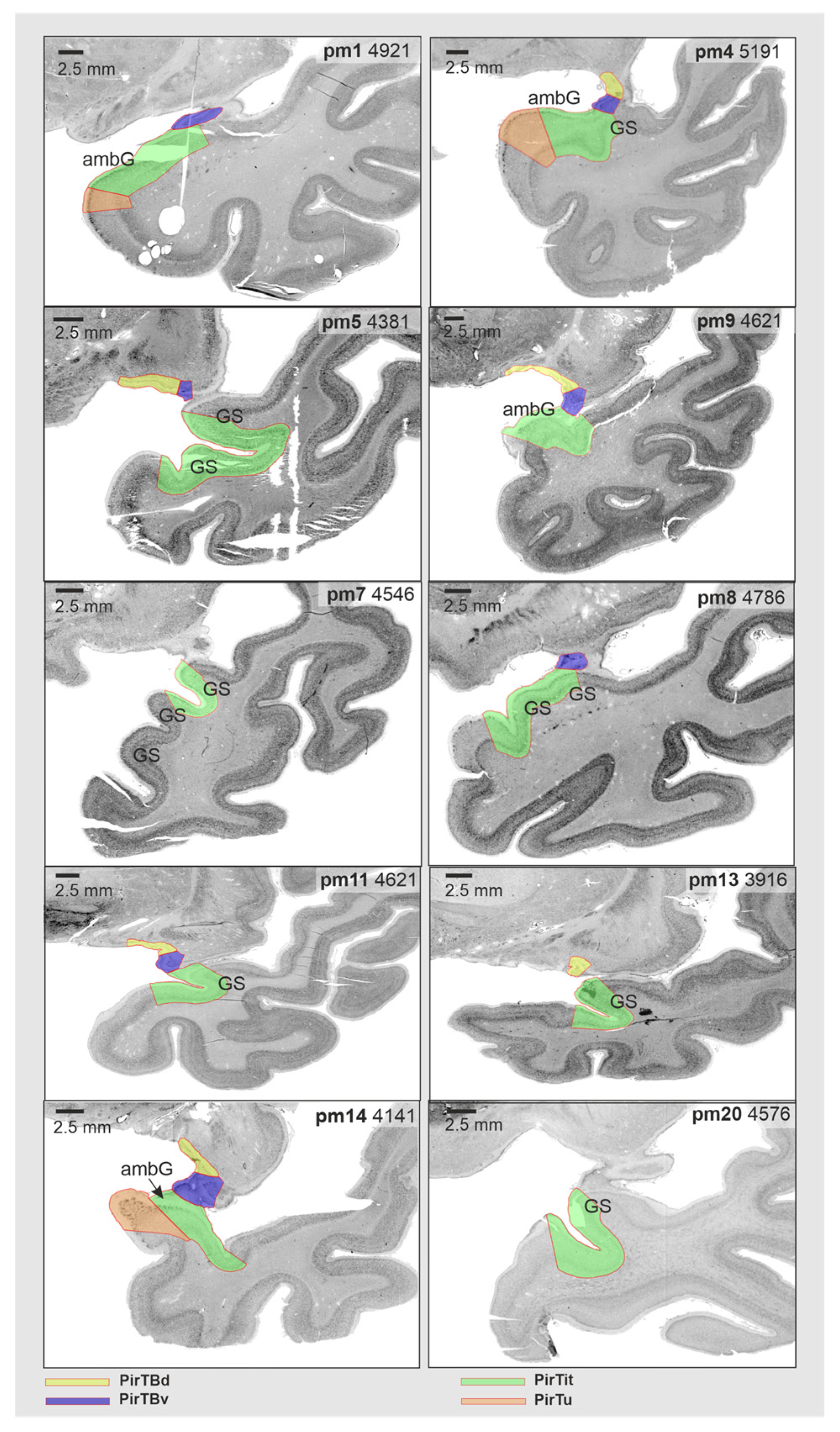


| Case | Brain | Cause of Death | Gender | Weight 1, Grams | Age at Death, Years |
|---|---|---|---|---|---|
| 1 | pm 1 | Carcinoma of the bladder | female | 1350 | 79 |
| 2 | pm 4 | Toxic glomerulonephritis | male | 1349 | 75 |
| 3 | pm 5 | Cardiorespiratory insufficiency | female | 1142 | 59 |
| 4 | pm 7 | Right heart failure | male | 1437 | 37 |
| 5 | pm 8 | Renal failure | female | 1216 | 72 |
| 6 | pm 9 | Cardiorespiratory insufficiency | female | 1110 | 79 |
| 7 | pm 11 | Cardiac infarction | male | 1381 | 74 |
| 8 | pm 13 | Drowning | male | 1234 | 39 |
| 9 | pm 14 | Cardiorespiratory insufficiency | female | 1113 | 86 |
| 10 | pm 20 * | Cardiorespiratory insufficiency | male | 1392 | 65 |
| Classification | Area | Localization | |
|---|---|---|---|
| In Sections | In Relation to the Landmarks | ||
| Allocortex | PirTBd | caudal | within the Sulcus endorhinalis |
| rostral | medial side of the Limen insulae or basal brain surface | ||
| PirTBv | caudal | dorsomedial surface of temporal lobe | |
| rostral | medial side of the Limen insulae or basal brain surface | ||
| Periallocortex | PirTit | caudal limit | on the rostral ambient gyrus |
| middle | Incisura temporalis, incipient gyri of Schwalbe | ||
| rostral | Temporal pole: dorsomedial surface, gyri of Schwalbe, temporopolar sulcus | ||
| rostral limit | Fundus and/medial bank of the temporopolar sulcus | ||
| PirTu | Full extent | Uncal apex | |
| Mean Volume (±SD), mm3 | PirTBd | PirTBv | PirTit | PirTu |
|---|---|---|---|---|
| Female (left and right) | 50.25 ± 9.71 | 48.14 ± 8.46 | 319.78 ± 96.53 | 92.15 ± 28.80 |
| Male (left and right) | 66.14 ± 23.12 | 61.43 ± 17.00 | 393.94 ± 99.02 | 87.01 ± 29.51 |
| Left (male and female) | 54.85 ± 18.30 | 52.28 ± 12.57 | 338.77 ± 63.79 | 95.75 ± 29.25 |
| Right (male and female) | 61.54 ± 20.23 | 57.29 ± 16.94 | 374.95 ± 131.84 | 83.41 ± 27.84 |
| Volume per Hemisphere/Entire Brain Volume (±SD), % per Hemisphere | PirTBd | PirTBv | PirTit | PirTu |
| Female (left and right) | 4.41 ± 1.01 | 4.20 ± 0.76 | 27.96 ± 8.66 | 7.94 ± 2.12 |
| Male (left and right) | 5.00 ± 1.72 | 4.65 ± 1.23 | 29.83 ± 6.65 | 6.61 ± 2.23 |
