A Network Pharmacology to Explore the Potential Targets of Canagliflozin and Dapagliflozin in Treating Atherosclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Prediction of Targets for Canagliflozin and Dapagliflozin
2.2. Collection of Disease Targets of Atherosclerosis
2.3. Venn Diagram Plotting
2.4. Protein-Protein Interaction (PPI)
2.5. Gene Functions and Pathway Enrichment Analysis with Potential Targets
2.6. Construction of Target Gene-Drug Network
2.7. Molecular Docking of the Target Gene
3. Results
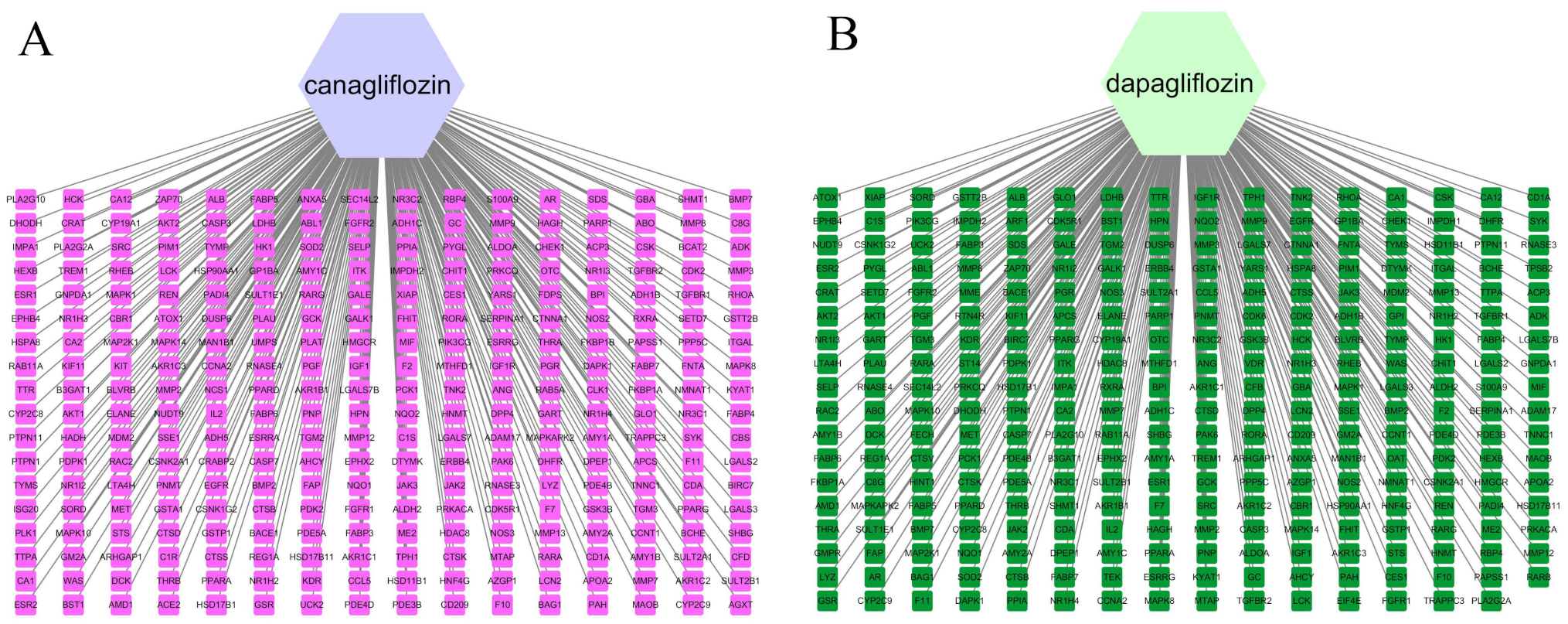
3.1. Network Construction of Drugs and Targets
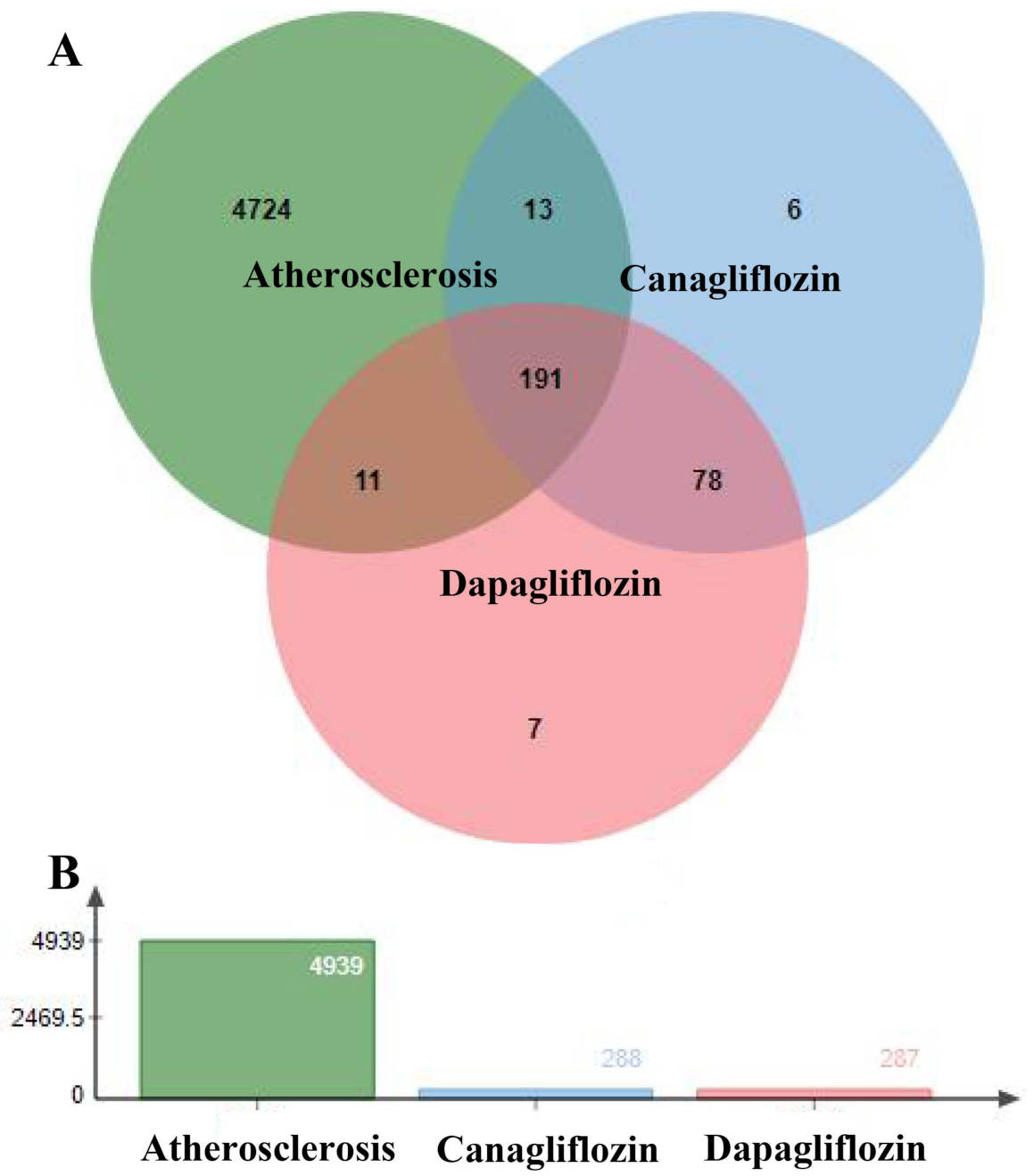
3.2. Targets of Atherosclerosis
3.3. Prediction of Canagliflozin and Dapagliflozin Targets in Atherosclerosis
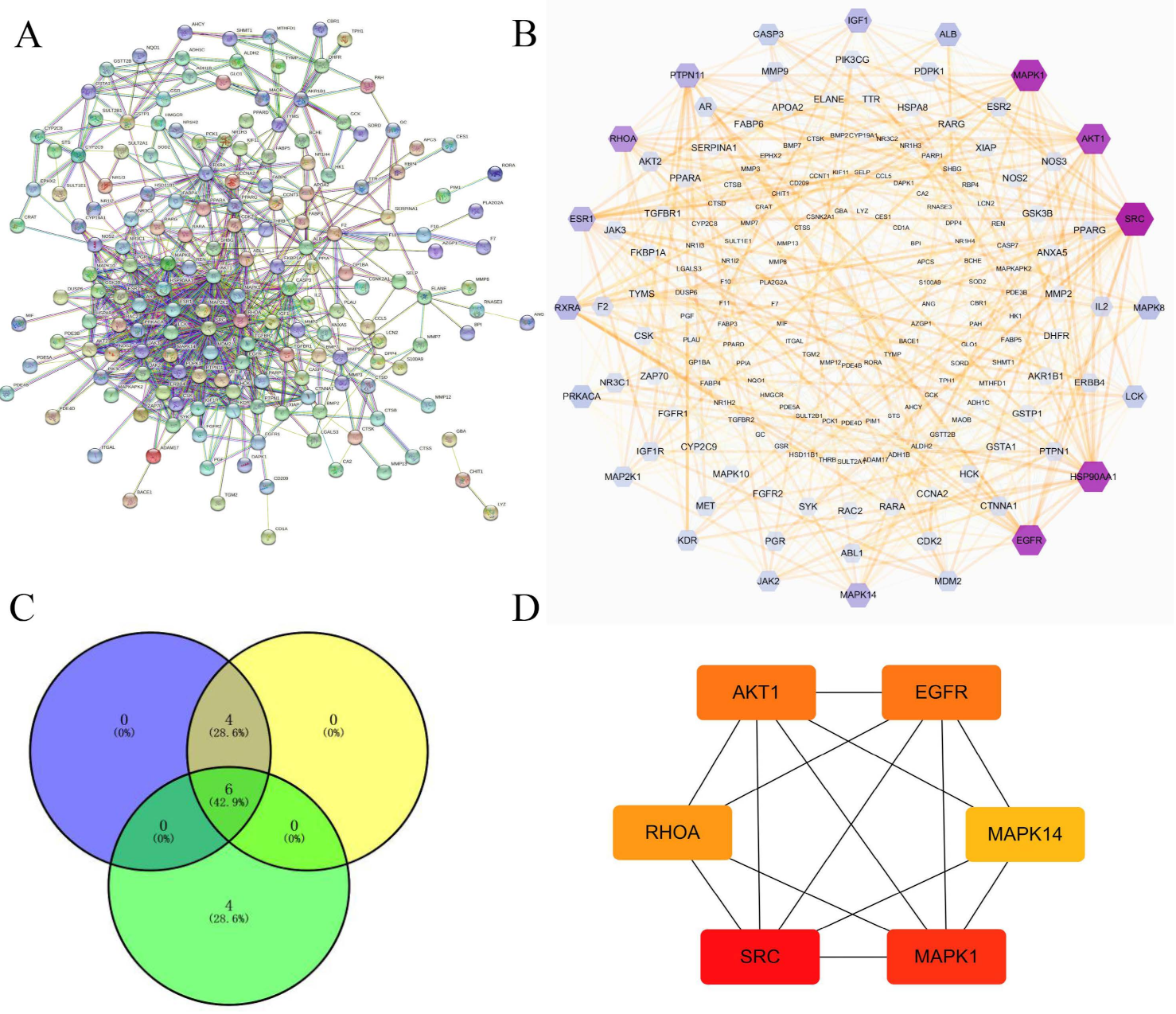
3.4. Construction of PPI Networks
3.5. GO Enrichment Analysis and KEGG Pathway Enrichment Analysis
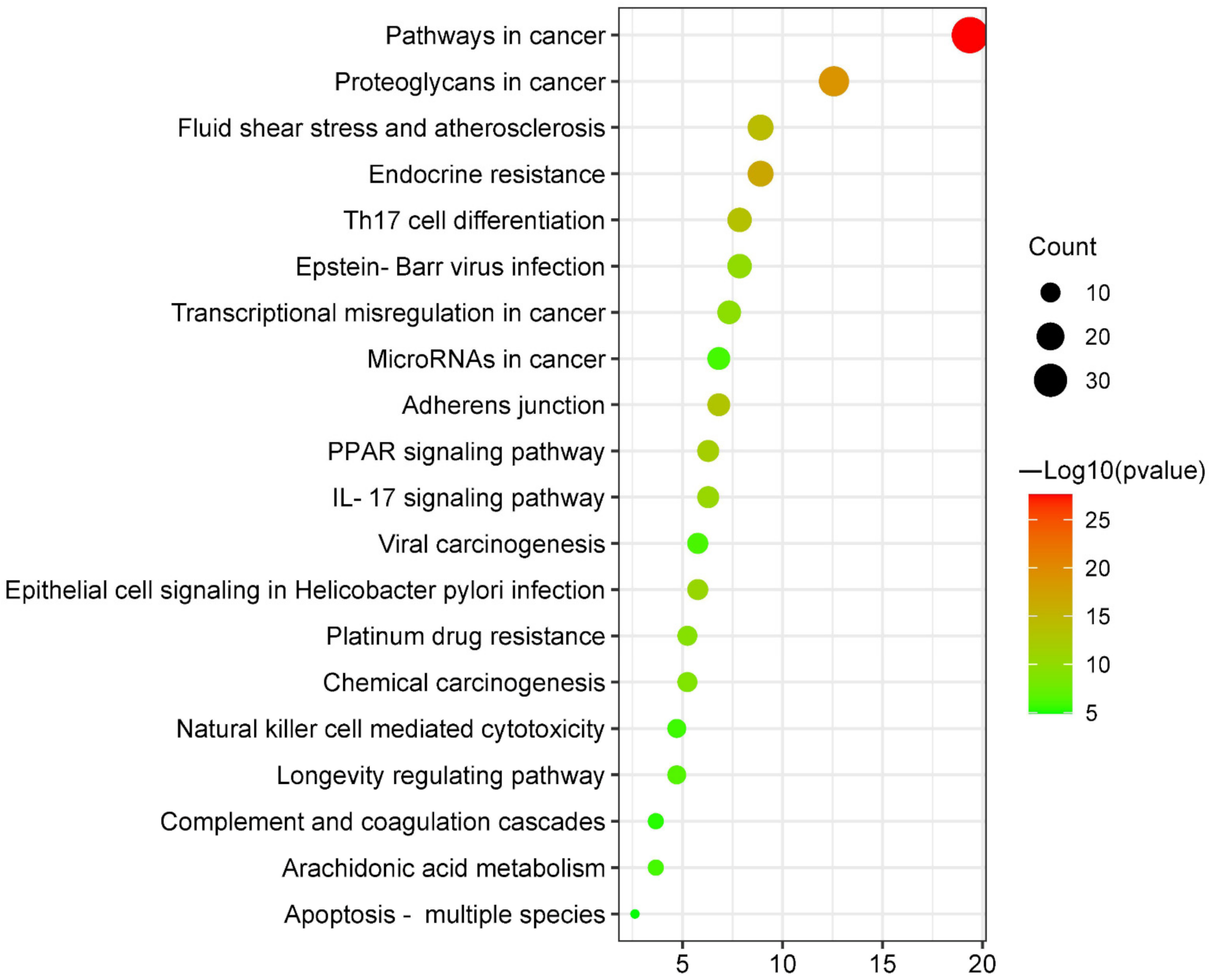
3.5.1. KEGG Pathway Enrichment Analysis
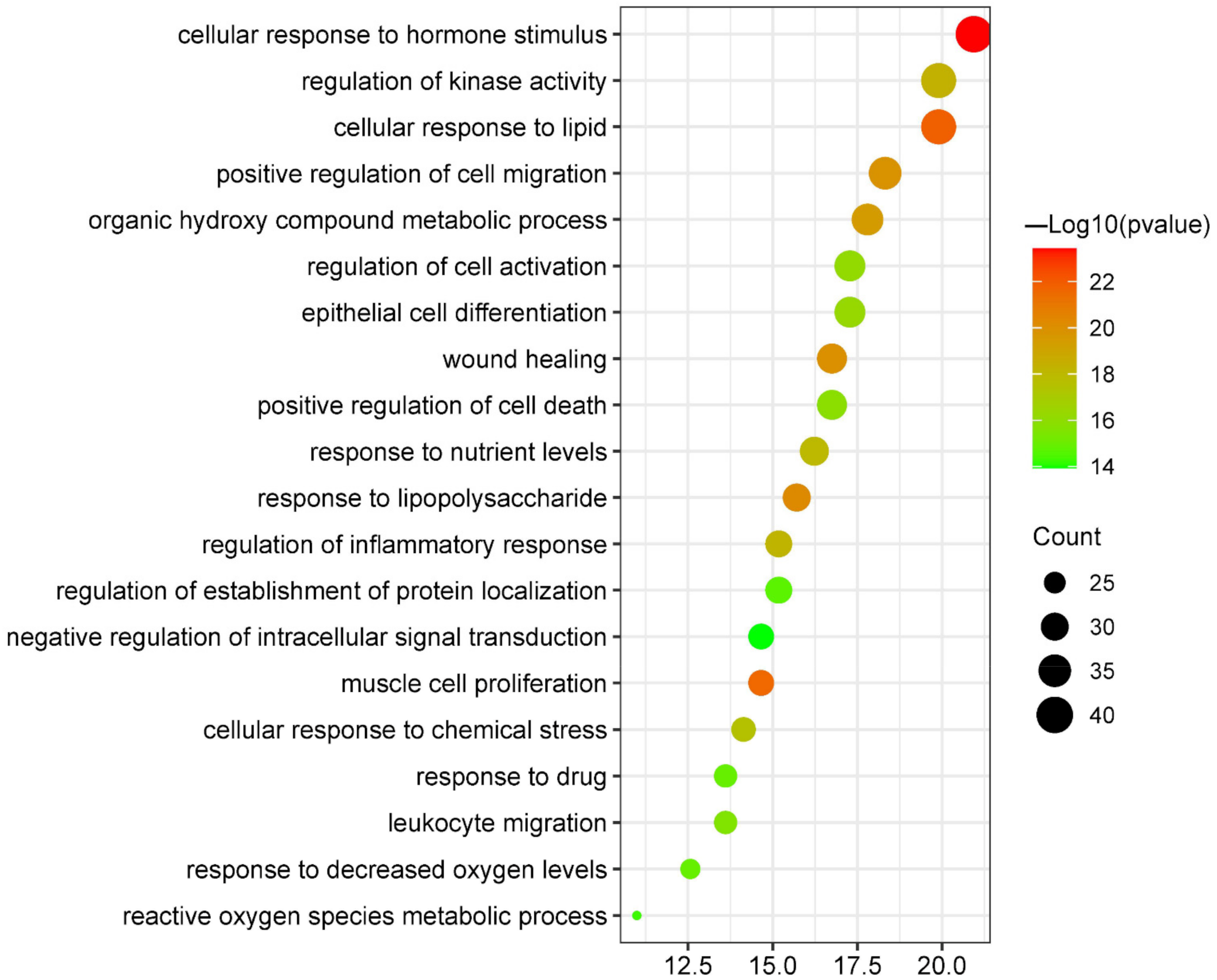
3.5.2. Biological Process Enrichment Analysis
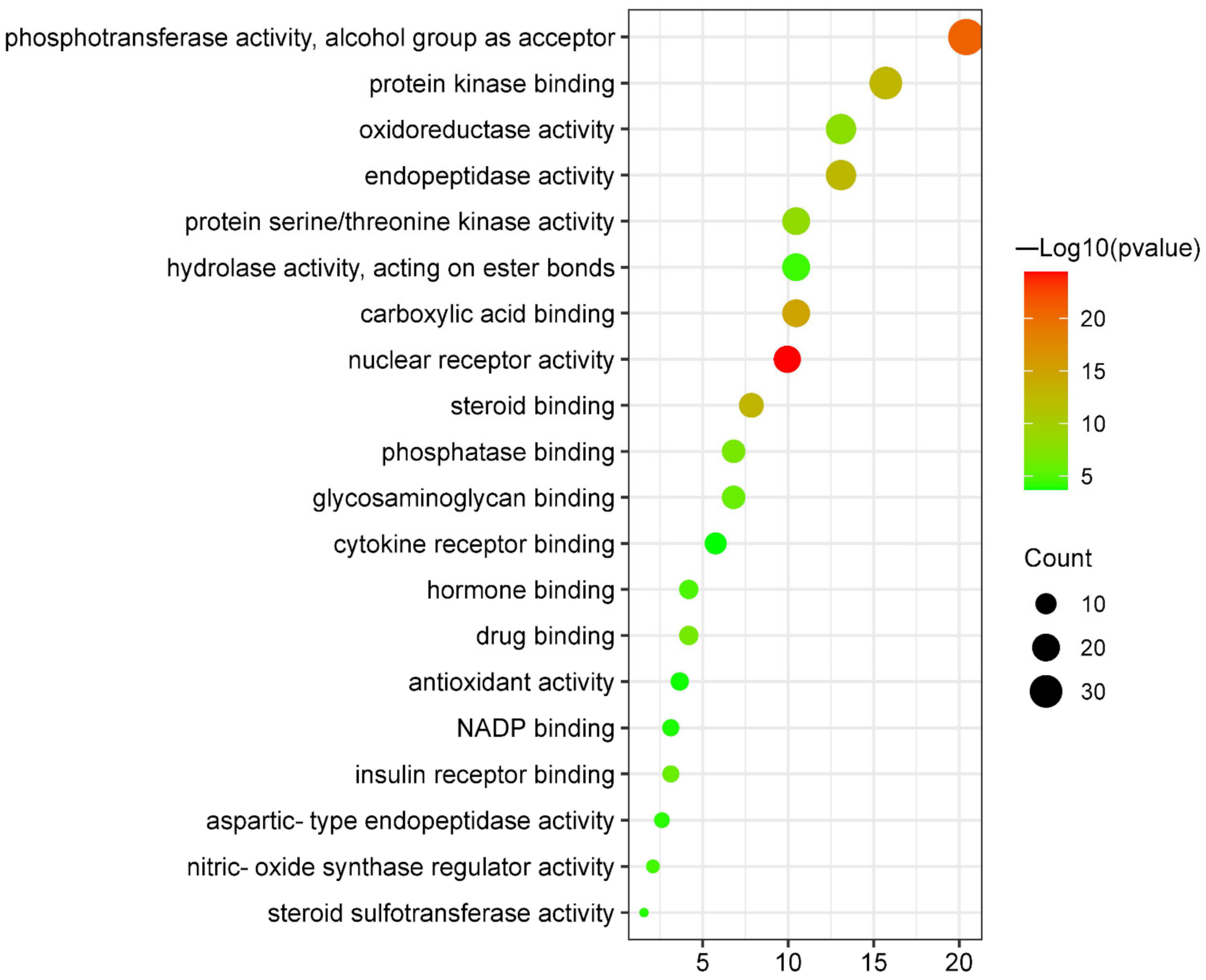
3.5.3. Molecular Functions Enrichment Analysis
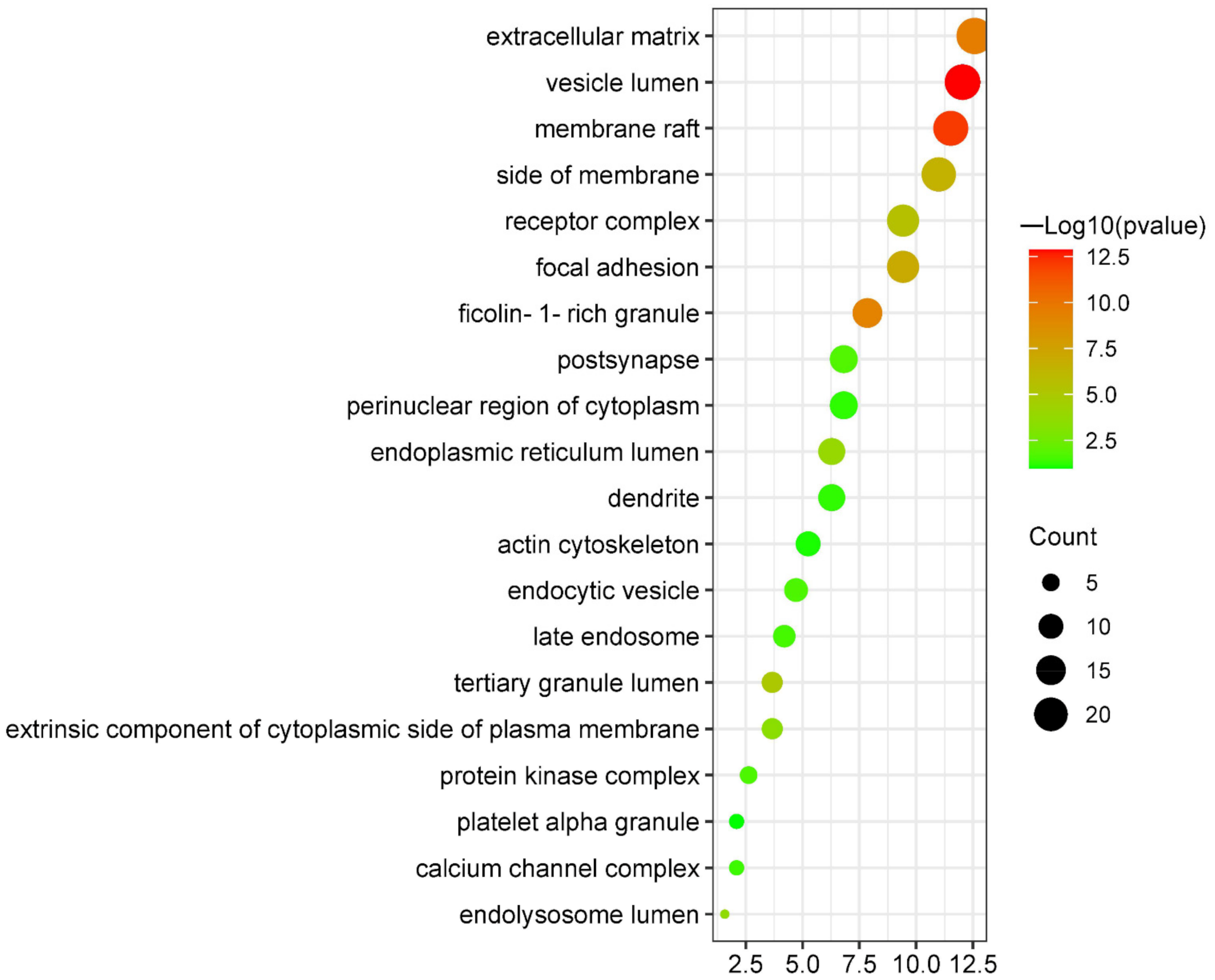
3.5.4. Cellular Components Enrichment Analysis
3.6. Network Construction of Targets-Pathways
3.7. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirichenko, T.V.; Sukhorukov, V.N.; Markin, A.M.; Nikiforov, N.G.; Liu, P.Y.; Sobenin, I.A.; Tarasov, V.V.; Orekhov, A.N.; Aliev, G. Medicinal Plants as a Potential and Successful Treatment Option in the Context of Atherosclerosis. Front. Pharmacol. 2020, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Shan, R.; Liu, N.; Yan, Y.; Liu, B. Apoptosis, autophagy and atherosclerosis: Relationships and the role of Hsp27. Pharmacol. Res. 2021, 166, 5169. [Google Scholar] [CrossRef] [PubMed]
- Poznyak, A.V.; Wu, W.K.; Melnichenko, A.A.; Wetzker, R.; Sukhorukov, V.; Markin, A.M.; Khotina, V.A.; Orekhov, A.N. Signaling Pathways and Key Genes Involved in Regulation of foam Cell Formation in Atherosclerosis. Cells 2020, 9, 584. [Google Scholar] [CrossRef] [PubMed]
- Muller, W.A. Mechanisms of leukocyte transendothelial migration. Annu. Rev. Pathol. 2011, 6, 23–44. [Google Scholar] [CrossRef]
- Moss, J.W.; Ramji, D.P. Nutraceutical therapies for atherosclerosis. Nat. Rev. Cardiol. 2016, 13, 13–32. [Google Scholar] [CrossRef]
