Abstract
Fluorescence lifetime imaging microscopy (FLIM) has emerged as a promising tool for all scientific studies in recent years. However, the utilization of FLIM data requires complex data modeling techniques, such as curve-fitting procedures. These conventional curve-fitting procedures are not only computationally intensive but also time-consuming. To address this limitation, machine learning (ML), particularly deep learning (DL), can be employed. This review aims to focus on the ML and DL methods for FLIM data analysis. Subsequently, ML and DL strategies for evaluating FLIM data are discussed, consisting of preprocessing, data modeling, and inverse modeling. Additionally, the advantages of the reviewed methods are deliberated alongside future implications. Furthermore, several freely available software packages for analyzing the FLIM data are highlighted.
1. Introduction
In 1845, Fredrick W. Herschel made the first official discovery of fluorescence [1]. During his experiments, he observed that a quinine solution, such as tonic water, could be excited by UV radiation and emit blue light. Building upon this discovery, Sir George G. Stokes, a British scientist, further investigated fluorescence and noted that the emitted light had a longer wavelength than the UV radiation that initially excited the object [1]. The first applications of fluorophores in biological research were found several decades later, in the early 1900s, when they were used to stain tissues, bacteria, and other pathogens. This allowed scientists to visualize and study specific components within biological samples. In 1911, the first working fluorescence microscope was developed by Oskar Heimstaedt. Later, among many individual contributors, companies Carl Zeiss and Carl Reichert played a significant role in advancing fluorescence microscopy [1]. In 1929, fluorescence labeling was first introduced by Ellinger and Hirt [2]. Their contributions were instrumental in transforming fluorescence microscopy into a powerful tool in every research field. In 1988/1989, the first fluorescence lifetime imaging microscopy (FLIM) microscopy based on an ultra-fast laser scanning microscope was introduced in Jena, Germany [3]. This introduction of ultrafast lasers, combined with the advent of semiconductor-based, very fast detection schemes such as SPAD (single photon avalanche diode) and TCSPC (time-correlated single-photon counting) detectors, have made FLIM on the pico to nanosecond timescale a readily available experimental technique. This microscopy technique offers thorough details and high-resolution images of cell shape, intracellular concentration of chemicals, etc., and it has broad applications in biology, chemistry, materials science, and pharmaceutical research [4]. As a result, FLIM has grown in recognition in recent years. It is often combined with Förster resonance energy transfer (FRET), enabling the investigation of molecular mechanisms, biosensor activities, and protein–protein interactions within live cells [4,5]. FRET is a non-radiative energy transfer process wherein an excited fluorescence molecule, known as the donor, transfers energy to a non-excited molecule called the acceptor [4,5,6]. This energy transfer occurs through dipole-dipole coupling when the emission spectrum of the donor overlaps with the excitation spectrum of the acceptor, and the molecules are at a small distance (within 10 nm) with suitable relative orientations (cf. Figure 1b–e) [6]. FLIM-FRET offers the advantage of generating high-resolution spatial and temporal images. However, FLIM itself records fluorescence decay profiles rather than directly measuring fluorescence lifetimes. Thus, the fluorescence decay rate, which we discuss in the following paragraph, serves as the fundamental principle for FLIM operation. The underlying physical background of FLIM, the measurement techniques, and the data acquisition process are covered in the subsequent paragraphs.
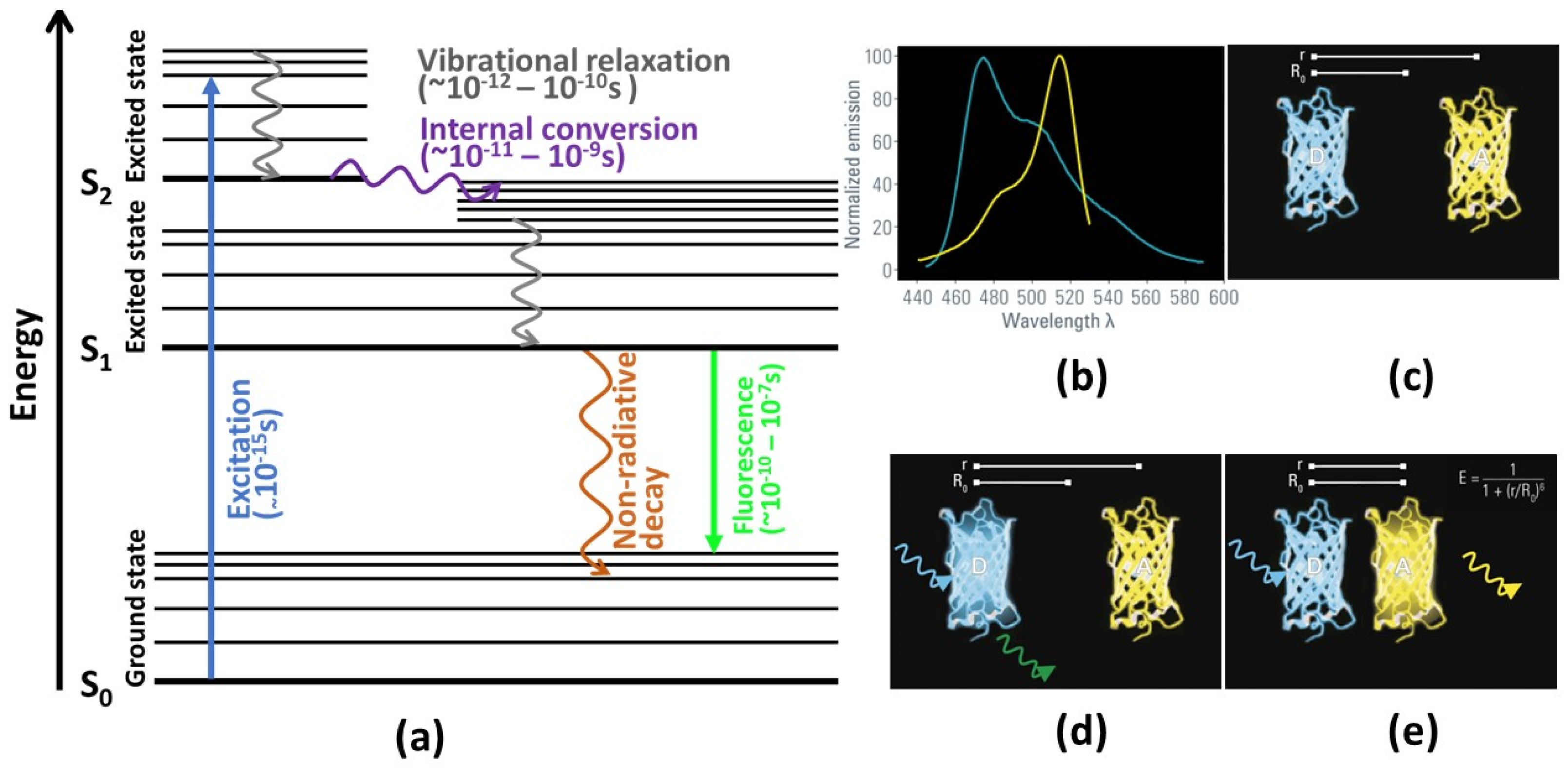
To understand the principles underlying FLIM, it is useful to refer to a Jablonski diagram (cf. Figure 1a), which shows the relevant processes observable in a typical photoexcited system. In Figure 1a, S0 refers to an electronic (singlet) ground state of a fluorophore system. Electrons in this ground state can be excited by absorption of light into various excited states (S1, S2 …) depending on the wavelength of the incident light. Notably, the electronic states are further divided into various vibrational (and rotational) states. Typically, excitation by visible light populates a vibrationally excited state of the excited states S>=1 governed by the Franck–Condon principle.
From this excited state, the electrons return to the ground state either by radiative or non-radiative processes. The non-radiative decay is governed by two main processes: ref. [7]. First, vibrational relaxation (also called vibrational cooling), by which energy is transferred from vibrationally excited states into kinetic modes of the molecule (or neighboring molecules), causes a transition between vibrational states of the same electronic state and decreases the energy of the system. Second, internal conversion couples different electronic states (e.g., S1 and S0) via their vibrational modes. Here, the system transitions from an electronically excited state into a highly vibrationally excited state of a lower electronic state without loss of energy, i.e., via a horizontal transition. Both of these non-radiative processes are usually very fast processes, happening in the femto-to-picosecond range.
Radiative decay is typically dominated by fluorescence. Here, the excited state is deactivated by emitting electromagnetic radiation in the form of a photon with a wavelength corresponding to the energy difference of the two states. However, due to the above mentioned speed of the non-radiative deactivation of the vibrationally excited state, this emission almost always happens from the vibrational ground state of the first electronically excited state (S0), an empirical observation known as Kasha’s rule [8]. Furthermore, due to the Franck–Condon principle, deactivation typically occurs in the vibrationally excited state of S0. Both of these effects cause the wavelength of the emitted photon to be red-shifted compared to the excitation light.
The easiest parameters to characterize the fluorescence process are the wavelength of the emitted photon, corresponding to the energy difference between the states and the intensity of the emitted light (directly related to the number of photons), which is governed by the absorption coefficient as well as the quantum efficiency of the fluorophore. The quantum efficiency, in turn, is directly related to the ratio of the radiative to non-radiative decay processes described above. However, in more complicated systems, these parameters are often not sufficient to discriminate between similar fluorophores, as their spectral profiles can overlap.
Here, FLIM can offer many advantages over intensity-based fluorescence techniques. By characterizing the lifetime of a fluorophore, the non-radiative processes can be characterized as the exact timescale of vibrational cooling to the vibrational ground state of S0, and the efficiency of internal conversion between S1 and S0 directly influences the lifetime of the fluorophore [4]. As these processes are extremely sensitive, even to slight changes in the chemical composition or even environment, the fluorescence lifetime can be used to discriminate between highly similar structures.
For example, nicotinamide adenine dinucleotide (NAD(P)H) and flavin adenine dinucleotide (FAD) play a vital role in cellular oxidation–reduction reactions [4,9]. These co-enzymes possess autofluorescence properties, enabling non-invasive imaging of living cell metabolic activity. However, their spectral properties in distinct cellular environments exhibit considerable similarity. In contrast, FLIM can effectively distinguish between these two fluorophores due to the disparity in their lifetimes, with NAD(P)H having a shorter lifetime compared to FAD [9]. See Table 1 for applications of FLIM.

Table 1.
Experimental applications of FLIM.
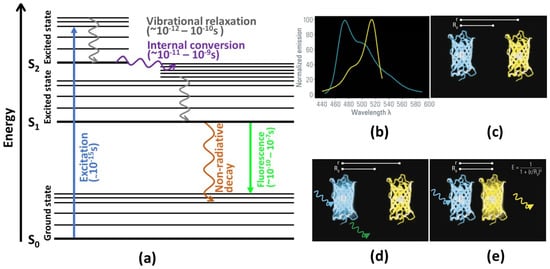
Figure 1.
(a) Illustration of Jablonski’s diagram. A molecule in S0 level absorbs energy, leading to an electronic excitation to a higher energy level for a short period of time. By internal conversion and vibrational relaxation processes, the electron moves to the lowest vibrational level of excited state. From the S1 electronic state, the electron returns to the ground state in either a radiative or non-radiative way (adapted from [4]). (b) The emission spectrum of the donor (blue line) must overlap with the excitation spectrum of the acceptor (yellow line). (c) The distance between donor and the acceptor molecule is important for the FLIM-FRET process. (d) If the distance is larger than the threshold value R0, no FRET is occurring. (e) If both molecules are in very close proximity, the donor’s energy can be transferred to the acceptor, and the acceptor molecule emits a photon. (b–e) reprinted with permission from [10] © Leica Microsystems GmbH.
Figure 1.
(a) Illustration of Jablonski’s diagram. A molecule in S0 level absorbs energy, leading to an electronic excitation to a higher energy level for a short period of time. By internal conversion and vibrational relaxation processes, the electron moves to the lowest vibrational level of excited state. From the S1 electronic state, the electron returns to the ground state in either a radiative or non-radiative way (adapted from [4]). (b) The emission spectrum of the donor (blue line) must overlap with the excitation spectrum of the acceptor (yellow line). (c) The distance between donor and the acceptor molecule is important for the FLIM-FRET process. (d) If the distance is larger than the threshold value R0, no FRET is occurring. (e) If both molecules are in very close proximity, the donor’s energy can be transferred to the acceptor, and the acceptor molecule emits a photon. (b–e) reprinted with permission from [10] © Leica Microsystems GmbH.
FLIM can be performed using various imaging modalities, such as laser scanning microscopy (LSM) and wide-field illumination (WFI) microscopy [4,11]. Depending on the excitation-detection technique used, LSM systems are classified as either confocal (CLSM) or multiphoton (MP-LSM) systems. These microscopic techniques provide 3D FLIM data. It is important to note that WFI has the advantages of higher frame rates and less photodamage than LSM [4].
