The Fungal Biorevolution: A Trifecta of Genome Mining, Synthetic Biology, and RNAi for Next-Generation Fungicides
Abstract
1. Introduction: The Imperative for a Paradigm Shift in Fungal Disease Management
1.1. The Twilight of the Chemical Fungicide Era
1.2. The Promise and Pitfalls of First-Generation Biological Control Agents
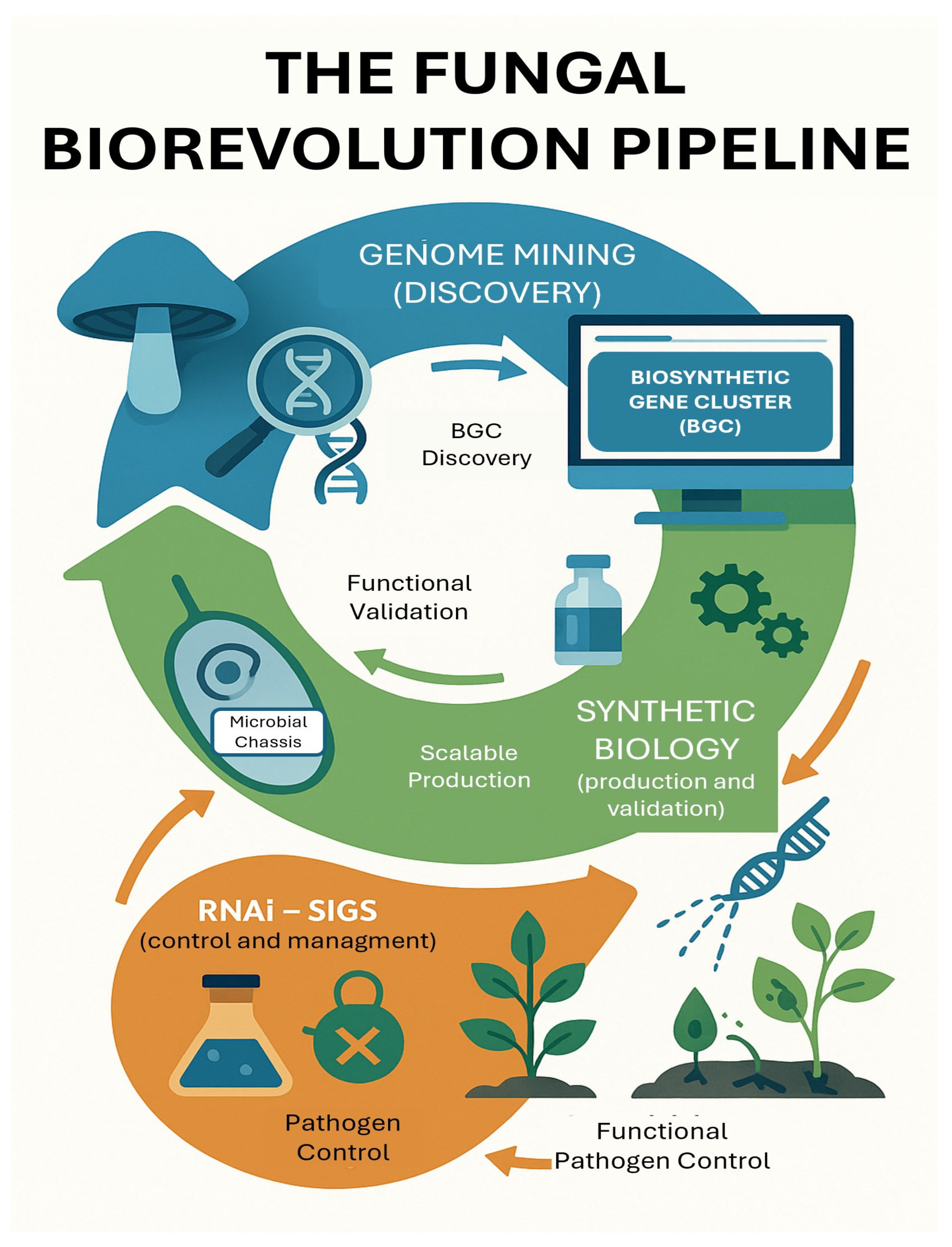
1.3. The Dawn of a New Technological Trifecta
- -
- Genome Mining: For the rational and targeted discovery of new antifungal natural products.