- Ghosh, R.K.; Ghosh, S.M.; Chawla, S.; Jasdanwala, S.A. SGLT2 inhibitors: A new emerging therapeutic class in the treatment of type 2 diabetes mellitus. J. Clin. Pharmacol. 2012, 52, 57–63. [Google Scholar] [CrossRef]
- Ghosh-Swaby, O.R.; Goodman, S.G.; Leiter, L.A.; Cheng, A.; Connelly, K.A.; Fitchett, D.; Jüni, P.; Farkouh, M.E.; Udell, J.A. Glucose-lowering drugs or strategies, atherosclerotic cardiovascular events, and heart failure in people with or at risk of type 2 diabetes: An updated systematic review and meta-analysis of randomised cardiovascular outcome trials. Lancet Diabetes Endocrinol. 2020, 8, 18–35. [Google Scholar] [CrossRef]
- Neal, B.; Perkovic, V.; Mahaffey, K.W.; de Zeeuw, D.; Fulcher, G.; Erondu, N.; Shaw, W.; Law, G.; Desai, M.; Matthews, D.R. Canagliflozin and Cardiovascular and Renal Events in Type 2 Diabetes. N. Engl. J. Med. 2017, 377, 44–57. [Google Scholar] [CrossRef]
- McMurray, J.J.V.; Solomon, S.D.; Inzucchi, S.E.; Køber, L.; Kosiborod, M.N.; Martinez, F.A.; Ponikowski, P.; Sabatine, M.S.; Anand, I.S.; Bělohlávek, J.; et al. Dapagliflozin in Patients with Heart Failure and Reduced Ejection Fraction. N. Engl. J. Med. 2019, 381, 995–2008. [Google Scholar] [CrossRef]
- Lee, S.G.; Lee, S.J.; Lee, J.J.; Kim, J.S.; Lee, O.H.; Kim, C.K.; Kim, D.; Lee, Y.H.; Oh, J.; Park, S.; et al. Anti-Inflammatory Effect for Atherosclerosis Progression by Sodium-Glucose Cotransporter 2 (SGLT-2) Inhibitor in a Normoglycemic Rabbit Model. Korean Circ. J. 2020, 50, 43–57. [Google Scholar] [CrossRef]
- Leng, W.; Ouyang, X.; Lei, X.; Wu, M.; Chen, L.; Wu, Q.; Deng, W.; Liang, Z. The SGLT-2 Inhibitor Dapagliflozin Has a Therapeutic Effect on Atherosclerosis in Diabetic ApoE(-/-) Mice. Mediat. Inflamm. 2016, 2016, 305735. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Huai, Y.; Miao, Z.; Qian, A.; Wang, Y. Systems Pharmacology for Investigation of the Mechanisms of Action of Traditional Chinese Medicine in Drug Discovery. Front. Pharmacol. 2019, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.T.; Hajduk, P.J. Rational approaches to targeted polypharmacology: Creating and navigating protein-ligand interaction networks. Curr. Opin. Chem. Biol. 2010, 14, 98–504. [Google Scholar] [CrossRef]
- UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D89–D480. [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database J. Biol. Databases Curation 2010, 2010, baq020. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Piñero, J.; Queralt-Rosinach, N.; Bravo, À.; Deu-Pons, J.; Bauer-Mehren, A.; Baron, M.; Sanz, F.; Furlong, L.I. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database J. Biol. Databases Curation 2015, 2015, bav028. [Google Scholar]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef]
- Liu, F.; Li, L.; Chen, J.; Wu, Y.; Cao, Y.; Zhong, P. A Network Pharmacology to Explore the Mechanism of Calculus Bovis in the Treatment of Ischemic Stroke. BioMed Res. Int. 2021, 2021, 611018. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 85. [Google Scholar] [CrossRef]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; DeJesus, P.; Che, J.; Mulder, L.C.; et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015, 18, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Dan, W.; Liu, J.; Ha, P.; Zhou, T.; Guo, X.; Hou, W. The Use of Traditional Chinese Medicine in Relieving EGFR-TKI-Associated Diarrhea Based on Network Pharmacology and Data Mining. Evid.-Based Complementary Altern. Med. 2021, 2021, 530898. [Google Scholar] [CrossRef]
- Revathidevi, S.; Munirajan, A.K. Akt in cancer: Mediator and more. Semin. Cancer Biol. 2019, 59, 89–91. [Google Scholar] [CrossRef]
- Linton, M.F.; Moslehi, J.J.; Babaev, V.R. Akt Signaling in Macrophage Polarization, Survival, and Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 2703. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 93–604. [Google Scholar] [CrossRef] [PubMed]
- Hasan, R.; Lasker, S.; Hasan, A.; Zerin, F.; Zamila, M.; Parvez, F.; Rahman, M.M.; Khan, F.; Subhan, N.; Alam, M.A. Canagliflozin ameliorates renal oxidative stress and inflammation by stimulating AMPK-Akt-eNOS pathway in the isoprenaline-induced oxidative stress model. Sci. Rep. 2020, 10, 4659. [Google Scholar] [CrossRef]
- Sayour, A.A.; Korkmaz-Icöz, S.; Loganathan, S.; Ruppert, M.; Sayour, V.N.; Oláh, A.; Benke, K.; Brune, M.; Benkő, R.; Horváth, E.M.; et al. Acute canagliflozin treatment protects against in vivo myocardial ischemia-reperfusion injury in non-diabetic male rats and enhances endothelium-dependent vasorelaxation. J. Transl. Med. 2019, 17, 27. [Google Scholar] [CrossRef]
- Gong, L.; Wang, X.; Pan, J.; Zhang, M.; Liu, D.; Liu, M.; Li, L.; An, F. The co-treatment of rosuvastatin with dapagliflozin synergistically inhibited apoptosis via activating the PI3K/AKt/mTOR signaling pathway in myocardial ischemia/reperfusion injury rats. Open Med. 2021, 15, 7–57. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 22–27. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 997–2007. [Google Scholar] [CrossRef] [PubMed]