Fluorescence lifetimes are measured either in the time domain (TD-FLIM) or in the frequency domain (FD-FLIM) [4]. TD-FLIM measures the time delay between excitation and emission photons using time-correlated single-photon counting (TCSPC) or time-gated detection, while FD-FLIM analyzes the phase and amplitude of fluorescence signals modulated at different frequencies. Both techniques have their strengths and are suitable for different experimental setups and sample characteristics.
Among all FLIM measurement methods, TCSPC is probably the most often-used technique [4,12]. Generally, in TCSPC systems, a photon-over-time histogram is measured. The intensity is calculated by convolving the systems instrument response function () and the weighted sum of a number of fluorescence decays, typically modeled via simple first-order exponential functions, as shown in Equation (1).
To utilize FLIM data, the lifetimes () and abundances () need to be extracted, where the lifetimes represent the different types of molecules and abundances highlight the molecular concentration. For this purpose, it is important to know the exact lifetimes and abundances from the measured decay traces. There are a few traditional methods: curve-fitting techniques, such as the phasor approach, and deconvolution methods [4,13]. The curve-fitting method tries to fit the photon histograms by applying Equation (1) with algorithms like the Levenberg–Marquardt algorithm [14] or maximum likelihood estimation [15,16,17]. These algorithms afford ‘optimal’ (according to the utilized error function) parameters (lifetimes, abundances and offset, etc.) to describe the measured results but require prior knowledge of the source background fluorescence, the number of fluorophores, and the offset for convergence to the correct values. The method of curve fitting strongly depends on the number of photons, as it is highly sensitive to the signal-to-noise ratio, which increases with higher photon counts. This means that a higher photon number usually increases the fitting accuracy. There are two strategies for curve fitting—local fitting and global fitting. In local curve fitting, each pixel is fitted with different lifetime parameters. In the global fitting, all pixel decay traces are fitted at once, so all fluorophores are present in each pixel and show the same lifetime [18]. Here, each pixel represents a decay trace. The low implementation complexity is one of the main advantages of all fitting methods.
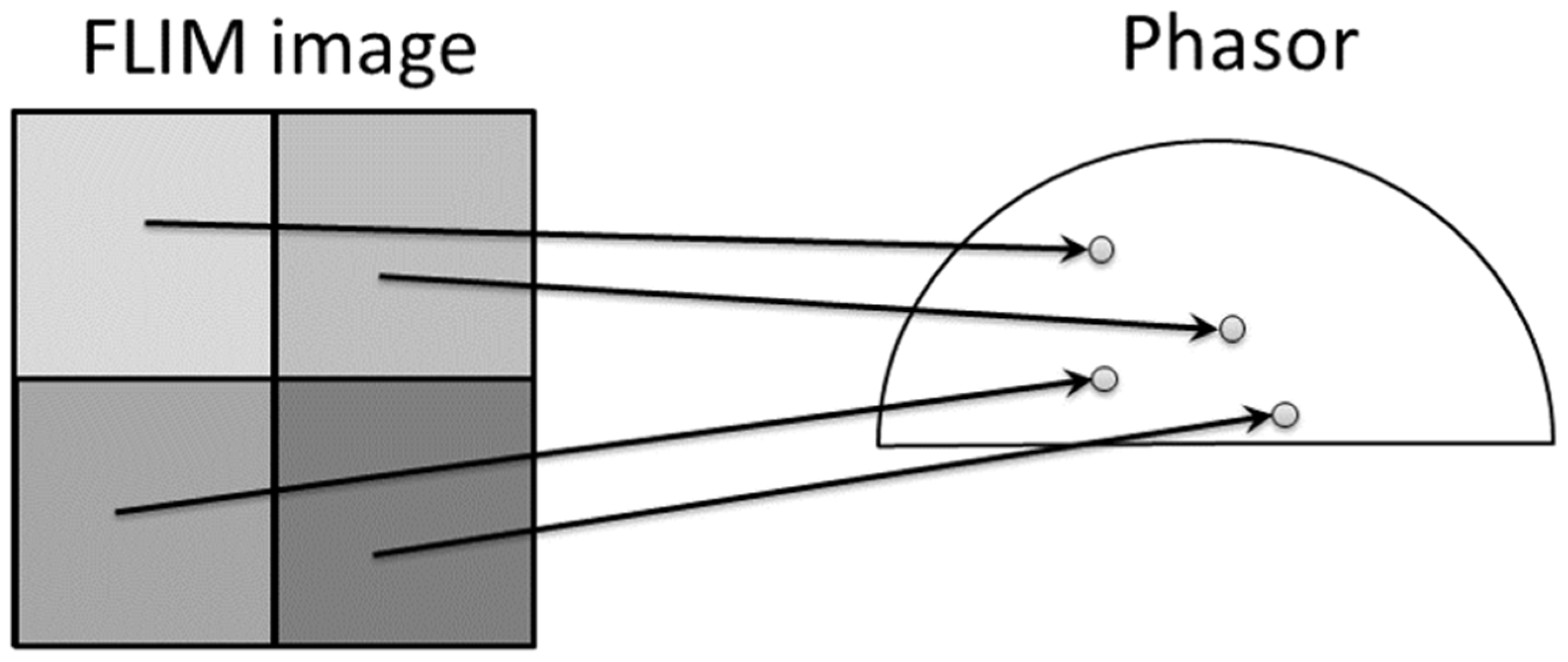
Another popular approach for analyzing lifetimes without fitting is the Phasor approach [6,19]. It provides a 2D graphical view of lifetime distributions. A phasor diagram is derived from the TCSPC data using a Fourier transform, and each pixel in the image corresponds to a point in the phasor diagram, as shown in Figure 2. The phasor space is constructed by two phasor vectors (G, S).
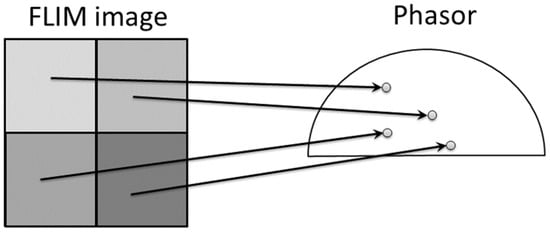
Figure 2.
This figure shows the relationship between FLIM image and phasor plot. The phasor distributions are calculated by Fourier transform after data acquisition from TCSPC. Each intensity pixel value in the image was converted as a point in the phasor plot. Adopted with permission from [6] © Leica Microsystems GmbH.
The time domain phasor plot is defined by Equations (2) and (3).
In these equations, and are the coordinates along the horizontal (G) and vertical (S) axis, is the modulation frequency, is the TCSPC data at the ith pixel.
In the frequency domain FLIM measurement, the phasor plot is defined by the following equations:
Here, and are the modulation ratio and the phase delay given a particular frequency at a pixel location . The average lifetime at the ith pixel is defined as the ratio of and .
The deconvolution-based method is another common form of decay trace analysis technique [4,20]. Deconvolution-based methods recover the lifetime decay from the measured fluorescence signal by deconvolution of the system response function (IRF). The Laguerre polynomial method is the most popular deconvolution method. In this method, the decay traces are represented by a Laguerre polynomial, which consists of the series expansion of decay and IRF. The main advantage of the Laguerre polynomial is that it is more precise than the phasor approach.
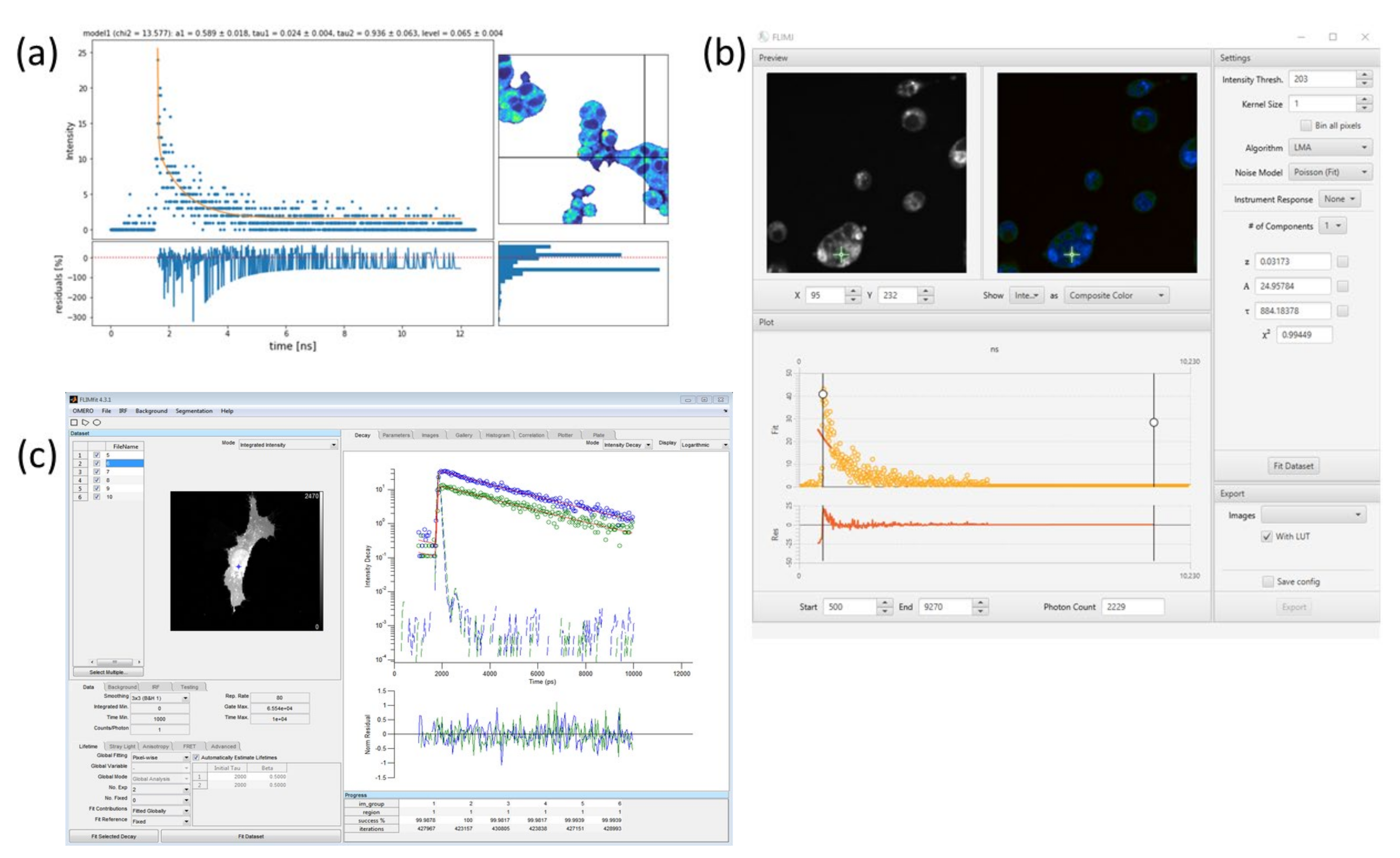
These techniques (or combinations of them) are implemented in various software packages, both commercial and freely available (cf. Figure 3). The commercial software SPCImage (all versions) [12,21] (Becker & Hickl) is widely used to estimate the lifetime parameters and is considered the standard method. It implements all of the above mentioned methods and defaults to using iterative decay fitting by applying first-, second-, and triple-order exponential decays to fit the data. Also, some free software packages can be used for lifetime extraction. For example, the Python package FLIMview [22] is based on the principle of curve fitting. FLIMJ, which is based on Fiji, allows for visualizing images and analyzing the fit using various methods [23]. Another MATLAB-based package is FLIMfit [24]. See Table 2 for an comparison of these software packages.
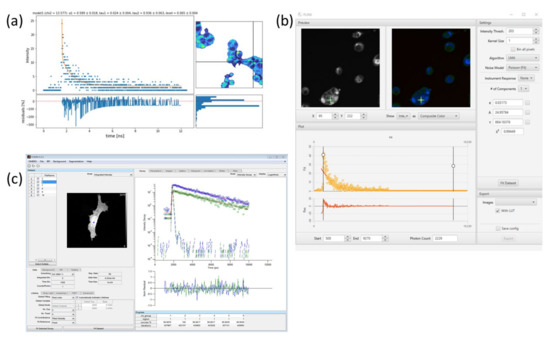
Figure 3.
Illustration showing different software packages for lifetime estimation: (a) Python-based FLIMview where fitted curve, residue, and pixel coordinates can be visualized together [22]. (b) Fiji FLIMJ package [23]. (c) FLIMfit package is connected with Omero for image analysis [24].

Table 2.
Comparison between different FLIM data analysis software packages.
Despite their many advantages, curve fitting, phasor method, and deconvolution have several disadvantages. All methods are time-consuming and error-prone. Recently, machine learning (ML), especially deep learning, has received a considerable boost in popularity due to its outstanding performance in this area. This review focuses on the application of ML and DL methods for FLIM data analysis. For this, the review is divided into four sections. The first section discusses the preprocessing of FLIM data. The second section deals with data modeling, followed by inverse modeling in the third section. The last part consists of a summary and an outlook.