- -
- Synthetic Biology: For the reliable, scalable, and cost-effective production of these discovered products.
- -
- RNA Interference (RNAi): For hyper-specific, non-chemical control and strategic resistance management.
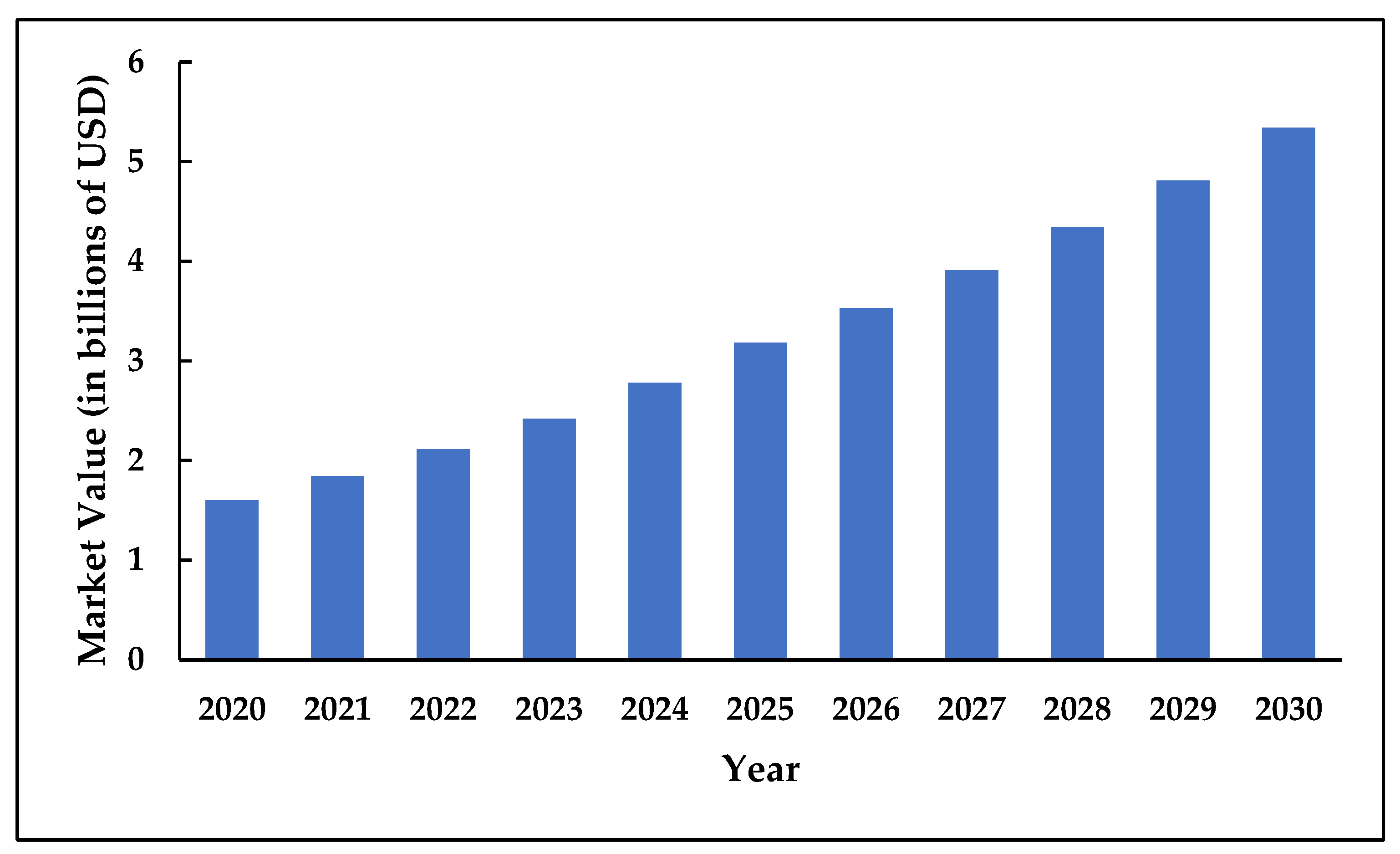
1.4. The Economic Imperative: Market Dynamics of Next-Generation Fungicides
2. Unlocking Nature’s Blueprint: Fungal Genome Mining for Novel Antifungal Chemotypes
2.1. From Random Screening to Rational Discovery
2.2. The Centrality of Biosynthetic Gene Clusters (BGCs)
2.3. Activating the Silent Majority: Strategies to Awaken Cryptic BGCs
2.4. Case Studies in Fungal Bioprospecting
2.5. The Characterization Bottleneck: From Sequence to Function
3. Engineering the Cellular Factory: Synthetic Biology for Scalable Biofungicide Production
3.1. The Microbial Chassis Concept
3.2. The Synthetic Biologist’s Toolkit for Pathway Engineering
3.3. Beyond Imitation: Creating “Better-Than-Nature” Molecules
4. Precision Warfare: RNAi-Based Biofungicides for Targeted Pathogen Neutralization
4.1. The Mechanism of RNAi as a Fungicide
4.2. SIGS: A Non-Transgenic Route for Crop Protection
4.3. Key Challenges and Emerging Solutions for Field Application
4.4. Successful Applications Against Relevant Fungal Pathogens
4.5. From Lab to Field: Overcoming the Hurdles of SIGS Application
| Target Pathogen | Host Plant | Target Gene(s) | dsRNA Delivery Method | Reported Efficacy | Reference |
|---|---|---|---|---|---|
| Botrytis cinerea | Various (tomato, strawberry) | Dicer-like genes (DCL1/2), virulence genes | Spraying of naked dsRNA, nanocarriers | Significant reduction in pre- and post-harvest disease | [50] |
| Fusarium graminearum | Barley, wheat | CYP51 genes (A, B, C) | Spraying of naked dsRNA | Reduction in disease and mycotoxin accumulation | [51] |
| Podosphaera xanthii | Cucumber | Chitin synthase genes | Spraying of naked dsRNA | Inhibition of powdery mildew growth | [20] |
| Sclerotinia sclerotiorum | Canola | Photolyase gene | Spraying of naked dsRNA | Reduction in disease severity | [57] |
| Phomopsis obscurans | Strawberry | Virulence genes | Bioautography with dsRNA | Demonstrated antifungal activity | [31] |
| Fusarium circinatum | Pine | Vesicle trafficking, signal transduction, cell wall biosynthesis genes | Spraying of naked dsRNA | Inhibition of pathogen virulence in pine seedlings | [48] |
5. The Integrated Biofungicide Pipeline: A Synergistic Framework for the Future
5.1. From Silos to Synergy