| Left (male and female) | 4.43 ± 1.33 | 4.22 ± 0.78 | 27.50 ± 4.41 | 7.80 ± 2.38 |
| Right (male and female) | 4.98 ± 1.49 | 4.63 ± 1.22 | 30.29 ± 9.87 | 6.75 ± 2.03 |
| Coordinates | H | PirTBd | PirTBv | PirTit | PirTu | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | x | y | z | x | y | z | ||
| Colin 27 | L | −26 | 6 | −15 | −27 | 6 | −19 | −29 | 9 | −27 | −22 | 6 | −29 |
| MNI ICBM 152 | L | −27 | 5 | −15 | −28 | 5 | −19 | −31 | 8 | −28 | −24 | 5 | −30 |
| Colin 27 | R | 30 | 7 | −14 | 31 | 8 | −18 | 33 | 10 | −26 | 26 | 9 | −27 |
| MNI ICBM 152 | R | 28 | 7 | −15 | 28 | 7 | −19 | 30 | 9 | −27 | 24 | 8 | −28 |
| Areas | Br [71] | vEK [22] | Brh [23] | F [24] | St [25] | I [68] | S [4] H [72] | D [43] | M [42] | |
|---|---|---|---|---|---|---|---|---|---|---|
| PirTB | PirTBd | 51 | TK | prpd, prpi | Pp2b, Pp2a | Pf | -- | PirF | Pir | PirF |
| PirTBv | prpv | Pp1 (p) | Pt | -- | PirT | PirT | ||||
| PirT | PirTu | Part of prA | eta1, eta0 (p) | -- | PRh (35+ 36) | TPpall (p) | LEC (EO) (vp) | PRC35 | ||
| PirTit | part of TI | mtm | Pper | LEC (EO) (dp), TI (mp) | PirT | |||||
| partly mti | eti | TPCl (p) | ||||||||
| eta0 (p) | e | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kedo, O.; Bludau, S.; Schiffer, C.; Mohlberg, H.; Dickscheid, T.; Amunts, K. Cytoarchitectonic Analysis and 3D Maps of the Mesial Piriform Region in the Human Brain. Anatomia 2024, 3, 68-92. https://doi.org/10.3390/anatomia3020007
Kedo O, Bludau S, Schiffer C, Mohlberg H, Dickscheid T, Amunts K. Cytoarchitectonic Analysis and 3D Maps of the Mesial Piriform Region in the Human Brain. Anatomia. 2024; 3(2):68-92. https://doi.org/10.3390/anatomia3020007
Chicago/Turabian StyleKedo, Olga, Sebastian Bludau, Christian Schiffer, Hartmut Mohlberg, Timo Dickscheid, and Katrin Amunts. 2024. "Cytoarchitectonic Analysis and 3D Maps of the Mesial Piriform Region in the Human Brain" Anatomia 3, no. 2: 68-92. https://doi.org/10.3390/anatomia3020007
APA StyleKedo, O., Bludau, S., Schiffer, C., Mohlberg, H., Dickscheid, T., & Amunts, K. (2024). Cytoarchitectonic Analysis and 3D Maps of the Mesial Piriform Region in the Human Brain. Anatomia, 3(2), 68-92. https://doi.org/10.3390/anatomia3020007