2. Preprocessing
The primary objective of preprocessing is to enhance the quality of images or data to enable proper analysis. In fluorescence microscopy, image preprocessing is necessary due to the limited number of photons captured by the detector, resulting in a weak signal [25,26]. This weak signal leads to fluorescence images exhibiting a significant contribution of Poisson–Gaussian noise [25]. To experimentally obtain improved images, two approaches can be employed: First, increasing the power of the excitation laser increases the emitted photon flux (albeit only up to a certain limit); second, increasing the exposure time increases the total number of captured photons emitted with a constant photon flux. However, both methods tend to cause photodamage and can thus not be utilized for all samples or unlimited signal increases [25,26]. Therefore, developing algorithms that effectively denoise fluorescence spectroscopy data and give high-quality images is necessary to analyze complex datasets.
A wide variety of denoising and image reconstruction algorithms for fluorescence images are known from the literature. Some recent approaches are listed in Table 3. In the next paragraph, we review several denoising algorithms.

Table 3.
Different denoising methods and their examples are shown.
First, a general problem in training and evaluating denoising methods was addressed by Zhang et al. [25]. They addressed the issue of simulating artificial datasets that accurately reflect the properties of real measured fluorescence datasets. They recognized that most datasets are generated using Gaussian noise, which does not accurately represent the Poisson noise typically present in fluorescence images. Therefore, their primary objective was to bridge this gap by creating a Poisson–Gaussian denoising dataset.
To achieve this, the authors developed a dataset called “Fluorescence microscopy denoising (FMD)”. The FMD dataset consists of 12,000 real noisy microscopic images obtained from confocal, multiphoton, and wide-field microscopes. The ground truth images were created through image averaging. By using this dataset, they compared the performance of ten traditional denoising algorithms with deep learning methods, demonstrating that deep learning approaches outperformed traditional methods on this dataset with more realistic noise.
In their paper [26], the authors proposed a two-step generative adversarial network (GAN)-based denoising model called the global noise modeling denoiser (GNMD). In the first step, the GAN-based model was trained using a combination of binary masked images and real images. This trained model could then generate synthetic images by taking a binary mask as input, which combined a synthetic foreground signal (gamma distributed intensities) with synthetic background noise (global noise generated by Pix2Pix). In the second step, the output from the first step and a clean image were fed into the same network, which was then trained using both clean and noisy images. This trained model aimed to enhance the denoising capacity. To validate the denoising capabilities of their model, the authors tested it using real fluorescence images of mitochondria acquired through a wide-field fluorescence microscope. The performance of their model was also compared to three traditional denoising models (PURE-LET, VST-BM3D, Noise2self [33]), and the GNMD model outperformed the others. In summary, Zhong et al. [26] addressed the need for realistic simulations by creating a Poisson–Gaussian denoising dataset (FMD) [25]. They introduced the GNMD model, a two-step GAN-based denoising approach, and demonstrated its effectiveness using real fluorescence images. Their study highlighted the superiority of deep learning methods over traditional denoising algorithms in the context of fluorescence microscopy denoising.
Another deep-learning-based denoising technique was demonstrated by Mannam et al. [34]. The authors [34] demonstrated a convolutional neural network (CNN) based denoising techniques to achieve a high signal-to-ratio (SNR). This paper was divided into two parts. Phasor-based denoising techniques were described in part 1, and in the second part, the segmentation technique was mentioned. For denoising, they used the Noise2Noise and DnCNN [31] models, which were pretrained with 12,000 fluorescence intensity images. The authors used this denoising technique as an ImageJ plugin. Additionally, they compare their results with traditional denoising methods (mean or median filter). To prove their network efficiency, they claimed that traditional denoising filters should be used several times to obtain a clear image, whereas CNN can eliminate noise by using it one time.
Although the deep-learning-based denoising approach has gained huge popularity in recent years, it still faces certain challenges [35]. For example, deep learning algorithms are data-driven and data-hungry processes. For training a deep network, thousands of noisy and clean data pairs are needed, and this data acquisition process is laborious and error-prone. On the other hand, supervised learning methods can easily be overfit to the training dataset (“memorize the training data”). This can lead the model to predict clean images similar to the training data independent from its input, a phenomenon known as hallucination [36].
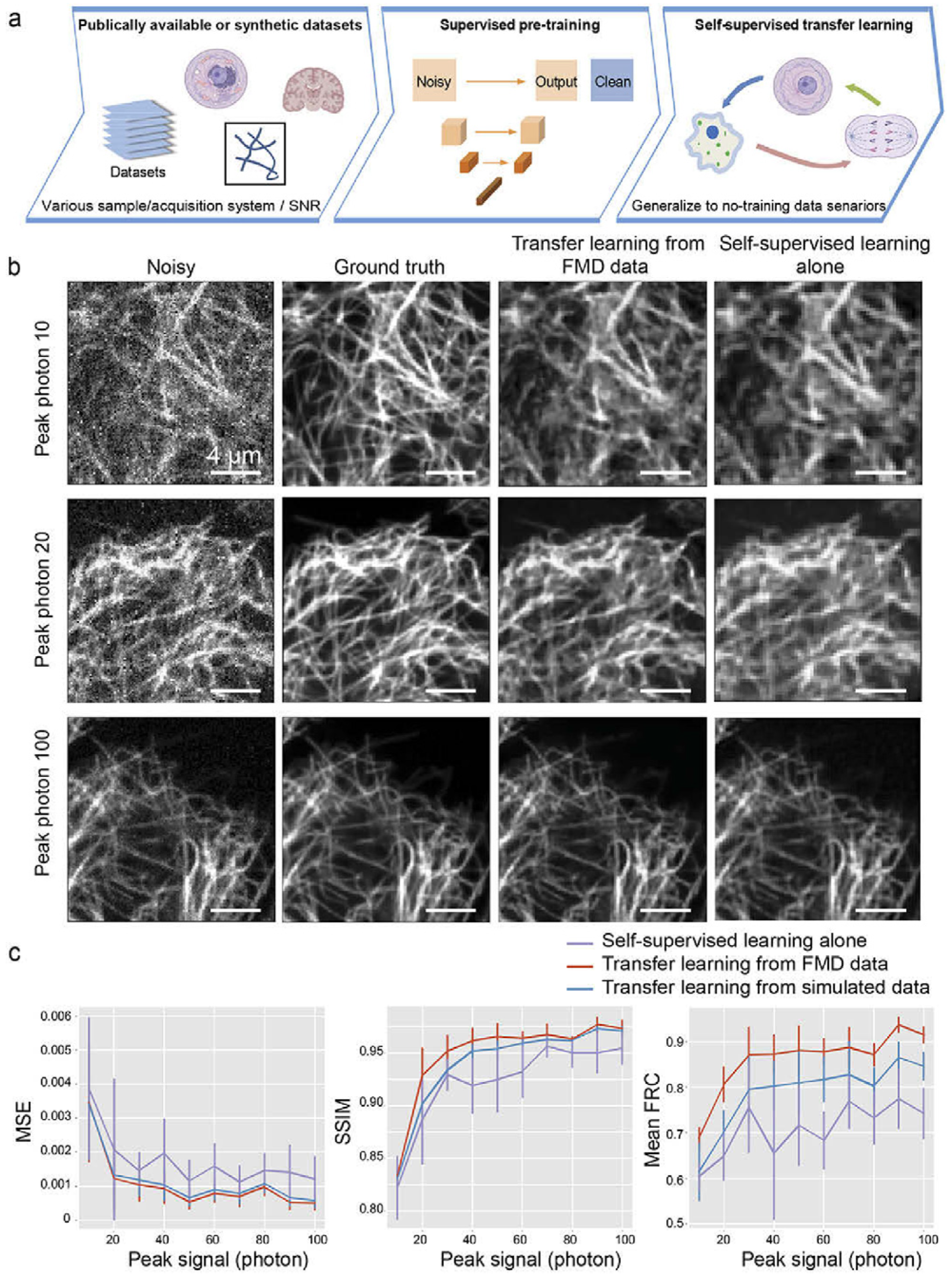
To handle the above two problems, Wang et al. [36] described a transfer learning-based denoising technique. In the beginning, they trained the network by supervised learning with a U-net architecture and used generic and synthetic noisy or clean images for training. They then transferred the weights to another U-net model and trained this model with a self-supervised training framework, Noise2self [33], where only noisy data are required for training. Finally, they showed the results for only the self-supervised method and a combination of the supervised and self-supervised methods. They used the same FMD [25] dataset, and the comparison of the result is shown in Figure 4.
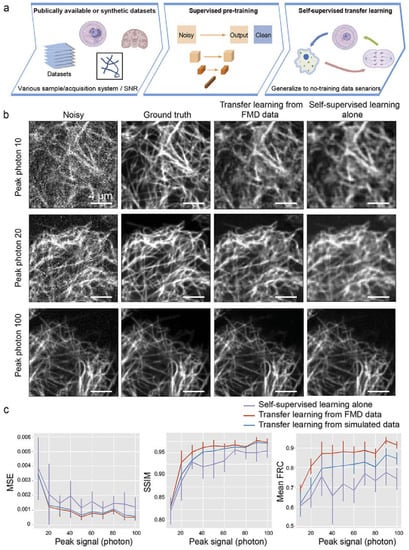
Figure 4.
Performance of transfer learning in denoising: (a) Schematic of the transfer learning method. (b) Synthetic noisy images from microtube confocal images and corresponding denoised images with transfer learning denoising from pre-training using FMD dataset and compared to self-supervised denoising without pre-training. (c) The denoising performance, in terms of “Mean square error”, “Structure similarity index”, and “Mean Fourier ring correlation as a function of the synthetic noisy image”. Reprinted with permission from [36] under the terms of the OSA Open Access Publishing Agreement.
Although all proposed methods successfully have removed noise from the fluorescence image dataset, some improvements need to be made. For example, these methods need to be tested with various datasets of different tissue samples or any other biomedical dataset.
3. Data Modeling
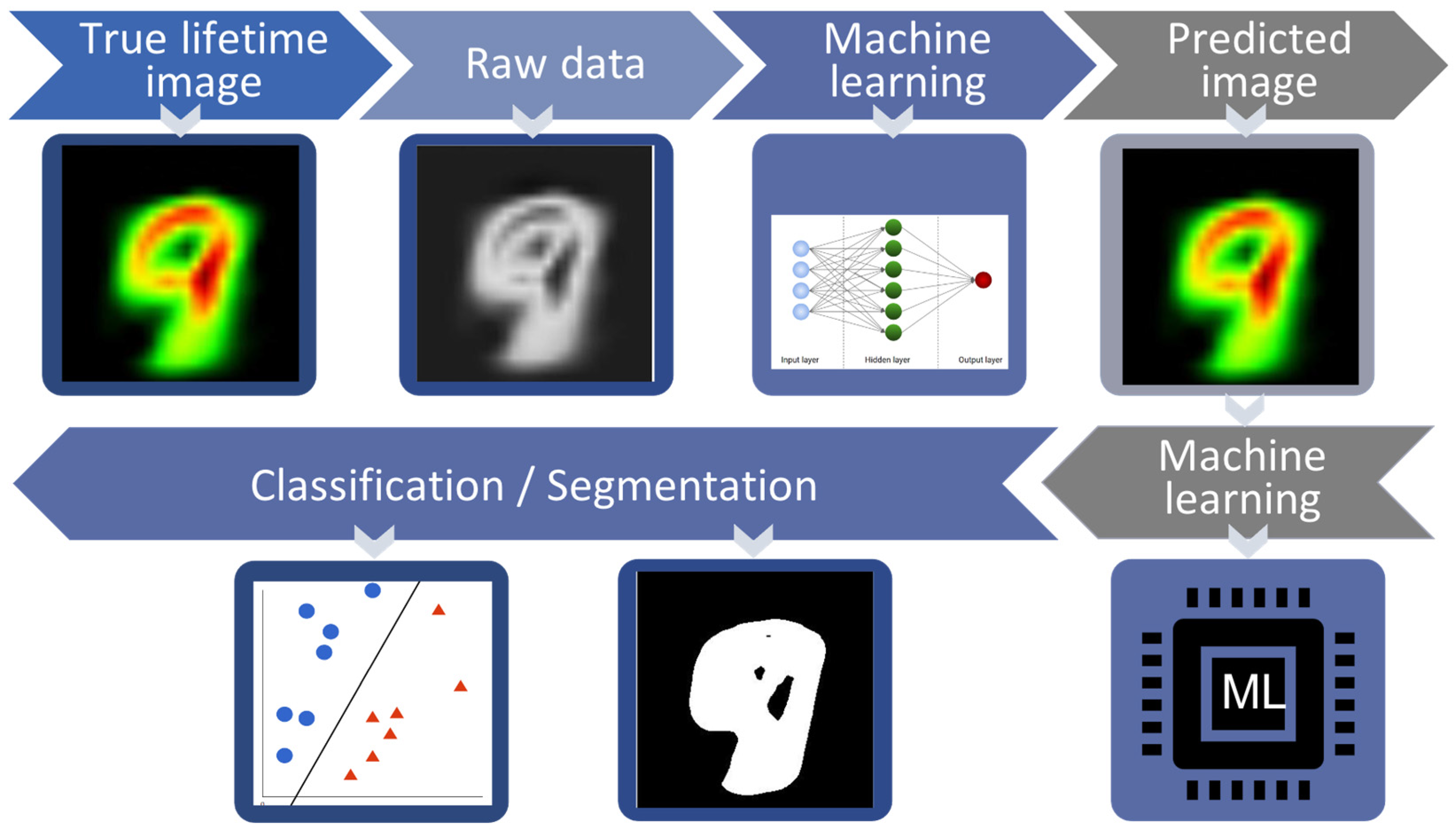
Fluorescence lifetime imaging microscopy (FLIM) has gained significant recognition in the biomedical field due to its label-free nature and high sensitivity. However, the conventional approach to FLIM data analysis usually involves curve-fitting procedures that require manually tuning parameters for the extraction of the lifetime values, which becomes inefficient when dealing with large amounts of sample data. Moreover, when there are subtle differences among different data points, the distribution of fluorescence lifetime values can be wide or very small, making it challenging to manually differentiate them into multiple classes. Fortunately, the recent popularity of ML or DL has greatly contributed to the advancement of data classification and data analysis in FLIM as shown in Figure 5.