- Motta, M.; Pannone, L.; Pantaleoni, F.; Bocchinfuso, G.; Radio, F.C.; Cecchetti, S.; Ciolfi, A.; Di Rocco, M.; Elting, M.W.; Brilstra, E.H.; et al. Enhanced MAPK1 Function Causes a Neurodevelopmental Disorder within the RASopathy Clinical Spectrum. Am. J. Hum. Genet. 2020, 107, 99–513. [Google Scholar] [CrossRef] [PubMed]
- Corre, I.; Paris, F.; Huot, J. The p38 pathway, a major pleiotropic cascade that transduces stress and metastatic signals in endothelial cells. Oncotarget 2017, 8, 5684–5714. [Google Scholar] [CrossRef]
- Cheng, M.; Yang, L.; Fan, M.; An, S.; Li, J. Proatherogenic stimuli induce HuR in atherosclerosis through MAPK/ErK pathway. Am. J. Transl. Res. 2019, 11, 317–327. [Google Scholar]
- Reustle, A.; Torzewski, M. Role of p38 MAPK in Atherosclerosis and Aortic Valve Sclerosis. Int. J. Mol. Sci. 2018, 19, 3761. [Google Scholar] [CrossRef]
- Madkour, M.M.; Anbar, H.S.; El-Gamal, M.I. Current status and future prospects of p38α/MAPK14 kinase and its inhibitors. Eur. J. Med. Chem. 2021, 213, 13216. [Google Scholar] [CrossRef]
- Rahadian, A.; Fukuda, D.; Salim, H.M.; Yagi, S.; Kusunose, K.; Yamada, H.; Soeki, T.; Sata, M. Canagliflozin Prevents Diabetes-Induced Vascular Dysfunction in ApoE-Deficient Mice. J. Atheroscler. Thromb. 2020, 27, 141–151. [Google Scholar] [CrossRef]
- Yue, Z.; Li, L.; Fu, H.; Yin, Y.; Du, B.; Wang, F.; Ding, Y.; Liu, Y.; Zhao, R.; Zhang, Z.; et al. Effect of dapagliflozin on diabetic patients with cardiovascular disease via MAPK signalling pathway. J. Cell. Mol. Med. 2021, 25, 500–512. [Google Scholar] [CrossRef]
- El-Sayed, N.; Mostafa, Y.M.; AboGresha, N.M.; Ahmed, A.A.M.; Mahmoud, I.Z.; El-Sayed, N.M. Dapagliflozin attenuates diabetic cardiomyopathy through erythropoietin up-regulation of AKT/JAK/MAPK pathways in streptozotocin-induced diabetic rats. Chem.-Biol. Interact. 2021, 347, 09617. [Google Scholar] [CrossRef] [PubMed]
- De Kock, L.; Freson, K. The (Patho)Biology of SRC Kinase in Platelets and Megakaryocytes. Medicina 2020, 56, 633. [Google Scholar] [CrossRef] [PubMed]