5.2. A Hypothetical Case Study: Designing a Next-Generation Control Strategy for Botrytis cinerea

6. Ecological Compatibility and Regulatory Horizons
6.1. Designing for the Holobiont
6.2. Assessing Off-Target Effects on Soil Fauna
6.3. Navigating the Regulatory Landscape
7. Conclusions and Future Perspectives
7.1. Summary of the Integrated Vision
7.2. The Role of AI and Machine Learning
7.3. On-Demand and in Situ Production
7.4. A Call to Action
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| BCAs | Biological Control Agents |
| BGC | Biosynthetic Gene Cluster |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats associated protein 9 |
| DMIs | Demethylation Inhibitors |
| dsRNA | double-stranded RNA |
| GRAS | Generally Recognized as Safe |
| HIGS | Host-Induced Gene Silencing |
| IPM | Integrated Pest Management |
| MBCs | Methyl Benzimidazole Carbamates |
| NRPS | Non-Ribosomal Peptide Synthetase |
| OSMAC | One Strain, Many Compounds |
| PKS | Polyketide Synthase |
| PTGS | Post-Transcriptional Gene Silencing |
| QoIs | Quinone outside Inhibitors |
| RNAi | RNA interference |
| SDHIs | Succinate Dehydrogenase Inhibitors |
| SIGS | Spray-Induced Gene Silencing |
| siRNAs | small interfering RNAs |
| SynBio | Synthetic Biology |
References
- Zubrod, J.P.; Bundschuh, M.; Arts, G.; Brühl, C.A.; Imfeld, G.; Knäbel, A.; Payraudeau, S.; Rasmussen, J.J.; Rohr, J.; Scharmüller, A.; et al. Fungicides: An Overlooked Pesticide Class? Environ. Sci. Technol. 2019, 53, 3347–3365. [Google Scholar] [CrossRef]
- Steinberg, G.; Gurr, S.J. Fungi, fungicide discovery and global food security. Fungal Genet. Biol. 2020, 144, 103476. [Google Scholar] [CrossRef] [PubMed]
- Fenta, L.; Mekonnen, H. Microbial Biofungicides as a Substitute for Chemical Fungicides in the Control of Phytopathogens: Current Perspectives and Research Directions. Scientifica 2024, 2024, 5322696. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.-Y.; Patil, A.T.; Huo, D.; Lei, Q.; Kao-Kniffin, J.; Koch, P. Fungicide use intensity influences the soil microbiome and links to fungal disease suppressiveness in amenity turfgrass. Appl. Environ. Microbiol. 2025, 91, e01771-24. [Google Scholar] [CrossRef] [PubMed]
- Bakker, L.; van der Werf, W.; Tittonell, P.; Wyckhuys, K.A.G.; Bianchi, F.J.J.A. Neonicotinoids in global agriculture: Evidence for a new pesticide treadmill? Ecol. Soc. 2020, 25, art26. [Google Scholar] [CrossRef]