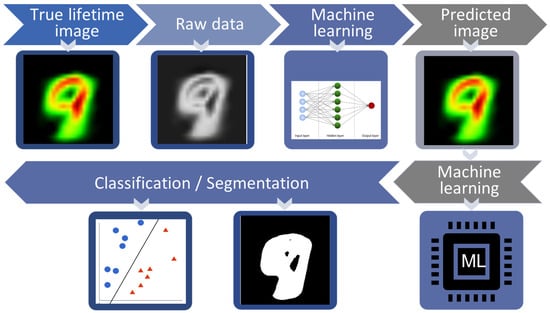
Figure 5.
Schematic diagram of data modeling: First, true lifetime images were extracted from raw data by conventional methods like maximum likelihood method and least-square fitting. The machine learning model was then used to predict lifetime parameters. Afterward, machine learning was used for classification and segmentation. By segmentation region of interest (ROI) has been separated from background and in classification hyperplane divides data into different classes.
We divided data modeling into two sub parts: classification and segmentation (see Figure 5). Classification and segmentation are two related concepts but are used for different tasks and can complement each other in various applications. Classification involves assigning a label or category to an entire input based on its characteristics. Segmentation, on the other hand, involves dividing an image into meaningful regions or segments, typically on a pixel basis. However, segmentation and classification can be used together to provide a more comprehensive understanding of an image. In the next section, we discuss the segmentation and classification methods used in FLIM data modeling.
3.1. Segmentation
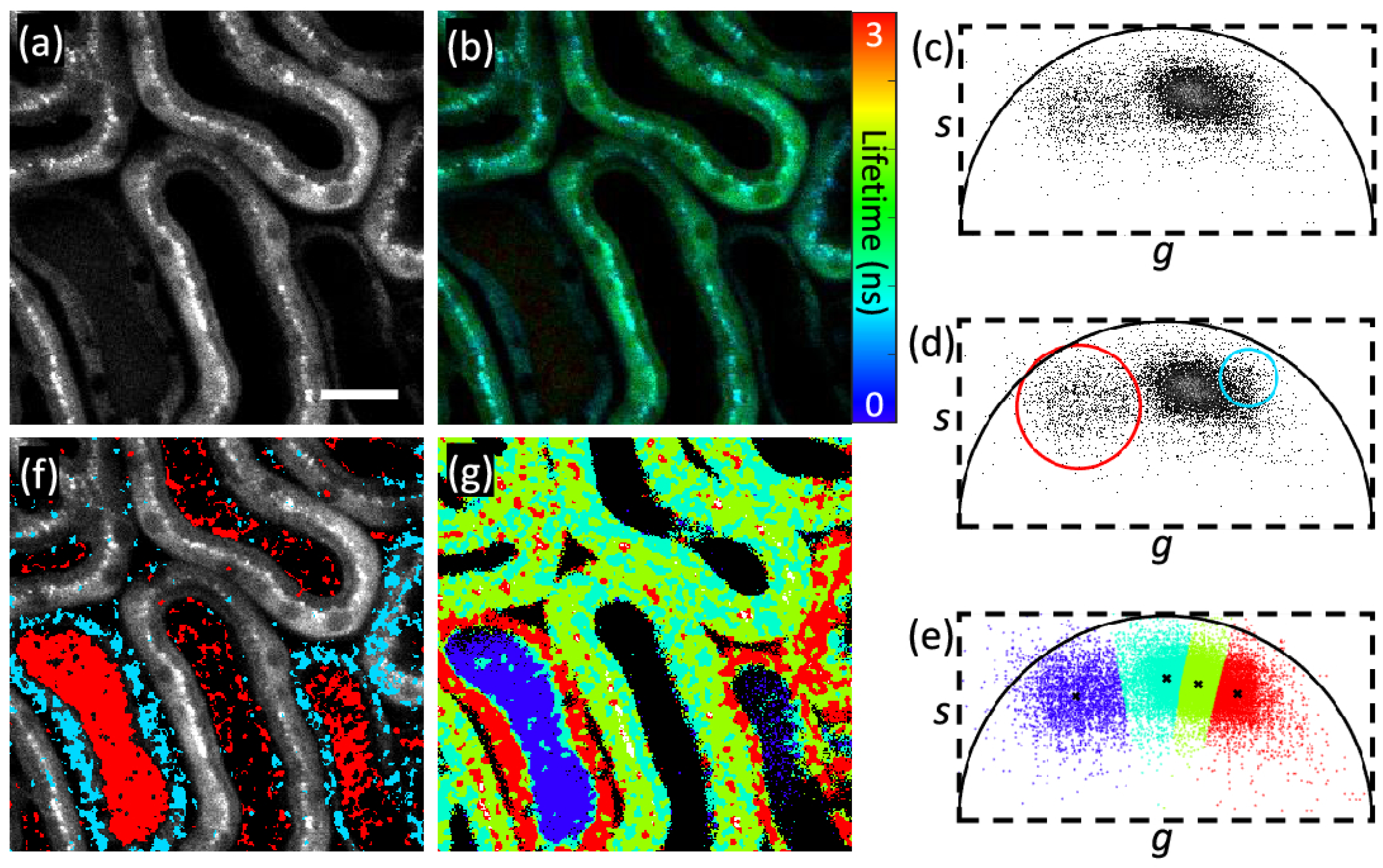
Zhang et al. [37] used K-means clustering to segment lifetime images by using a phasor plot (cf. Figure 6). In the phasor plot, pixels with similar decay phasors were sorted into the same cluster. This feature is useful for segmenting pixels based on the similarity of their fluorescence decays. Therefore, the lifetime segmentation technique is simplified using phasors as the problem is transformed into a classical clustering problem of points in a 2D plane. This method achieved success in segmentation with greater speed than traditional methods. In reference [38], the authors applied Otsu’s thresholding-based segmentation method to separate the background pixels from the foreground pixels [39]. They then used a morphological operation to remove the non-cellular region. This work is discussed in detail in the classification part. Here, the authors used segmentation as the initial step for classification. Also, in reference [40], the authors used a simple thresholding-based segmentation to remove background before training an ML model on the dataset. This work is also discussed in detail in the next section.
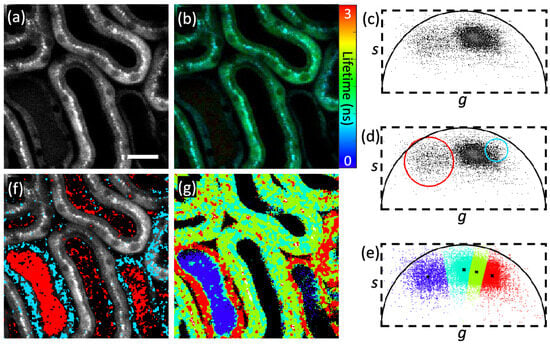
Figure 6.
This image shows segmentation method by phasor plot: (a) two-photon intensity image; (b) fluorescence lifetime image; (c) phasor plot; (d,f) phasor labeling. Similar fluorescence decays have same features, so they cluster together. Here, red and blue color clusters are selected manually by users and this label their corresponding pixels in the image with certain colors; (e,g) clustered images (each color in g represents the one cluster). Reprinted with permission from [37] © The Optical Society.
3.2. Classification
3.2.1. Lung Cancer Classification
Lung cancer ranks among the top causes of cancer-related deaths globally, and the 5-year survival rate after pneumonectomy is less than 14% [41]. Surgery can improve survival rates; however, the success is strongly dependent on the stage of detection. Currently, the gold standard for assessing and diagnosing diseases is hematoxylin and eosin (H&E) stained histopathology, which takes 20–30 min even under ideal conditions with expensive and laborious interoperative cryosectioning [41].
Nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) play a significant role in cellular energy metabolism [41]. Cancer cells have higher rates of glycolysis for rapid cell division compared to normal cells. These metabolic changes are often accompanied by alterations in fluorophores, which can be detected using fluorescence imaging and spectroscopy. Recent studies have demonstrated the potential of autofluorescence imaging and spectroscopy as diagnostic methods for various cancers, including oral, cervical, and breast cancers [41]. However, due to the irregular shape of the tissue, as well as various concentrations of perturbing absorbers in the tissue, intensity-based fluorescence techniques are challenging to apply. Here, FLIM, which is insensitive to these perturbations, can be applied to clearly differentiate between cancerous and healthy tissue.
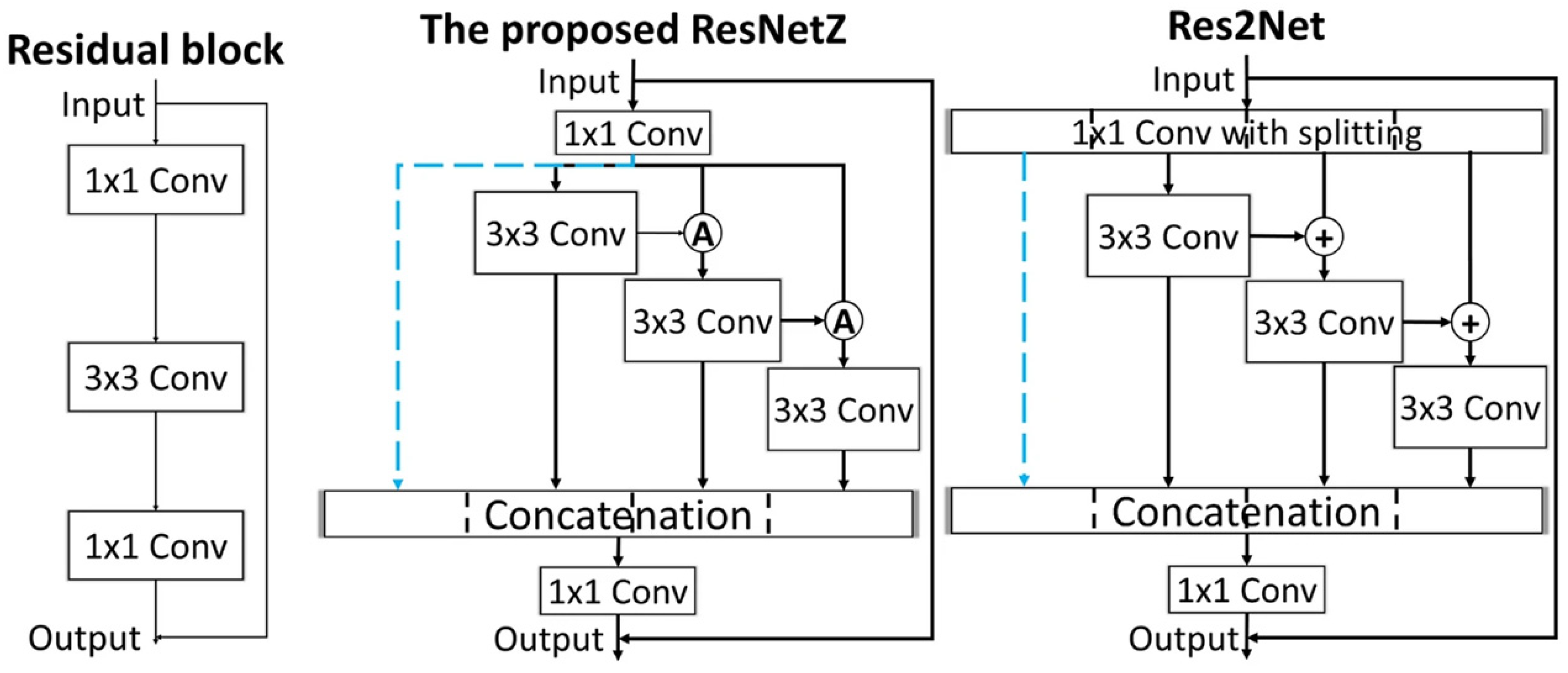
Wang et al. [40] aimed to classify healthy and cancerous lung tissue by four different ML methods (K-nearest neighbor (KNN), support vector classifier (SVC), neural network (NN), random forest (RF)). First, almost 20,000 fluorescence image frames (each frame contains one intensity and corresponding lifetime images) with a dimension of 128×128 px were collected from 10 patients (cancerous and non-cancerous). The measurements were performed by a fiber-based fluorescence lifetime imaging endomicroscope. In the preprocessing phase, the images went through several steps: thresholding, normalization, and Gaussian smoothing. Afterward, a dimension reduction was performed by principal component analysis (PCA) to remove zero values, boundary, and zero lifetime pixels. Therefore, the number of features decreases from 16,384 (128 × 128) to 2100. These features were then used to classify the data into two classes by four different ML methods. Among these methods, RF performed the best. Though all these methods successfully distinguish cancer and non-cancer images, there are still some points that need to be improved; e.g., this paper used PCA-based approaches that require flattening of the input from 2D to 1D, and the employed ML methods use single-pixel values. This means that correlations between adjacent pixels were lost. To fill this gap, the authors from [40] extended their work, which is mentioned in paper [42], and they used deep learning for classification. The authors collected 70,000 images with a dimension of 128 × 128. These images then went through many preprocessing steps, with an important step being a thresholding step that removed all pixel values for which the measured intensity was smaller than the square root of the mean intensity over the whole image. Thereafter, a normalization was applied. For the CNN model, they used different architectures and compared their results. For the performance evaluation, the authors created three types of combined image datasets—only lifetime images, two-channel images, and three-channel images. Three channels mean a pair of intensity and lifetime images were filled into two different channels of an RGB image, leaving the remaining channel with zero values. On the other hand, a two-channel image refers to a stack of images where intensity and lifetime images are merged. In this paper, the authors compared the performance among different DL models with the said three types of datasets, where DenseNet121′s accuracy is highest with approximately 86%. Also, they compare their results with other ML methods and CNN results surpassed the ML in almost every aspect. In conclusion, the authors conclude that using the three-channel dataset gave the best performance. To improve this CNN performance another work was performed by Wang et al. [43]. They replaced the bottleneck residual block of ResNet50 with a multiscale concatenated dilation (MSCD) block. The accuracy of MSCD was almost 87% among all other CNN methods. Though all these DL architectures, particularly ResNet [44] and DenseNet [45], performed well in classification problems, there are still some generalized problems. For example, ResNet produces some redundant features but is unable to create new features. To handle such a problem, Wang et al. [46] proposed a ResNetZ-based lung cancer classification method. Also, they compared the performance and architecture of their proposed network with ResNet [44] and Res2Net [47]. During the first stage, they collected 100,000 FLIM images from 18 patients. The preprocessing step was the same as the paper [34], and they chose three-channel images for lung cancer discrimination. The architecture of three different networks is shown in Figure 7. From Figure 7, we can see that the authors replaced a 1D convolution splitting block with a 1D convolution block. However, the Res2Net model outperforms, while the ResNetZ and the Res2Net performances are quite similar.