- Byeon, S.E.; Yi, Y.S.; Oh, J.; Yoo, B.C.; Hong, S.; Cho, J.Y. The role of Src kinase in macrophage-mediated inflammatory responses. Mediat. Inflamm. 2012, 2012, 12926. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.; Jung, H.; Chen, J.; Kim, Y.C.; Kim, D.H.; Kong, B.; Guo, G.; Kemper, B.; Kemper, J.K. Phosphorylation of hepatic farnesoid X receptor by FGF19 signaling-activated Src maintains cholesterol levels and protects from atherosclerosis. J. Biol. Chem. 2019, 294, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Linton, M.F.; Babaev, V.R.; Huang, J.; Linton, E.F.; Tao, H.; Yancey, P.G. Macrophage Apoptosis and Efferocytosis in the Pathogenesis of Atherosclerosis. Circ. J. 2016, 80, 259–268. [Google Scholar] [CrossRef] [PubMed]
- El-Daly, M.; Pulakazhi Venu, V.K.; Saifeddine, M.; Mihara, K.; Kang, S.; Fedak, P.W.M.; Alston, L.A.; Hirota, S.A.; Ding, H.; Triggle, C.R.; et al. Hyperglycaemic impairment of PAR2-mediated vasodilation: Prevention by inhibition of aortic endothelial sodium-glucose-co-Transporter-2 and minimizing oxidative stress. Vasc. Pharmacol. 2018, 109, 6–71. [Google Scholar] [CrossRef]
- Sabbah, D.A.; Hajjo, R.; Sweidan, K. Review on Epidermal Growth Factor Receptor (EGFR) Structure, Signaling Pathways, Interactions, and Recent Updates of EGFR Inhibitors. Curr. Top. Med. Chenistry 2020, 20, 15–34. [Google Scholar] [CrossRef]
- Zeboudj, L.; Maître, M.; Guyonnet, L.; Laurans, L.; Joffre, J.; Lemarie, J.; Bourcier, S.; Nour-Eldine, W.; Guérin, C.; Friard, J.; et al. Selective EGF-Receptor Inhibition in CD4(+) T Cells Induces Anergy and Limits Atherosclerosis. J. Am. Coll. Cardiol. 2018, 71, 60–72. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Z.; Huang, W.; Chen, X.; Shan, P.; Zhong, P.; Khan, Z.; Wang, J.; Fang, Q.; Liang, G.; et al. Inhibition of epidermal growth factor receptor attenuates atherosclerosis via decreasing inflammation and oxidative stress. Sci. Rep. 2017, 8, 5917. [Google Scholar] [CrossRef]
- Hahn, C.; Schwartz, M.A. Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 3–62. [Google Scholar] [CrossRef]
- Chiu, J.J.; Chien, S. Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 2011, 91, 27–87. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.F.; Civelek, M.; Fang, Y.; Fleming, I. The atherosusceptible endothelium: Endothelial phenotypes in complex haemodynamic shear stress regions in vivo. Cardiovasc. Res. 2013, 99, 15–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| GO | Description | Gene Ratio | p | Count |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 19.37 | 2.45471 × 10−28 | 37 |