- Mikaberidze, A.; Gokhale, C.S.; Bargués-Ribera, M.; Verma, P. The cost of fungicide resistance evolution in multi-field plant epidemics. PLoS Sustain. Transform. 2025, 4, e0000178. [Google Scholar] [CrossRef]
- Lucas, J.A.; Hawkins, N.J.; Fraaije, B.A. The Evolution of Fungicide Resistance. Adv. Appl. Microbiol. 2015, 90, 29–92. [Google Scholar]
- Deising, H.B.; Reimann, S.; Pascholati, S.F. Mechanisms and significance of fungicide resistance. Braz. J. Microbiol. 2008, 39, 286–295. [Google Scholar] [CrossRef]
- Wu, P.-H.; Chang, H.-X.; Shen, Y.-M. Effects of synthetic and environmentally friendly fungicides on powdery mildew management and the phyllosphere microbiome of cucumber. PLoS ONE 2023, 18, e0282809. [Google Scholar] [CrossRef]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.G.; Meis, J.F. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef]
- McLaughlin, M.S.; Yurgel, S.N.; Abbasi, P.A.; Ali, S. The effects of chemical fungicides and salicylic acid on the apple microbiome and fungal disease incidence under changing environmental conditions. Front. Microbiol. 2024, 15, 1342407. [Google Scholar] [CrossRef]
- Cenobio-Galindo, A.d.J.; Hernández-Fuentes, A.D.; González-Lemus, U.; Zaldívar-Ortega, A.K.; González-Montiel, L.; Madariaga-Navarrete, A.; Hernández-Soto, I. Biofungicides Based on Plant Extracts: On the Road to Organic Farming. Int. J. Mol. Sci. 2024, 25, 6879. [Google Scholar] [CrossRef]
- Villavicencio-Vásquez, M.; Espinoza-Lozano, F.; Espinoza-Lozano, L.; Coronel-León, J. Biological control agents: Mechanisms of action, selection, formulation and challenges in agriculture. Front. Agron. 2025, 7, 1578915. [Google Scholar] [CrossRef]
- Bonaterra, A.; Badosa, E.; Cabrefiga, J.; Francés, J.; Montesinos, E. Prospects and limitations of microbial pesticides for control of bacterial and fungal pomefruit tree diseases. Trees 2012, 26, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Galindo, E.; Serrano-Carreón, L.; Gutiérrez, C.R.; Allende, R.; Balderas, K.; Patiño, M.; Trejo, M.; Wong, M.A.; Rayo, E.; Isauro, D.; et al. The challenges of introducing a new biofungicide to the market: A case study. Electron. J. Biotechnol. 2013, 16, 5. [Google Scholar] [CrossRef]
- David, F.; Davis, A.M.; Gossing, M.; Hayes, M.A.; Romero, E.; Scott, L.H.; Wigglesworth, M.J. A Perspective on Synthetic Biology in Drug Discovery and Development—Current Impact and Future Opportunities. SLAS Discov. 2021, 26, 581–603. [Google Scholar] [CrossRef]
- Sellamuthu, G.; Chakraborty, A.; Vetukuri, R.R.; Sarath, S.; Roy, A. RNAi-biofungicides: A quantum leap for tree fungal pathogen management. Crit. Rev. Biotechnol. 2025, 45, 1131–1158. [Google Scholar] [CrossRef]