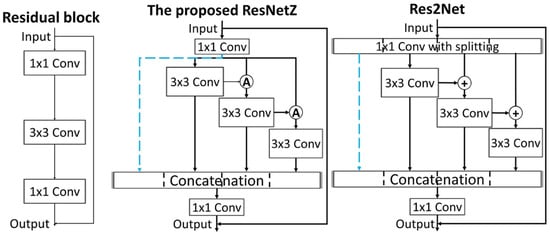
Figure 7.
Schematic diagram shows the different architectures of ResNet, ResNetZ, and Res2Net. The proposed network and Res2Net have a shortcut connection. Reprinted with permission from [46] under the terms of the CC-BY 4. license.
3.2.2. Skin Cancer Classification
The standard procedure for determining the stage and extent of skin cancer is biopsy [48]. However, due to its lack of specificity, this method’s accuracy is greatly reliant on the dermatologists’ experience, which could result in misdiagnosis. Recent advancements in biomedical studies could lead to rapid and proper diagnosis, but all these processes are time-consuming. We already discussed how small changes in cell metabolism can be identified by FLIM in the previous section. In this section, we discuss the detection of skin cancer by FLIM.
In paper [48], skin biopsies suspected to be skin cancer were classified into cancerous and normal tissue by machine learning methods. The authors collected FLIM images of 24 normal cases and 138 cancer cases for this study. Before classification, data were partitioned into two samples: training and testing. Validation was performed by three methods: (1) bootstrapping, (2) hold-out method, and (3) k-fold cross-validation. Then, four classification methods (RF, KNN, support vector machine (SVM), and linear discriminant analysis (LDA)) were used, and the results were compared. Using the bootstrapping sample partitioning scheme, all tested ML methods achieved accuracies over 80%.
Chen et al. [49] presented a study on a linear-kernel support vector machine (LSVM) model to distinguish basal cell carcinoma (BCC) from actinic keratosis (AK) and Bowen’s disease (BD). The input parameters of the LSVM model consist of lifetime components and lifetime entropies, which were extracted from two-photon fluorescence lifetime imaging of H&E-stained biopsy sections. In constructing the SVM models, features obtained from the lifetime (τ2) of the second component were found to be significantly more predictive than the average fluorescence lifetime (τm) in terms of diagnostic accuracy, sensitivity, and specificity. The above findings were confirmed based on the receiver operating characteristic (ROC) curves of diagnostic models. Furthermore, the results showed that adding Shannon entropy as an independent feature could further improve the diagnostic accuracy. The establishment of the SVM training model involved the extraction of fluorescence lifetime features and the calculation of the information entropy. The fluorescence lifetime data were fitted with triple-exponential decays. The SVM model was optimized by leave-one-out 5-fold cross-validation of the training datasets. Lifetime calculations and fitting were performed using SPCImage software (Becker & Hickl GmbH, Berlin, Germany). From this study, the authors conclude that using the τm feature, the LSVM model effectively distinguished BCC from the precancerous lesions (AK and BD) with a prediction accuracy of 90.4%. However, this model failed to distinguish between the AK and BD subcategories. On the other hand, the LSVM model using the lifetime component of τ2 as a training feature achieved better classification performance and was able to classify AK and BD. The prediction accuracy was 95.6 % for BCC vs. (AK and BD) and 91.1 % for AK vs. BD. Also, prediction accuracy increased by adding entropy as a feature.
3.2.3. Cervical Cancer Classification
Cervical cancer is the fourth most common cancer in women, leading to over 300,000 deaths a year [50]. Again, the prognosis of the diagnosis is highly dependent on the stage of detection, with early-stage detection significantly improving the outcome. To detect cancer, biopsies are currently performed, which are invasive, painful, and time-consuming procedures. Consequently, there is a need for a non-invasive and highly sensitive screening method. In contrast, it is known that cell metabolism changes during cancer, and it can be detected by the activity of a co-enzyme, namely NAD(P)H [51]. We discuss cervical cancer classification using FLIM in this section.
In paper [51], the authors showed cervical tissue classification using FLIM. Lifetime information was extracted with a bi-exponential model using expectation–maximization and the Bayesian information criterion algorithm (EM-BIC). Then, they applied the extreme learning machine (ELM) method to classify cancerous and non-cancerous, and the classification gave more than 80% specificity.
In another work performed by the authors [52], tissue was classified into normal and cervical intraepithelial neoplasia (CIN) based on the lifetime information [52]. In the images, epithelium and stroma regions were annotated, then calculated lifetimes were considered as the feature vector of the ELM method. Here, they used commercial SPCImage for lifetime calculation. Before applying the ML method, they used manual segmentation which was performed by pathologists. Their ELM method, which the authors say has better generalizability than traditional ML methods like SVM and back-propagation (BP), was able to differentiate between the tissue types with accuracies of up to 94%.
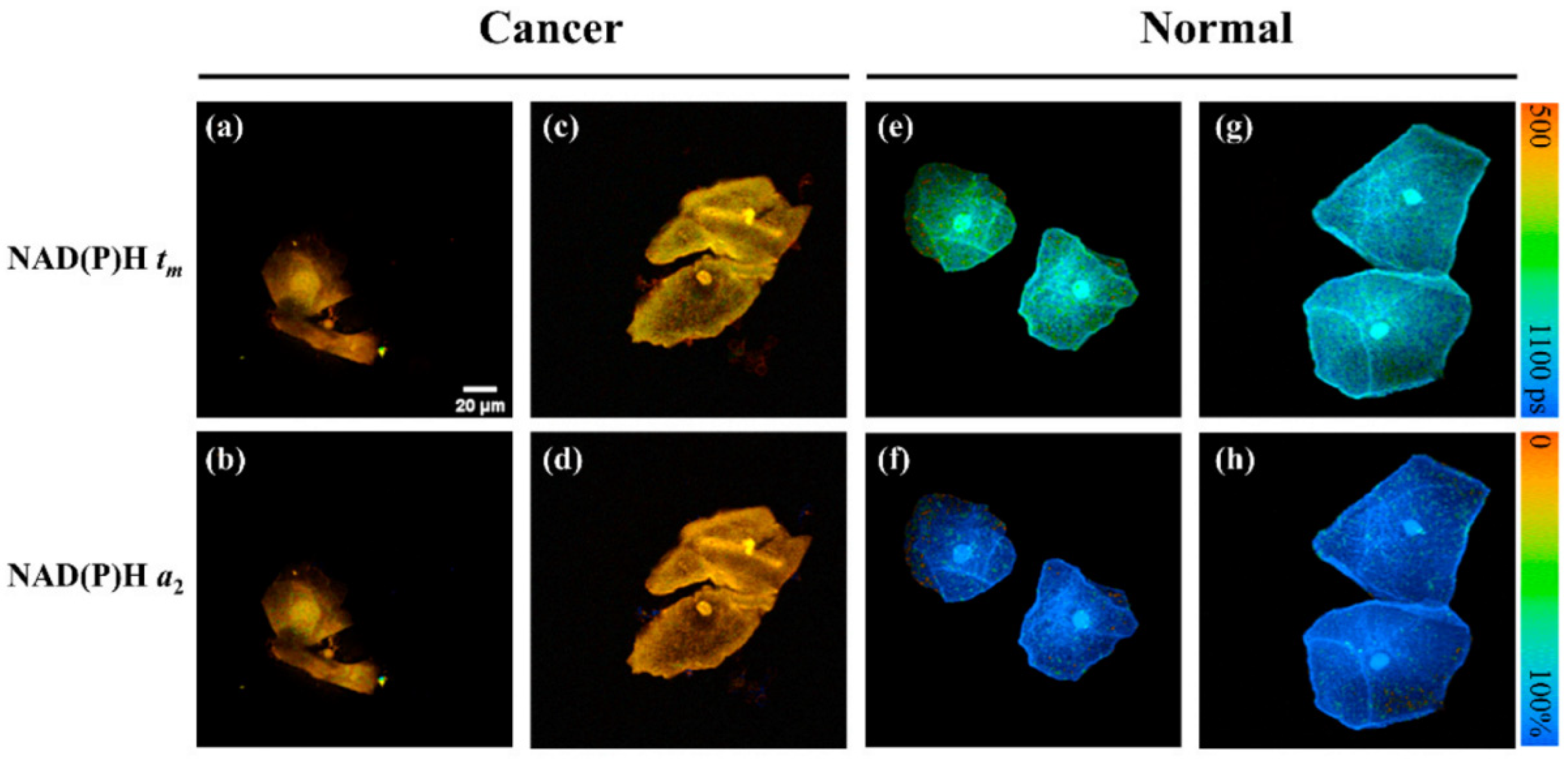
The authors of the paper [38] used an unsupervised machine learning method for cancer classification. In total, FLIM images of 71 patient samples (cf. Figure 8) were taken for this study. Some preprocessing steps were applied to increase the classification accuracy. Before classification with k-means, they used a pretrained Alexnet [53] for feature extraction and PCA for dimension reduction. This method gave 90.9% sensitivity and 100% specificity, which is higher than the traditional liquid-based cytology (LBC) method. The authors used three kinds of FLIM images as input mean lifetime (tm), second abundance component (a2), and tm and a2 together. They also showed a case study on real diagnosis, where tm images gave better accuracy than other image types. They also showed in the case study that FLIM gave a promising result against conventional diagnosis.
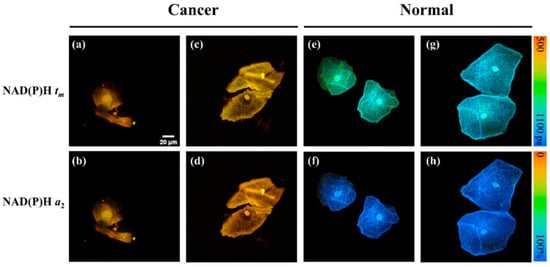
Figure 8.
Each column shows the FLIM images of cervical cancer from four participants: One row represents tm value from the NAD(P)H, and another row shows the a2 values from NAD(P)H. Lifetime values are shorter in cancerous cells than in normal cells because cancer cells tend to undergo glycolysis rather than oxidative phosphorylation. (a−d) samples are from cervical cancer patients and (e−h) samples are from normal patient. Reprinted with permission from [38] under the terms of the CC BY 4.0 license.
