A Pilot Study to Investigate the Antimicrobial Activity of Pulsed UVA and UVC
Abstract
:1. Introduction
2. Materials and Methods
2.1. Safety Consideration
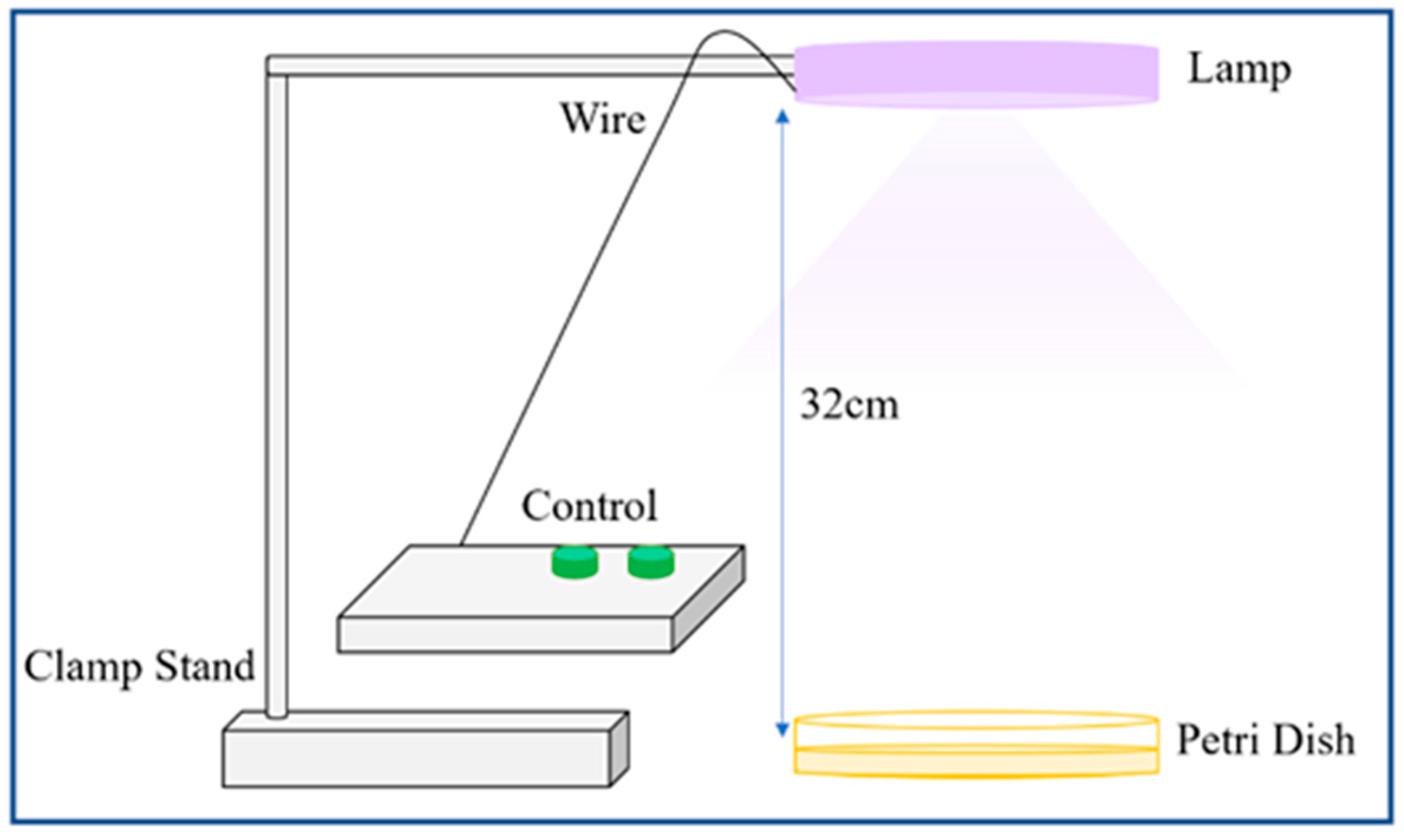
2.2. Materials
2.3. Antibacterial Assessment of UVA and UVC Exposure
2.3.1. Microorganisms and Culture Methods
2.3.2. Protocol I
2.3.3. Protocol II
2.3.4. ATP Measurement
2.4. Antiviral Assessment of UVA and UVC Exposure
2.4.1. Infecting with HCoV-229E
2.4.2. Protocol 1
2.4.3. Protocol 2
2.4.4. Protocol 2a
2.4.5. Detection of Viral Infectivity
2.4.6. Analysis of Viral Infectivity by Using the TCID50 Assay
2.4.7. Modelling for Cross-Contamination Risks
3. Results
3.1. Effect of UVA and UVC Light on Bacteria
3.1.1. Protocol I
3.1.2. Protocol II
Distance Measurements
Iteration Measurements
3.1.3. Response Modelling for Cross-Contamination Risks
3.2. Effect of UVA and UVC Exposure on Human Coronavirus
3.2.1. Investigation of Reduction in Viral Infectivity
3.2.2. Effect of Protocol 1 on HCoV-229E
3.2.3. Effect of Protocol 2a on HCoV-229E
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chatzisymeon, E. Inactivation of bacteria in seafood processing water by means of UV treatment. J. Food Eng. 2016, 173, 1–7. [Google Scholar] [CrossRef]
- Jelden, K.C.; Gibbs, S.G.; Smith, P.W.; Hewlett, A.L.; Iwen, P.C.; Schmid, K.K.; Lowe, J.J. Ultraviolet (UV)-reflective paint with ultraviolet germicidal irradiation (UVGI) improves decontamination of nosocomial bacteria on hospital room surfaces. J. Occup. Environ. Hyg. 2017, 14, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, L.; Xu, M.; Zhou, H.; Wang, S.; Wang, Y.; Bai, M.; Zhang, C. Selective antibiotic resistance genes in multiphase samples during biofilm growth in a simulated drinking water distribution system: Occurrence, correlation and low-pressure ultraviolet removal. Sci. Total Environ. 2019, 649, 146–155. [Google Scholar] [CrossRef] [PubMed]
- McLeod, A.; Liland, K.H.; Haugen, J.-E.; Serheim, O.; Myhrer, K.S.; Jolck, A.L. Chicken fillets subjected to UV-C and pulsed UV light: Reduction of pathogenic and spoilage bacteria, and changes in sensory quality. J. Food Saf. 2018, 38, e12421. [Google Scholar] [CrossRef]
- Vollmerhausen, T.L.; Conneely, A.; Bennett, C.; Wagner, V.E.; Victor, J.C.; O’Byrne, C.P. Visible and UVA light as a potential means of preventing Escherichia coli biofilm formation in urine and on materials used in urethral catheters. J. Photochem. Photobiol. B Biol. 2017, 170, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.K.; Mitchell, H.A. Realigning the conventional routes of transmission: An improved model for occupational exposure assessment and infection prevention. J. Infect. Dis. 2020, 105, 17–23. [Google Scholar] [CrossRef]