- Mattern, D.J.; Valiante, V.; Unkles, S.E.; Brakhage, A.A. Synthetic biology of fungal natural products. Front. Microbiol. 2015, 6, 775. [Google Scholar] [CrossRef]
- Zhao, J.; Liang, D.; Li, W.; Yan, X.; Qiao, J.; Caiyin, Q. Research Progress on the Synthetic Biology of Botanical Biopesticides. Bioengineering 2022, 9, 207. [Google Scholar] [CrossRef]
- Ray, P.; Sahu, D.; Aminedi, R.; Chandran, D. Concepts and considerations for enhancing RNAi efficiency in phytopathogenic fungi for RNAi-based crop protection using nanocarrier-mediated dsRNA delivery systems. Front. Fungal Biol. 2022, 3, 977502. [Google Scholar] [CrossRef]
- Siddiqui, M.S.; Thodey, K.; Trenchard, I.; Smolke, C.D. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012, 12, 144–170. [Google Scholar] [CrossRef]
- Chen, C.; Imran, M.; Feng, X.; Shen, X.; Sun, Z. Spray-induced gene silencing for crop protection: Recent advances and emerging trends. Front. Plant Sci. 2025, 16, 1527944. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Li, P.; Jain, R.G.; Taochy, C.; Fletcher, S.J.; Carroll, B.J.; Lu, G.Q.; Xu, Z.P. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 2017, 3, 16207. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. The Impact of Genetically Engineered Crops on Farm Sustainability in the United States; National Academies Press: Washington, WA, USA, 2010; pp. 1–270. [Google Scholar] [CrossRef]
- Mandel, G.N.; Marchant, G.E. The Living Regulatory Challenges of Synthetic Biology. Iowa Law Rev. 2014, 100, 155–200. [Google Scholar] [CrossRef]
- Rinaldi, A.; Mat Jalaluddin, N.S.; Mohd Hussain, R.B.; Abdul Ghapor, A. Building public trust and acceptance towards spray-on RNAi biopesticides: Lessons from current ethical, legal and social discourses. GM Crops Food 2025, 16, 398–412. [Google Scholar] [CrossRef]
- Biopesticides Market Size, Trends, Growth, Industry Report Forecast. Available online: https://www.marketsandmarkets.com/Market-Reports/biopesticides-267.html (accessed on 30 September 2025).
- Bio fungicides Market Size, Report 2030F. Available online: https://www.techsciresearch.com/report/bio-fungicides-market/4156.html (accessed on 30 September 2025).
- United States Bio-Fungicide Market Size & Share Analysis—Industry Research Report—Growth Trends. Available online: https://www.mordorintelligence.com/industry-reports/us-biofungicide-market (accessed on 30 September 2025).