3.2.4. Microglia Classification
The authors from [54] studied a FLIM-based artificial neural network (ANN) approach to identify microglia position. They divided their studies into two parts. In the first part of the paper, they trained the ANN with lifetime parameters (lifetime values), and in the second part of the paper, they trained the ANN directly with the exponential decay traces. In the first case, lifetimes were extracted with SPCImage. The resulting lifetime data were split into training/validation/testing sets in a 70/15/15 split regime. The performance index was mean square error (MSE). In the second approach, they used 256 times bin histograms for ANN training. From the above two cases, lifetime-based classification performed better (almost 40% better sensitivity for one test case), which the authors attribute to the experimental setup used. They suggest that the direct classification of the decay traces could be significantly improved with a more varied training dataset.
3.2.5. Other Classification
Jo et al. [55] demonstrated FLIM-based early-stage detection of oral cancer and dysplasia. In this experiment, the authors collected tissue samples from 73 patients. They developed a computer-aided multispectral FLIM endoscope. First, a deconvolution method was applied to fluorescence decay traces for each pixel. From the deconvolved decay traces, spectral intensities, normalized fluorescence intensity, and average lifetime were used as features to design a quadratic discriminant analysis (QDA) classifier to discriminate cancerous oral tissue and mild dysplasia. This statistical model achieved 95% accuracy and 87% specificity.
Walsh et al. used a random forest (RF) classifier to monitor the activation of T-cells using NAD(P)H and FAD autofluorescence [56]. Uniform Manifold Approximation and Projection (UMAP) was used for dimension reduction. The authors achieved a classification accuracy of almost 98%. Additionally, the authors of [57] used a new technology to distinguish the parathyroid gland from other glands using FLIM images. Twenty-one patients underwent parathyroid surgery, and three ML models (NN, (RF), SVM) were used to distinguish parathyroid glands from other glands in the pharynx. The RF model showed the highest sensitivity and specificity. After classification, the Laguerre deconvolution method was used to predict lifetime values. A significant difference was found in the average lifetime of the parathyroid gland compared to other glands, such as thyroid, adipose tissue, and lymphoid tissue, enabling discrimination among them.
4. Inverse Modeling
As laid out in the introduction, FLIM measurements are carried out by obtaining fluorescence decay curves by means of TD or FD measurements. To use these data for further analysis, the relevant parameters describing the observed system, i.e., the fluorescence lifetimes, need to be extracted from these curves. However, this is an ill-posed problem [58]. Curve fitting, the traditional extraction technique, is a difficult procedure with numerous software options offered by various businesses and research organizations (as shown in the introduction).
Some authors suggested that lifetime extraction approaches based on machine learning (ML), especially deep learning (DL), can solve this issue. This is based on the assumption that extraction of lifetime parameters from decay curves is essential for an inverse modeling problem of the measurement procedure (see Table 4 for examples).
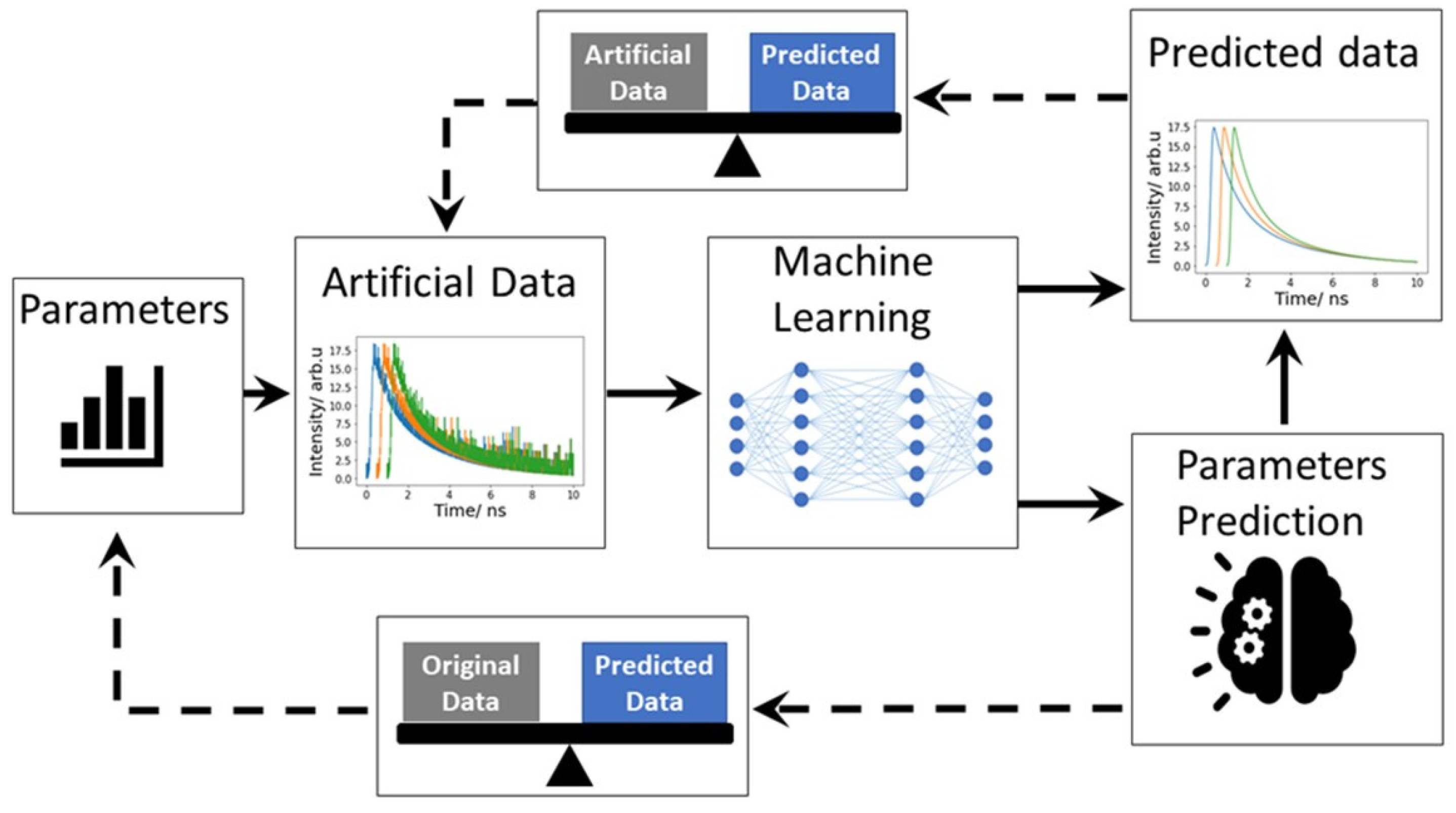
There are several software packages to solve general inverse modeling problems. However, the application of these programs to FLIM remains challenging. One challenge is the multi-exponential nature of the decay curves: fluorescence decay in biological samples is often characterized by contributions of multiple fluorophores, which means that multiple lifetimes contribute to the overall fluorescence signal. Choosing an appropriate model or number of decay constants for fitting is challenging. On the other hand, the low photon counts in FLIM often cause high noise contributions in comparison with the signal of interest, which can affect lifetime parameter estimation. This section focuses on inverse modeling with ML/DL for lifetime feature extraction (see Figure 9) and their advantages and disadvantages.
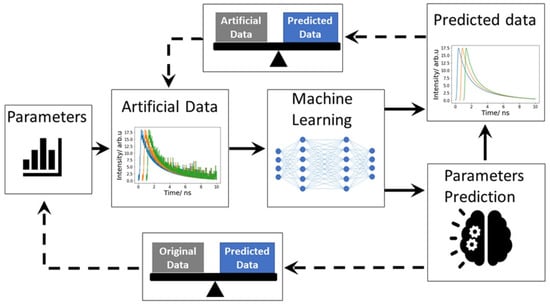
Figure 9.
This is the workflow of inverse modeling. First, a machine learning model was trained with artificial FLIM data. Artificial data mimic the real experimental data. To generate the artificial data, system response function (IRF) is convolved with an exponential function. Predicted parameters were compared with original artificial data. Finally, the machine learning model is tested with real data, measured data. Here, thick line represents the data generation and training method and dotted line represents the evolution process.
Wu et al. [59] employed an ANN approach based on a bi-exponential model to estimate lifetime parameters from TCSPC raw data. The objective was to train an ANN model to approximate the function that maps the TCSPC raw data into the unknown lifetime parameters. The ANN model used in the study consisted of two hidden fully connected (FC) layers using the time bins and their respective photon counts as input values and outputting estimated lifetimes and abundance ratio. The results from the study demonstrated that the ANN-based method enabled the estimation of a lifetime image (256 × 256 size) in just 0.9 s, which was 180 times faster compared to the curve-fitting technique based on the least-squares method (LSM). The authors also observed that the LSM method struggled to accurately estimate lifetime parameters due to its sensitivity to initial conditions. The success rate for accurately estimating lifetime parameters from real experimental data using the ANN method was reported as 99.93%, while the LSM method achieved a success rate of 95.93%. This indicates that the ANN method significantly improved the accuracy of lifetime estimation compared to conventional curve-fitting tools.
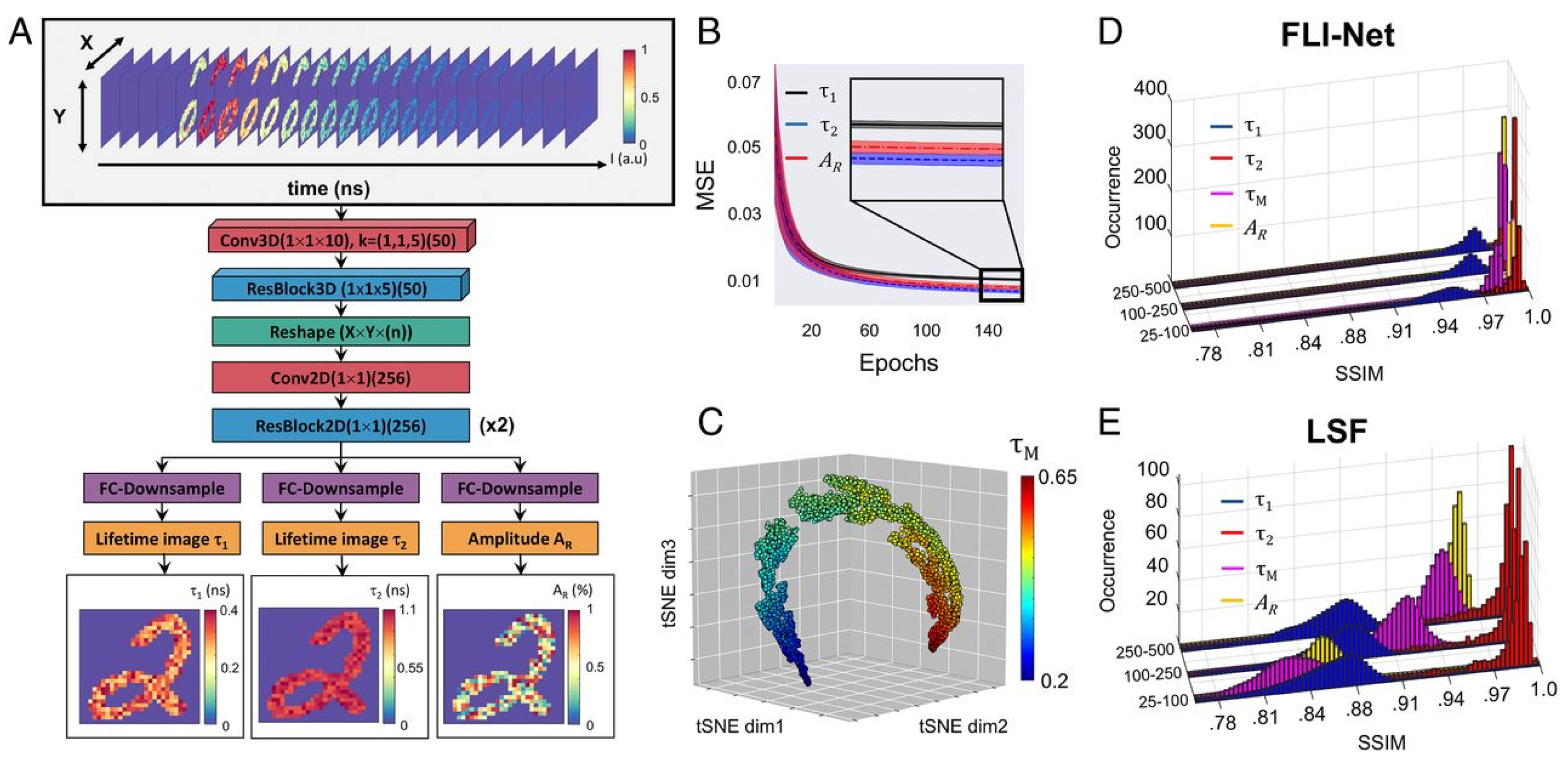
However, this ANN failed for low photon counts. To overcome this problem, Smith et al. [60] proposed an innovative approach for estimating lifetime parameters from a complete three-dimensional TCSPC dataset using a CNN. Their architecture, called FLI-Net (Fluorescence Lifetime Imaging Network, cf. Figure 10), was trained with synthetic data produced from simulating biexponential decays and IRFs for each pixel in the popular MNIST handwritten digits dataset. FLI-Net consists of a shared branch for temporal feature extraction and separate branches for reconstructing lifetime images and fractional amplitudes of short lifetimes. To enable spatially independent feature extraction and capture of each temporal point spread function (TPSF), 3D convolutions (Conv3D) are applied along the temporal dimension for each pixel location. By using a Conv3D layer (with a kernel size 1 × 1 × 10), unwanted artifacts from neighboring pixels in the spatial dimensions were minimized during both the training and testing phases. Additionally, a residual block (ResBlock) with a reduced kernel length allows for further extraction of temporal information. This network has three output branches. Each branch employs a sequence of convolutions for down-sampling. By comparing the results obtained from FLI-Net with those from the conventional least-squares fitting (LSF) method, Smith et al. [60] found that FLI-Net gave high accuracy and was approximately 30 times faster than the SPCImage. However, this method’s performance depends on spatial information of the FLIM data, which means the model needs more datasets to train.