| hsa05205 | Proteoglycans in cancer | 12.57 | 1.51356 × 10−19 | 24 |
| hsa01522 | Endocrine resistance | 8.9 | 1.44544 × 10−17 | 17 |
| hsa05418 | Fluid shear stress and atherosclerosis | 8.9 | 6.76083 × 10−15 | 17 |
| hsa04659 | Th17 cell differentiation | 7.85 | 3.23594 × 10−14 | 15 |
| hsa04520 | Adherens junction | 6.81 | 7.58578 × 10−14 | 13 |
| hsa03320 | PPAR signaling pathway | 6.28 | 1.58489 × 10−12 | 12 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 5.76 | 1.86209 × 10−11 | 11 |
| hsa04657 | IL-17 signaling pathway | 6.28 | 2.63027 × 10−11 | 12 |
| hsa05169 | Epstein–Barr virus infection | 7.85 | 1.09648 × 10−10 | 15 |
| hsa05202 | Transcriptional misregulation in cancer | 7.33 | 2.81838 × 10−10 | 14 |
| hsa01524 | Platinum drug resistance | 5.24 | 6.60693 × 10−10 | 10 |
| hsa05204 | Chemical carcinogenesis | 5.24 | 1.90546 × 10−9 | 10 |
| hsa04211 | Longevity regulating pathway | 4.71 | 3.63078 × 10−7 | 9 |
| hsa05203 | Viral carcinogenesis | 5.76 | 8.12831 × 10−7 | 11 |
| hsa05206 | MicroRNAs in cancer | 6.81 | 1.14815 × 10−6 | 13 |
| hsa00590 | Arachidonic acid metabolism | 3.66 | 1.14815 × 10−6 | 7 |
| hsa04650 | Natural killer cell-mediated cytotoxicity | 4.71 | 1.7378 × 10−6 | 9 |
| hsa04610 | Complement and coagulation cascades | 3.66 | 5.49541 × 10−6 | 7 |
| hsa04215 | Apoptosis-multiple species | 2.62 | 1.25893 × 10−5 | 5 |
| GO | Description | Gene Ratio | p | Count |
|---|---|---|---|---|
| GO:0032870 | cellular response to hormone stimulus | 20.94 | 3.71535 × 10−24 | 40 |
| GO:0071396 | cellular response to lipid | 19.9 | 1.38038 × 10−22 | 38 |
| GO:0033002 | muscle cell proliferation | 14.66 | 2.75423 × 10−22 | 28 |
| GO:0032496 | response to lipopolysaccharide | 15.71 | 5.88844 × 10−21 | 30 |
| GO:0042060 | wound healing | 16.75 | 1.07152 × 10−20 | 32 |
| GO:0030335 | positive regulation of cell migration | 18.32 | 1.38038 × 10−20 | 35 |
| GO:1901615 | organic hydroxy compound metabolic process | 17.8 | 3.89045 × 10−20 | 34 |
| GO:0043549 | regulation of kinase activity | 19.9 | 3.71535 × 10−19 | 38 |
| GO:0050727 | regulation of inflammatory response | 15.18 | 7.24436 × 10−19 | 29 |
| GO:0031667 | response to nutrient levels | 16.23 | 1.02329 × 10−18 | 31 |
| GO:0062197 | cellular response to chemical stress | 14.14 | 3.01995 × 10−18 | 27 |
| GO:0030855 | epithelial cell differentiation | 17.28 | 5.49541 × 10−17 | 33 |
| GO:0050865 | regulation of cell activation | 17.28 | 7.24436 × 10−17 | 33 |