- Li, Z.; Zhu, D.; Shen, Y. Discovery of novel bioactive natural products driven by genome mining. Drug Discov. Ther. 2018, 12, 318–328. [Google Scholar] [CrossRef]
- Tamang, P.; Upadhaya, A.; Paudel, P.; Meepagala, K.; Cantrell, C.L. Mining Biosynthetic Gene Clusters of Pseudomonas vancouverensis Utilizing Whole Genome Sequencing. Microorganisms 2024, 12, 548. [Google Scholar] [CrossRef]
- Singh, G.; Dal Grande, F.; Schmitt, I. Genome mining as a biotechnological tool for the discovery of novel biosynthetic genes in lichens. Front. Fungal Biol. 2022, 3, 993171. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Nickles, G.R.; Oestereicher, B.; Keller, N.P.; Drott, M.T. Mining for a new class of fungal natural products: The evolution, diversity, and distribution of isocyanide synthase biosynthetic gene clusters. Nucleic Acids Res. 2023, 51, 7220–7235. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; Van Der Hooft, J.J.J.; Van Santen, J.A.; Tracanna, V.; Duran, H.G.S.; Andreu, V.P.; et al. MIBiG 2.0: A repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Zhang, A.; Xu, X.; Yin, W.-B. Genome Mining of Epicoccum dendrobii Reveals Diverse Antimicrobial Natural Products. J. Agric. Food Chem. 2025, 73, 6691–6701. [Google Scholar] [CrossRef]
- Klug, K.; Zhu, P.; Pattar, P.; Mueller, T.; Safari, N.; Sommer, F.; Valero-Jiménez, C.A.; van Kan, J.A.L.; Huettel, B.; Stueber, K.; et al. Genome Comparisons between Botrytis fabae and the Closely Related Gray Mold Fungus Botrytis cinerea Reveal Possible Explanations for Their Contrasting Host Ranges. J. Fungi 2024, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Song, A.; He, Y.; Yang, L.; Chen, L.; Dai, W.; Wu, Q.; Yuan, S. Genome mining and biosynthetic pathways of marine-derived fungal bioactive natural products. Front. Microbiol. 2024, 15, 1520446. [Google Scholar] [CrossRef] [PubMed]
- Richman, E.K.; Hutchison, J.E. The Nanomaterial Characterization Bottleneck. ACS Nano 2009, 3, 2441–2446. [Google Scholar] [CrossRef] [PubMed]
- Schüller, A.; Studt-Reinhold, L.; Strauss, J. How to Completely Squeeze a Fungus—Advanced Genome Mining Tools for Novel Bioactive Substances. Pharmaceutics 2022, 14, 1837. [Google Scholar] [CrossRef]
- Wang, D.; Jin, S.; Lu, Q.; Chen, Y. Advances and Challenges in CRISPR/Cas-Based Fungal Genome Engineering for Secondary Metabolite Production: A Review. J. Fungi 2023, 9, 362. [Google Scholar] [CrossRef]
- Moreno-Giménez, E.; Gandía, M.; Sáez, Z.; Manzanares, P.; Yenush, L.; Orzáez, D.; Marcos, J.F.; Garrigues, S. FungalBraid 2.0: Expanding the synthetic biology toolbox for the biotechnological exploitation of filamentous fungi. Front. Bioeng. Biotechnol. 2023, 11, 1222812. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Blount, B.A.; Weenink, T.; Vasylechko, S.; Ellis, T. Rational Diversification of a Promoter Providing Fine-Tuned Expression and Orthogonal Regulation for Synthetic Biology. PLoS ONE 2012, 7, e33279. [Google Scholar] [CrossRef]
- De Schutter, K.; Taning, C.N.T.; Van Daele, L.; Van Damme, E.J.M.; Dubruel, P.; Smagghe, G. RNAi-Based Biocontrol Products: Market Status, Regulatory Aspects, and Risk Assessment. Front. Insect Sci. 2021, 1, 818037. [Google Scholar] [CrossRef] [PubMed]
- Welch, K.; Pierce, A.; Mendelsohn, M. Regulation of RNAi in Pesticidal Products in the United States. In RNA Interference in Agriculture: Basic Science to Applications; Springer Nature: Cham, Switzerland, 2025; pp. 633–645. [Google Scholar]