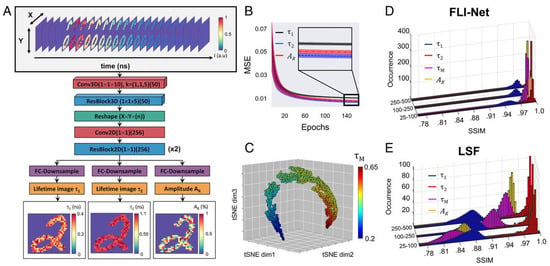
Figure 10.
(A) The architecture of FLI–Net. The input of the FLI–Net is 3D data cube. (B) MSE vs. epochs for lifetime 1, lifetime 2, abundance ratio. (C) t–SNE projection from the last activation map. (D) FLI–Net performance concerning structure similarity index (SSIM). (E) LSF performance concerning SSIM. From the plot, it is clear that FLI–Net performance is better than LSF. Reprinted with permission from [60] under the terms of the PNAS license.
Guo et al. [20] described a new method where lifetimes and abundances were inverse modeled from decay traces through ML. They trained a random forest (RF) model with 3000 artificial traces, which was tested with experimental data to estimate lifetimes and abundances. The performances of the ML model were verified based on two things: First, the predicted values were compared with their true values for artificial data, which showed good prediction performance. Secondly, model performances are verified on real-world data (SPCImage software for lifetime estimation), and results roughly match each other. In summary, ML performed better than the traditional method. The authors also compared their model with the FLI-Net [60]. Their RF-based model performs better than FLI-Net within three aspects: First, the RF model works on a pixel basis and thus does not need retraining when spatial dimensions change. Second, the employed Laguerre polynomial approach and a large span of training values make the model highly generalizable. Third, compared to FLI-Net, decay traces with more than two components can be analyzed.
Yao et al. [61] proposed a novel optical instrument for compressive macroscopic fluorescence lifetime images (MFLI). They demonstrated a method to enhance the resolution of fluorescence lifetime images, which is mainly based on a CNN model called Net-FLICS trained on a modified (with simulated decay traces in each pixel) EMNIST dataset. One block aims to recover sparsity information from compressive data with a 1D CNN layer. The second segment is responsible for revealing the intensity of images through a 2D convolution layer, and the third segment utilizes 1D convolution layers to reconstruct lifetime images directly from raw data.
Another ANN technique to retrieve lifetimes from raw FLIM data was introduced in [62]. The ANN takes the raw decay trace acquired from a SPAD (single photon avalanche diode) pixel, and, using three hidden layers, it directly outputs the associated lifetime. The model, trained with simulated data, was evaluated in comparison to results obtained from least-squares (LSQ) deconvolution and also tested with real experimental data. The authors showed that the ANN successfully estimates lifetime values from synthetic data as well as real data and is 1000 times faster than LSQ.
In general, most convolutional neural networks are designed to handle 2-, 3-, or multidimensional data. However, these high-dimensional CNNs have a higher number of trainable parameters and thus increase the training and calculation complexity. To overcome this problem, Xiao et al. [63] used fluorescence data in a 1D CNN instead of 2D or 3D CNNs. In this paper, the 1D CNN was mainly divided into two parts. In the first part, the decay features were extracted, and in the second part, containing n + 1 branches for an n-exponential decay fit, the lifetimes and abundancies for the components were reconstructed. For training and testing, the authors simulated two different datasets, each containing 40,000 decay samples, and performances were compared with the traditional trust-region-reflective algorithm (TRRA) method, FLI-Net, and DenseNet architecture. This model can resolve the multi-exponential decay model, and it is 8 and 300 times faster than other CNN models and TRRA, respectively. These are some important advantages of using 1D CNN. Also, the authors showed that their network successfully estimated fluorescence lifetime from experimental data.
Xiao et al. [64] proposed a DL-based lifetime image estimation method for few-photon fluorescence lifetime imaging (FPFLI). They showed that traditional methods, like LSF, MLE, Bayesian analysis (BA), or phasor methods, need high photon counts for FLIM analysis, while DL models can deal with this problem. Here, they increased the training speed by training this model with large synthetic data. They showed that FPFLI performed where MLE failed to estimate lifetimes in low photon counts. Ochoa et al. [65] proposed NetFLICS-CR, where a compressed ratio (CR) block is added to the NetFLICS [61] model. This CR block reduces the input dimension. They compared their model performance with TVRecon. They trained the network with a modified (simulated decay traces for each pixel) EMNIST dataset. Before training, they performed data augmentation by rotation and combining two different datasets. Mean absolute error (MAE) was considered as an evolution matrix between predicted and reconstructed. The trained Net-FLICS took approximately 2.2 s in total to reconstruct 800 samples, whereas TVRecon took 15 s. In conclusion, Net-FLICS performs better than TVRecon.

Table 4.
Comparison between some different inverse modeling methods.
Table 4.
Comparison between some different inverse modeling methods.
| Network Used | Dataset Used | |
|---|---|---|
| Guo et al. [20] | Random forest | Cell sample |
| Wu et al. [59] | ANN with 4 layers. 1—input 1—output 2—hidden | Daisy pollens |
| FLI-Net [60] | 10 layers (combination with convolution and ResNet block) | Mice liver and bladder sample |
| NetFLICS [61] | ResNet | Mice liver and bladder |
| Xiao et al. [63] | Mainly 4 layers. In combination with CNN and ResNetBlock | Human prostate cancer tissue |
In conclusion, we can confirm that all DL or ML methods can estimate lifetimes from experimental data, usually faster and more accurate than conventional methods.
5. Discussion and Conclusions
In our comprehensive review, we have summarized recent investigations on the utilization of machine learning (ML) and deep learning (DL) techniques for fluorescence lifetime imaging microscopy (FLIM) data analysis and modeling. Traditional methods for extracting fluorescence lifetimes from FLIM data can be complex and time-consuming. However, ML and DL methods have emerged as promising alternatives that offer faster and more accurate extraction of fluorescence lifetimes. Combining different inverse modeling approaches or combining inverse modeling with forward modeling to take advantage of each process can be a new direction in the FLIM inverse modeling technique.
Among the ML and DL approaches, convolutional neural network (CNN)-based techniques were found to be prevalent in the majority of the reviewed papers. These techniques have demonstrated their effectiveness in various aspects of FLIM analysis. For instance, CNNs have been successfully applied for denoising and enhancing fluorescence microscopic images, thereby improving the quality of fluorescence data. Additionally, novel DL techniques have been developed, such as the Net-FLICS network for fluorescence lifetime imaging using compressive sensing data and the 3D CNN-based FLI-Net network for lifetime extraction.
In this review, we highlighted several areas of further research and development. One key aspect is addressing the challenge of limited data availability. Strategies to overcome this limitation could involve data augmentation techniques or transfer learning approaches. Another important aspect is the generation of simplified and easily deployable models for extracting lifetime images from FLIM data. Additionally, developing neural network models capable of distinguishing overlapping fluorescence lifetimes would be valuable.
Furthermore, we provided a concise overview of different freely available software tools for lifetime extraction, such as FLIMJ, FLIMview, and FLIMfit. These software options can significantly reduce the cost, complexity, and time required for FLIM data analysis.
In conclusion, ML and DL-based techniques have demonstrated great potential in the field of FLIM data modeling. They offer faster and more accurate lifetime extraction, denoising capabilities, and improved image enhancement. Future research should focus on addressing data limitations, simplifying model generation, handling overlapping lifetimes, and exploring open-source software solutions for FLIM analysis.
Author Contributions
Writing—original draft preparation, M.A.; writing—review and editing, R.H., J.H. and T.B.; visualization, M.A.; supervision, R.H. and T.B.; funding acquisition, T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Federal Ministry of Education and Research of Germany (BMBF), funding program Photonics Research Germany (FKZ: 13N15706, 13N15466, 13N15710) and is integrated into the Leibniz Center for Photonics in Infection Research (LPI).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Renz, M. Fluorescence Microscopy—A Historical and Technical Perspective. Cytom. Part A 2013, 83, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, P.; Hirt, A. Mikroskopische Untersuchungen an lebenden Organen. Z. Anat. Entwickl. Gesch. 1929, 90, 791–802. [Google Scholar] [CrossRef]
- König, K. 1 Brief History of Fluorescence Lifetime Imaging. In 1 Brief History of Fluorescence Lifetime Imaging; De Gruyter: Berlin, Germany, 2018; pp. 3–16. ISBN 978-3-11-042998-5. [Google Scholar]
- Datta, R.; Heaster, T.M.; Sharick, J.T.; Gillette, A.A.; Skala, M.C. Fluorescence Lifetime Imaging Microscopy: Fundamentals and Advances in Instrumentation, Analysis, and Applications. J. Biomed. Opt. 2020, 25, 071203. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, J.W.; Conchello, J.-A. Fluorescence Microscopy. Nat. Methods 2005, 2, 910–919. [Google Scholar] [CrossRef]
- Ossato, G. Phasor Analysis for FLIM (Fluorescence Lifetime Imaging Microscopy). Available online: https://www.leica-microsystems.com/science-lab/phasor-analysis-for-flim-fluorescence-lifetime-imaging-microscopy/ (accessed on 26 May 2023).
- Bixon, M.; Jortner, J. Intramolecular Radiationless Transitions. J. Chem. Phys. 2003, 48, 715–726. [Google Scholar] [CrossRef]
- Kasha, M. Characterization of Electronic Transitions in Complex Molecules. Discuss. Faraday Soc. 1950, 9, 14–19. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Datta, R.; Gratton, E.; Hochbaum, A.I. Metabolic Fingerprinting of Bacteria by Fluorescence Lifetime Imaging Microscopy. Sci. Rep. 2017, 7, 3743. [Google Scholar] [CrossRef]
- Kappel, C.; Kuschel, L.; DeRose, J. Was ist FRET mit FLIM (FLIM-FRET)? Available online: https://www.leica-microsystems.com/de/science-lab/life-science/was-ist-fret-mit-flim-flim-fret/ (accessed on 28 August 2023).
- Verveer, P.J.; Gemkow, M.J.; Jovin, T.M. A Comparison of Image Restoration Approaches Applied to Three-Dimensional Confocal and Wide-Field Fluorescence Microscopy. J. Microsc. 1999, 193, 50–61. [Google Scholar] [CrossRef]
- Becker, W. Fluorescence Lifetime Imaging–Techniques and Applications: Fluorescence Lifetime Imaging. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Datta, R.; Gillette, A.; Stefely, M.; Skala, M.C. Recent Innovations in Fluorescence Lifetime Imaging Microscopy for Biology and Medicine. J. Biomed. Opt. 2021, 26, 070603. [Google Scholar] [CrossRef]
- Gavin, H.P. The Levenberg-Marquardt Algorithm for Nonlinear Least Squares Curve-Fitting Problems. 2022. Available online: https://people.duke.edu/~hpgavin/ExperimentalSystems/lm.pdf (accessed on 26 May 2023).
- Chessel, A.; Waharte, F.; Salamero, J.; Kervrann, C. A Maximum Likelihood Method for Lifetime Estimation in Photon Counting-Based Fluorescence Lifetime Imaging Microscopy. In Proceedings of the 21st European Signal Processing Conference (EUSIPCO 2013), Marrakech, Morocco, 9–13 September 2013; pp. 1–5. [Google Scholar]
- Maximum Likelihood Estimation|Theory, Assumptions, Properties. Available online: https://www.statlect.com/fundamentals-of-statistics/maximum-likelihood (accessed on 26 May 2023).