| GO:0010942 | positive regulation of cell death | 16.75 | 1.38038 × 10−16 | 32 |
| GO:0050900 | leukocyte migration | 13.61 | 2.51189 × 10−16 | 26 |
| GO:0036293 | response to decreased oxygen levels | 12.57 | 1.28825 × 10−15 | 24 |
| GO:0042493 | response to drug | 13.61 | 1.28825 × 10−15 | 26 |
| GO:0070201 | regulation of establishment of protein localization | 15.18 | 2.0893 × 10−15 | 29 |
| GO:0072593 | reactive oxygen species metabolic process | 10.99 | 5.24807 × 10−15 | 21 |
| GO:1902532 | negative regulation of intracellular signal transduction | 14.66 | 1.1749 × 10−14 | 28 |
| GO | Description | Gene Ratio | p | Count |
|---|---|---|---|---|
| GO:0004879 | nuclear receptor activity | 9.95 | 3.80 × 10−25 | 19 |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 20.42 | 1.45 × 10−21 | 39 |
| GO:0031406 | carboxylic acid binding | 10.47 | 1.41 × 10−15 | 20 |
| GO:0005496 | steroid binding | 7.85 | 1.26 × 10−13 | 15 |
| GO:0019901 | protein kinase binding | 15.71 | 1.58 × 10−13 | 30 |
| GO:0004175 | endopeptidase activity | 13.09 | 2.00 × 10−13 | 25 |
| GO:0004674 | protein serine/threonine kinase activity | 10.47 | 4.47 × 10−9 | 20 |
| GO:0016491 | oxidoreductase activity | 13.09 | 1.02 × 10−8 | 25 |
| GO:0019902 | phosphatase binding | 6.81 | 1.91 × 10−7 | 13 |
| GO:0008144 | drug binding | 4.19 | 2.57 × 10−7 | 8 |
| GO:0005539 | glycosaminoglycan binding | 6.81 | 1.17 × 10−6 | 13 |
| GO:0005158 | insulin receptor binding | 3.14 | 1.26 × 10−6 | 6 |
| GO:0042562 | hormone binding | 4.19 | 1.48 × 10−5 | 8 |
| GO:0030235 | nitric-oxide synthase regulator activity | 2.09 | 2.63 × 10−5 | 4 |
| GO:0016788 | hydrolase activity, acting on ester bonds | 10.47 | 3.80 × 10−5 | 20 |
| GO:0004190 | aspartic-type endopeptidase activity | 2.62 | 9.12 × 10−5 | 5 |
| GO:0050294 | steroid sulfotransferase activity | 1.57 | 1.05 × 10−4 | 3 |
| GO:0050661 | NADP binding | 3.14 | 1.35 × 10−4 | 6 |
| GO:0016209 | antioxidant activity | 3.66 | 1.78 × 10−4 | 7 |
| GO:0005126 | cytokine receptor binding | 5.76 | 2.14 × 10−4 | 11 |
| GO | Description | Gene Ratio | p | Count |
|---|---|---|---|---|
| GO:0031983 | vesicle lumen | 12.04 | 1.32 × 10−13 | 23 |
| GO:0045121 | membrane raft | 11.52 | 7.76 × 10−13 | 22 |
| GO:0031012 | extracellular matrix | 12.57 | 2.95 × 10−10 | 24 |
| GO:0101002 | ficolin-1-rich granule | 7.85 | 5.75 × 10−10 | 15 |