- Wang, C.; Lu, S.; Zhang, Z. Inactivation of airborne bacteria using different UV sources: Performance modelling, energy utilisation and endotoxin degradation. Sci. Total Environ. 2019, 655, 787–795. [Google Scholar] [CrossRef]
- Zou, X.Y.; Lin, Y.L.; Xu, B.; Coa, T.C.; Tang, Y.L.; Pan, Y.; Gao, N.Y. Enhanced inactivation of E. coli by pulsed UV-LED irradiation during water disinfection. Sci. Total Environ. 2019, 650, 210–215. [Google Scholar] [CrossRef]
- Xiao, Y.; Chu, X.N.; He, M.; Liu, X.C.; Hu, J.Y. Impact of UVA pre-radiation on UVC disinfection performance: Inactivation repair and mechanism study. Water Res. 2018, 141, 279–288. [Google Scholar] [CrossRef]
- Darnell, M.E.R.; Subbarao, K.; Feinstone, S.M.; Taylor, D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods 2004, 121, 85–91. [Google Scholar] [CrossRef]
- Bolton, J.R.; Linden, K.G. Standardization of Methods for Fluence (UV Dosage) Determination in Bench-Scale UV Experiments. J. Environ. Eng. 2003, 129, 209–215. [Google Scholar] [CrossRef]
- Aubert, A.C.B. Inventor: Waterdown; Combination Ultra-Violet A (UVA) and Ultra-Violet C (UVC) System for Reduction and Inhibition of Growth of Pathogens. U.S. Patent US20210275705, 9 September 2020. [Google Scholar]
- Aubert, A.C.B. Inventor: Waterdown (Ontario); Ultra-Violet A (UVA) and Ultra-Violet C (UVC) System and Methods for Inactivation, Reduction and Inhibition of Growth of Coronavirus. U.S. Patent US20230063654, 2 March 2023. [Google Scholar]
- Camargo, C.; Lupien, A.; McIntosh, F.; Menzies, D.; Behr, M.A.; Sagan, S.M. Effectiveness of germicidal ultraviolet light to inactivate coronaviruses on personal protective equipment to reduce nosocomial transmission. Infect. Control Hosp. Epidemiol. 2022, 43, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Yang, J.; Hu, J.; Sun, X. On the Calculation of TCID50 for Quantitation of Virus Infectivity. Virol. Sin. 2021, 36, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Bartrand, T.A.; Weir, M.H.; Omura, T.; Haas, C.N. Development of a dose-response model for SARS coronavirus. Risk Anal. 2010, 30, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- DuPont, H.L.; Formal, S.B.; Hornick, R.B.; Snyder, M.J.; Libonati, J.P.; Sheahan, D.G.; LaBrec, E.H.; Kalas, J.P. Pathogenesis of Escherichia coli diarrhea. N. Engl. J. Med. 1971, 285, 1–9. [Google Scholar] [CrossRef]
- Sinha, R.P.; Häder, D.-P. UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 2002, 1, 225–236. [Google Scholar] [CrossRef]