- Maus, M.; Cotlet, M.; Hofkens, J.; Gensch, T.; De Schryver, F.C.; Schaffer, J.; Seidel, C.A.M. An Experimental Comparison of the Maximum Likelihood Estimation and Nonlinear Least-Squares Fluorescence Lifetime Analysis of Single Molecules. Anal. Chem. 2001, 73, 2078–2086. [Google Scholar] [CrossRef] [PubMed]
- Pelet, S.; Previte, M.J.R.; Laiho, L.H.; So, P.T.C. A Fast Global Fitting Algorithm for Fluorescence Lifetime Imaging Microscopy Based on Image Segmentation. Biophys. J. 2004, 87, 2807–2817. [Google Scholar] [CrossRef] [PubMed]
- Digman, M.A.; Caiolfa, V.R.; Zamai, M.; Gratton, E. The Phasor Approach to Fluorescence Lifetime Imaging Analysis. Biophys. J. 2008, 94, L14–L16. [Google Scholar] [CrossRef]
- Guo, S.; Silge, A.; Bae, H.; Tolstik, T.; Meyer, T.; Matziolis, G.; Schmitt, M.; Popp, J.; Bocklitz, T. FLIM Data Analysis Based on Laguerre Polynomial Decomposition and Machine-Learning. J. Biomed. Opt. 2021, 26, 022909. [Google Scholar] [CrossRef] [PubMed]
- Tcspc Hand Book, 9th ed.; Becker & Hickl GmbH:: Berlin, Germany, 2021; Available online: https://www.becker-hickl.com/literature/documents/flim/the-bh-tcspc-handbook/ (accessed on 14 September 2023).
- Carrasco Kind, M.; Zurauskas, M.; Alex, A.; Marjanovic, M.; Mukherjee, P.; Doan, M.; Spillman, D.R., Jr.; Hood, S.; Boppart, S.A. Flimview: A Software Framework to Handle, Visualize and Analyze FLIM Data. F1000Research 2020, 9, 574. [Google Scholar] [CrossRef]
- Gao, D.; Barber, P.R.; Chacko, J.V.; Kader Sagar, M.A.; Rueden, C.T.; Grislis, A.R.; Hiner, M.C.; Eliceiri, K.W. FLIMJ: An Open-Source ImageJ Toolkit for Fluorescence Lifetime Image Data Analysis. PLoS ONE 2020, 15, e0238327. [Google Scholar] [CrossRef]
- FLIMfit. Available online: https://flimfit.org/ (accessed on 26 May 2023).
- Zhang, Y.; Zhu, Y.; Nichols, E.; Wang, Q.; Zhang, S.; Smith, C.; Howard, S. A Poisson-Gaussian Denoising Dataset with Real Fluorescence Microscopy Images. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 11702–11710. [Google Scholar]
- Zhong, L.; Liu, G.; Yang, G. Blind Denoising of Fluorescence Microscopy Images Using GAN-Based Global Noise Modeling. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 863–867. [Google Scholar]
- Lebrun, M. An Analysis and Implementation of the BM3D Image Denoising Method. Image Process. Line 2012, 2, 175–213. [Google Scholar] [CrossRef]
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Trans. Image Process. 2017, 26, 3142–3155. [Google Scholar] [CrossRef]
- Aharon, M.; Elad, M.; Bruckstein, A. K-SVD: An Algorithm for Designing Overcomplete Dictionaries for Sparse Representation. IEEE Trans. Signal Process. 2006, 54, 4311–4322. [Google Scholar] [CrossRef]
- Li, J.; Luisier, F.; Blu, T. PURE-LET Image Deconvolution. IEEE Trans. Image Process. 2018, 27, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Belthangady, C.; Royer, L.A. Applications, Promises, and Pitfalls of Deep Learning for Fluorescence Image Reconstruction. Nat. Methods 2019, 16, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Weigert, M.; Schmidt, U.; Boothe, T.; Müller, A.; Dibrov, A.; Jain, A.; Wilhelm, B.; Schmidt, D.; Broaddus, C.; Culley, S.; et al. Content-Aware Image Restoration: Pushing the Limits of Fluorescence Microscopy. Nat. Methods 2018, 15, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Batson, J.; Royer, L. Noise2Self: Blind Denoising by Self-Supervision. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 524–533. [Google Scholar]
- Mannam, V.; Zhang, Y.; Yuan, X.; Hato, T.; Dagher, P.C.; Nichols, E.L.; Smith, C.J.; Dunn, K.W.; Howard, S. Convolutional Neural Network Denoising in Fluorescence Lifetime Imaging Microscopy (FLIM). In Proceedings of the Multiphoton Microscopy in the Biomedical Sciences XXI, Online. 5–12 March 2021; p. 43. [Google Scholar]
- Suykens, J.A.K.; Vandewalle, J.P.L.; De Moor, B.L.R. Artificial Neural Networks: Architectures and Learning Rules. In Artificial Neural Networks for Modelling and Control of Non-Linear Systems; Suykens, J.A.K., Vandewalle, J.P.L., De Moor, B.L.R., Eds.; Springer: Boston, MA, USA, 1996; pp. 19–35. ISBN 978-1-4757-2493-6. [Google Scholar]
- Wang, Y.; Pinkard, H.; Khwaja, E.; Zhou, S.; Waller, L.; Huang, B. Image Denoising for Fluorescence Microscopy by Supervised to Self-Supervised Transfer Learning. Opt. Express 2021, 29, 41303–41312. [Google Scholar] [CrossRef]
- Zhang, Y.; Hato, T.; Dagher, P.C.; Nichols, E.L.; Smith, C.J.; Dunn, K.W.; Howard, S.S. Automatic Segmentation of Intravital Fluorescence Microscopy Images by K-Means Clustering of FLIM Phasors. Opt. Lett. 2019, 44, 3928. [Google Scholar] [CrossRef]
- Ji, M.; Zhong, J.; Xue, R.; Su, W.; Kong, Y.; Fei, Y.; Ma, J.; Wang, Y.; Mi, L. Early Detection of Cervical Cancer by Fluorescence Lifetime Imaging Microscopy Combined with Unsupervised Machine Learning. Int. J. Mol. Sci. 2022, 23, 11476. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Wang, Q.; Vallejo, M.; Hopgood, J. Fluorescence Lifetime Endomicroscopic Image-Based Ex-Vivo Human Lung Cancer Differentiation Using Machine Learning. TechRxiv Preprint 2020. [Google Scholar] [CrossRef]
- Wang, M.; Tang, F.; Pan, X.; Yao, L.; Wang, X.; Jing, Y.; Ma, J.; Wang, G.; Mi, L. Rapid Diagnosis and Intraoperative Margin Assessment of Human Lung Cancer with Fluorescence Lifetime Imaging Microscopy. BBA Clin. 2017, 8, 7–13. [Google Scholar] [CrossRef]
- Wang, Q.; Hopgood, J.R.; Finlayson, N.; Williams, G.O.S.; Fernandes, S.; Williams, E.; Akram, A.; Dhaliwal, K.; Vallejo, M. Deep Learning in Ex-Vivo Lung Cancer Discrimination Using Fluorescence Lifetime Endomicroscopic Images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1891–1894. [Google Scholar]
- Wang, Q.; Hopgood, J.R.; Vallejo, M. Fluorescence Lifetime Imaging Endomicroscopy Based Ex-Vivo Lung Cancer Prediction Using Multi-Scale Concatenated-Dilation Convolutional Neural Networks. In Proceedings of the Medical Imaging 2021: Computer-Aided Diagnosis, Online. 15–20 February 2021; Drukker, K., Mazurowski, M.A., Eds.; SPIE: Bellingham, WA, USA, 2021; p. 94. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar] [CrossRef]
- Wang, Q.; Hopgood, J.R.; Fernandes, S.; Finlayson, N.; Williams, G.O.S.; Akram, A.R.; Dhaliwal, K.; Vallejo, M. A Layer-Level Multi-Scale Architecture for Lung Cancer Classification with Fluorescence Lifetime Imaging Endomicroscopy. Neural Comput. Appl. 2022, 34, 18881–18894. [Google Scholar] [CrossRef]
- Gao, S.-H.; Cheng, M.-M.; Zhao, K.; Zhang, X.-Y.; Yang, M.-H.; Torr, P. Res2Net: A New Multi-Scale Backbone Architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Qi, M.; Wu, Z.; Liu, L.; Gao, P.; Qu, J. Classification of Skin Cancer Based on Fluorescence Lifetime Imaging and Machine Learning. In Proceedings of the Optics in Health Care and Biomedical Optics X, Online. 11–16 October 2020; Luo, Q., Li, X., Gu, Y., Zhu, D., Eds.; SPIE: Bellingham, WA, USA, 2020; p. 71. [Google Scholar]
- Chen, B.; Lu, Y.; Pan, W.; Xiong, J.; Yang, Z.; Yan, W.; Liu, L.; Qu, J. Support Vector Machine Classification of Nonmelanoma Skin Lesions Based on Fluorescence Lifetime Imaging Microscopy. Anal. Chem. 2019, 91, 10640–10647. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Jun, G.; Koon, N.B.; Yaw, F.C.; s/o Gulam Razul, S.; Kim, L.S. Fluorescence Lifetime Diagnosis of Cervical Cancer Based on Extreme Learning Machine. In Proceedings of the 2010 Photonics Global Conference, Orchard, Singapore, 14–16 December 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 1–3. [Google Scholar]
- Gu, J.; Fu, C.Y.; Ng, B.K.; Liu, L.B.; Lim-Tan, S.K.; Lee, C.G.L. Enhancement of Early Cervical Cancer Diagnosis with Epithelial Layer Analysis of Fluorescence Lifetime Images. PLoS ONE 2015, 10, e0125706. [Google Scholar] [CrossRef]
- Wei, J. AlexNet: The Architecture That Challenged CNNs. Available online: https://towardsdatascience.com/alexnet-the-architecture-that-challenged-cnns-e406d5297951 (accessed on 26 May 2023).
- Sagar, M.A.K.; Cheng, K.P.; Ouellette, J.N.; Williams, J.C.; Watters, J.J.; Eliceiri, K.W. Machine Learning Methods for Fluorescence Lifetime Imaging (FLIM) Based Label-Free Detection of Microglia. Front. Neurosci. 2020, 14, 931. [Google Scholar] [CrossRef]
- Jo, J.A.; Cheng, S.; Cuenca-Martinez, R.; Duran-Sierra, E.; Malik, B.; Ahmed, B.; Maitland, K.; Cheng, Y.-S.L.; Wright, J.; Reese, T. Endogenous Fluorescence Lifetime Imaging (FLIM) Endoscopy for Early Detection of Oral Cancer and Dysplasia. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 3009–3012. [Google Scholar]
- Walsh, A.J.; Mueller, K.; Jones, I.; Walsh, C.M.; Piscopo, N.; Niemi, N.N.; Pagliarini, D.J.; Saha, K.; Skala, M.C. Label-Free Method for Classification of T Cell Activation. bioRxiv 2019. [Google Scholar] [CrossRef]
- Marsden, M.; Weaver, S.S.; Marcu, L.; Campbell, M.J. Intraoperative Mapping of Parathyroid Glands Using Fluorescence Lifetime Imaging. J. Surg. Res. 2021, 265, 42–48. [Google Scholar] [CrossRef]
- Zang, Z.; Xiao, D.; Wang, Q.; Jiao, Z.; Chen, Y.; Li, D.D.U. Compact and Robust Deep Learning Architecture for Fluorescence Lifetime Imaging and FPGA Implementation. Methods Appl. Fluoresc. 2023, 11, 025002. [Google Scholar] [CrossRef]
- Wu, G.; Nowotny, T.; Zhang, Y.; Yu, H.-Q.; Li, D.D.-U. Artificial Neural Network Approaches for Fluorescence Lifetime Imaging Techniques. Opt. Lett. 2016, 41, 2561. [Google Scholar] [CrossRef]
- Smith, J.T.; Yao, R.; Sinsuebphon, N.; Rudkouskaya, A.; Un, N.; Mazurkiewicz, J.; Barroso, M.; Yan, P.; Intes, X. Fast Fit-Free Analysis of Fluorescence Lifetime Imaging via Deep Learning. Proc. Natl. Acad. Sci. USA 2019, 116, 24019–24030. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Ochoa, M.; Yan, P.; Intes, X. Net-FLICS: Fast Quantitative Wide-Field Fluorescence Lifetime Imaging with Compressed Sensing—A Deep Learning Approach. Light Sci. Appl. 2019, 8, 26. [Google Scholar] [CrossRef]
- Zickus, V.; Wu, M.-L.; Morimoto, K.; Kapitany, V.; Fatima, A.; Turpin, A.; Insall, R.; Whitelaw, J.; Machesky, L.; Bruschini, C.; et al. Fluorescence Lifetime Imaging with a Megapixel SPAD Camera and Neural Network Lifetime Estimation. Sci. Rep. 2020, 10, 20986. [Google Scholar] [CrossRef]
- Xiao, D.; Chen, Y.; Li, D.D.-U. One-Dimensional Deep Learning Architecture for Fast Fluorescence Lifetime Imaging. IEEE J. Sel. Top. Quantum Electron. 2021, 27, 1–10. [Google Scholar] [CrossRef]
- Xiao, D.; Sapermsap, N.; Chen, Y.; Li, D.D.U. Deep learning enhanced fast fluorescence lifetime imaging with a few photons. Optica 2023, 10, 944–951. [Google Scholar] [CrossRef]
- Ochoa, M.; Rudkouskaya, A.; Yao, R.; Yan, P.; Barroso, M.; Intes, X. Deep Learning Enhanced Hyperspectral Fluorescence Lifetime Imaging. bioRxiv 2020. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).