- Bocos-Asenjo, I.T.; Amin, H.; Mosquera, S.; Díez-Hermano, S.; Ginésy, M.; Diez, J.J.; Niño-Sánchez, J. Spray-Induced Gene Silencing (SIGS) as a Tool for the Management of Pine Pitch Canker Forest Disease. Plant Dis. 2025, 109, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Xu, H.; Liu, Y.; Hong, Y.; Yang, H.; Zhou, C.; Tao, L. Computational advances in biosynthetic gene cluster discovery and prediction. Biotechnol. Adv. 2025, 79, 108532. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Sherif, S.M. RNAi-Based Biofungicides as a Promising Next-Generation Strategy for Controlling Devastating Gray Mold Diseases. Int. J. Mol. Sci. 2020, 21, 2072. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-Based Control of Fusarium graminearum Infections Through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Parker, K.M.; Barragán Borrero, V.; van Leeuwen, D.M.; Lever, M.A.; Mateescu, B.; Sander, M. Environmental Fate of RNA Interference Pesticides: Adsorption and Degradation of Double-Stranded RNA Molecules in Agricultural Soils. Environ. Sci. Technol. 2019, 53, 3027–3036. [Google Scholar] [CrossRef]
- Galli, M.; Imani, J.; Kogel, K.-H. Labeling of dsRNA for Fungal Uptake Detection Analysis. In RNA Tagging: Methods and Protocols; Springer: New York, NY, USA, 2020; pp. 227–238. [Google Scholar]
- Wytinck, N.; Manchur, C.L.; Li, V.H.; Whyard, S.; Belmonte, M.F. dsRNA Uptake in Plant Pests and Pathogens: Insights into RNAi-Based Insect and Fungal Control Technology. Plants 2020, 9, 1780. [Google Scholar] [CrossRef]
- Verdonckt, T.-W.; Vanden Broeck, J. Methods for the Cost-Effective Production of Bacteria-Derived Double-Stranded RNA for in vitro Knockdown Studies. Front. Physiol. 2022, 13, 836106. [Google Scholar] [CrossRef]
- Ongvarrasopone, C.; Roshorm, Y.; Panyim, S. A Simple and Cost Effective Method to Generate dsRNA for RNAi Studies in Invertebrates. ScienceAsia 2007, 33, 035. [Google Scholar] [CrossRef]
- Mu, F.; Xie, J.; Cheng, S.; You, M.P.; Barbetti, M.J.; Jia, J.; Wang, Q.; Cheng, J.; Fu, Y.; Chen, T.; et al. Virome Characterization of a Collection of S. sclerotiorum from Australia. Front. Microbiol. 2018, 8, 2540. [Google Scholar] [CrossRef] [PubMed]
- Kogan, M. Integrated Pest Management: Historical Perspectives and Contemporary Developments. Annu. Rev. Entomol. 1998, 43, 243–270. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Todorović, I.; Moënne-Loccoz, Y.; Raičević, V.; Jovičić-Petrović, J.; Muller, D. Microbial diversity in soils suppressive to Fusarium diseases. Front. Plant Sci. 2023, 14, 1228749. [Google Scholar] [CrossRef]
- Whangbo, J.S.; Weisman, A.S.; Chae, J.; Hunter, C.P. SID-1 Domains Important for dsRNA Import in Caenorhabditis elegans. G3 Genes|Genomes|Genet. 2017, 7, 3887–3899. [Google Scholar] [CrossRef]
- Montgomery, M.K.; Xu, S.; Fire, A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1998, 95, 15502–15507. [Google Scholar] [CrossRef]
- Lundgren, J.G.; Duan, J.J. RNAi-Based Insecticidal Crops: Potential Effects on Nontarget Species. Bioscience 2013, 63, 657–665. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Lawrence, J.; Sawyer, A.; Manzie, N.; Gardiner, D.M.; Mitter, N.; Brosnan, C.A. dsRNAmax: A multi-target chimeric dsRNA designer for safe and effective crop protection. NAR Genom. Bioinform. 2025, 7, lqaf064. [Google Scholar] [CrossRef]