| GO:0005925 | focal adhesion | 9.42 | 1.12 × 10−7 | 18 |
| GO:0098552 | side of membrane | 10.99 | 2.69 × 10−7 | 21 |
| GO:0043235 | receptor complex | 9.42 | 2.82 × 10−6 | 18 |
| GO:1904724 | tertiary granule lumen | 3.66 | 9.55 × 10−6 | 7 |
| GO:0005788 | endoplasmic reticulum lumen | 6.28 | 1.35 × 10−4 | 12 |
| GO:0036021 | endolysosome lumen | 1.57 | 2.14 × 10−4 | 3 |
| GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | 3.66 | 4.07 × 10−4 | 7 |
| GO:0098794 | postsynapse | 6.81 | 1.95 × 10−2 | 13 |
| GO:1902911 | protein kinase complex | 2.62 | 2.19 × 10−2 | 5 |
| GO:0030139 | endocytic vesicle | 4.71 | 2.24 × 10−2 | 9 |
| GO:0005770 | late endosome | 4.19 | 3.09 × 10−2 | 8 |
| GO:0034704 | calcium channel complex | 2.09 | 3.89 × 10−2 | 4 |
| GO:0030425 | dendrite | 6.28 | 5.50 × 10−2 | 12 |
| GO:0048471 | perinuclear region of cytoplasm | 6.81 | 6.03 × 10−2 | 13 |
| GO:0015629 | actin cytoskeleton | 5.24 | 8.71 × 10−2 | 10 |
| GO:0031091 | platelet alpha granule | 2.09 | 1.07 × 10−1 | 4 |
| Gene | PDB ID | Affinity (kcal/mol) |
|---|---|---|
| AKT1 | 6HHG | −9.22 |
| EGFR | 5UG9 | −9.81 |
| MAPK1 | 4XJ0 | −7.79 |
| MAPK14 | 4L8M | −10.57 |
| RHOA | 1A2B | −5.59 |
| SRC | 2H8H | −9.22 |
| Gene | PDB ID | Affinity (kcal/mol) |
|---|---|---|
| AKT1 | 6HHG | −7.6 |
| EGFR | 5UG9 | −8.31 |
| MAPK1 | 4XJ0 | −7.31 |
| MAPK14 | 4L8M | −11.08 |
| RHOA | 1A2B | −6.90 |
| SRC | 2H8H | −8.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, D.; Ju, W.; Wang, H. A Network Pharmacology to Explore the Potential Targets of Canagliflozin and Dapagliflozin in Treating Atherosclerosis. J. Vasc. Dis. 2022, 1, 53-70. https://doi.org/10.3390/jvd1010007
Wang J, Li D, Ju W, Wang H. A Network Pharmacology to Explore the Potential Targets of Canagliflozin and Dapagliflozin in Treating Atherosclerosis. Journal of Vascular Diseases. 2022; 1(1):53-70. https://doi.org/10.3390/jvd1010007
Chicago/Turabian StyleWang, Jin, Dongning Li, Weiwei Ju, and Hongli Wang. 2022. "A Network Pharmacology to Explore the Potential Targets of Canagliflozin and Dapagliflozin in Treating Atherosclerosis" Journal of Vascular Diseases 1, no. 1: 53-70. https://doi.org/10.3390/jvd1010007
APA StyleWang, J., Li, D., Ju, W., & Wang, H. (2022). A Network Pharmacology to Explore the Potential Targets of Canagliflozin and Dapagliflozin in Treating Atherosclerosis. Journal of Vascular Diseases, 1(1), 53-70. https://doi.org/10.3390/jvd1010007