- Riedling, O.; Walker, A.S.; Rokas, A. Predicting fungal secondary metabolite activity from biosynthetic gene cluster data using machine learning. Microbiol. Spectr. 2024, 12, e03400-23. [Google Scholar] [CrossRef]
| Feature | Conventional Synthetic Fungicides | Botanical Extracts | Microbial BCAs | SynBio-Derived Molecules | RNAi-Based Fungicides | References |
|---|---|---|---|---|---|---|
| Specificity | Broad to Moderate | Broad to Moderate | Strain/Species-Specific | High (molecular target) | Exquisite (gene target) | [7,12,13,16,17] |
| Mode of Action | Single/multiple biochemical target | Multiple targets, often pleiotropic | Competition, antibiosis, parasitism | Specific, engineered biochemical target | Post-transcriptional gene silencing | [8,12,13,17,18] |
| Resistance Risk | High to Moderate | Low | Low to Moderate | Moderate to Low | Very Low (potential for multiple targets) | [7,12,13,16] |
| Environmental Impact | Persistence, non-target effects | Low persistence, possible non-target toxicity | Minimal, ecosystem-specific | Biodegradable, low non-target impact | Biodegradable, no non-target impact | [9,12,13,19,20] |
| Scalability/Consistency | High | Low to Moderate | Low | High | High (dsRNA production) | [3,12,14,21,22] |
| Development Cost | Very High | Moderate | Moderate to High | High (initially), then decreasing | High (initially), then decreasing | [3,6,12,23,24] |
| Regulatory Framework | Established | Variable, often simpler | Complex, strain-specific | Emerging, evolving | Emerging, evolving | [7,12,14,25,26] |
| Fungal Group | Key Genera | Classes of Secondary Metabolites | Noteworthy Bioactivity/Novelty | Reference |
|---|---|---|---|---|
| Endophytic Fungi | Aspergillus, Penicillium, Fusarium | Polyketides, NRPS, Terpenoids, Alkaloids | Prolific source of bioactive compounds with diverse applications | [33] |
| Marine-Derived Fungi | Aspergillus, Penicillium, Acremonium | Polyketides, Alkaloids (often halogenated) | Unique chemical structures adapted to extreme environments | [39] |
| Lichenized Fungi | Umbilicaria | PKS, NRPS | Highly divergent BGCs suggesting novel chemical scaffolds | [32] |
| Known Biocontrol Genera | Epicoccum, Trichoderma | Polyketides, Diketopiperazines (DKPs), Peptides | Proven antifungal activity against pathogens like B. cinerea | [37] |
| Extremophiles | Various | Compounds adapted to extreme conditions | Potential for novel, stable enzymes and molecules | [17] |
| Associated Bacteria | Pseudomonas, Streptomyces | Lipopeptides, Polyketides, Alkaloids | Rich source of antifungal compounds discovered through genome mining | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coca-Ruiz, V. The Fungal Biorevolution: A Trifecta of Genome Mining, Synthetic Biology, and RNAi for Next-Generation Fungicides. Agrochemicals 2025, 4, 18. https://doi.org/10.3390/agrochemicals4040018
Coca-Ruiz V. The Fungal Biorevolution: A Trifecta of Genome Mining, Synthetic Biology, and RNAi for Next-Generation Fungicides. Agrochemicals. 2025; 4(4):18. https://doi.org/10.3390/agrochemicals4040018
Chicago/Turabian StyleCoca-Ruiz, Víctor. 2025. "The Fungal Biorevolution: A Trifecta of Genome Mining, Synthetic Biology, and RNAi for Next-Generation Fungicides" Agrochemicals 4, no. 4: 18. https://doi.org/10.3390/agrochemicals4040018
APA StyleCoca-Ruiz, V. (2025). The Fungal Biorevolution: A Trifecta of Genome Mining, Synthetic Biology, and RNAi for Next-Generation Fungicides. Agrochemicals, 4(4), 18. https://doi.org/10.3390/agrochemicals4040018