- Hadi, J.; Dunowska, M.; Wu, S.; Brightwell, G. Control Measures for SARS-CoV-2: A Review on Light-Based Inactivation of Single-Stranded RNA Viruses. Pathogens 2020, 9, 737. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: London, UK, 2021; Volume 2, pp. 428–440. [Google Scholar]
- Dijkman, R.; Jebbink, M.F.; Gaunt, E.; Gaunt, J.W.A.; Templeton, K.E.; Kuijpers, T.W.; van der Hoek, L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012, 53, 135–139. [Google Scholar] [CrossRef]
- Villamil-Gómez, W.E.; Sánchez, Á.; Gelis, L.; Silvera, L.A.; Barbosa, J.; Otero-Nader, O.; Bonilla-Salgado, C.D.; Rodríguez-Morales, A.J. Fatal human coronavirus 229E (HCoV-229E) and RSV–Related pneumonia in an AIDS patient from Colombia. Travel. Med. Infect. Dis. 2020, 36, 101573. [Google Scholar] [CrossRef]
- Warnes, S.L.; Little, Z.R.; Keevil, W.C. Human Coronavirus 229E Remains Infectious on Common Touch Surface Materials. ASM 2015, 6, e01697-15. [Google Scholar] [CrossRef]
- Aboubakr, H.A.; Sharafeldin, T.A.; Goyal, S.M. Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the influence of climatic conditions: A review. Transbound. Emerg. Dis. 2021, 68, 296–312. [Google Scholar] [CrossRef]
- Buonanno, M.; Welch, D.; Shuryak, I.; Brenner, J.D. Far-UVC light (222 nm) efficiently and safely inactivates airborne human coronaviruses. Sci. Rep. 2020, 10, 10285. [Google Scholar] [CrossRef]
- Heilingloh, C.S.; Aufderhost, U.W.; Schipper, L.; Dittmer, U.; Witzke, O.; Yang, D.; Zheng, X.; Sutter, K.; Trilling, M.; Alt, M.; et al. Susceptibility of SARS-CoV-2 to UV irradiation. Am. J. Infect. Control 2020, 48, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, H.; Nimura, T.; Nazmui, T.; Shigemoto, N.; Sakaguchi, T.; Ohge, H. Effectiveness of 222-nm ultraviolet light on disinfecting SARS-CoV-2 surface contamination. Am. J. Infect. Control 2021, 49, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Bueckert, M.; Gupta, R.; Gupta, A.; Garg, M.; Mazumder, A. Infectivity of SARS-CoV-2 and Other Coronaviruses on Dry Surfaces: Potential for Indirect Transmission. Materials 2020, 13, 5211. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, A.; Leite, G.G.S.; Melmed, G.Y.; Mathur, R.; Villanueva-Millan, M.J.; Parodi, G.; Sin, J.; Germano, J.F.; Morales, W.; Weitsman, S.; et al. Ultraviolet A light effectively reduces bacteria and viruses including coronavirus. PLoS ONE 2020, 15, e023619. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Van de Poel, B.; De Coninc, B. UV-B light and its application potential to reduce disease and pest incidence in crops. Hortic. Res. 2021, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Koutchma, T. Advances in Ultraviolet Light Technology for Non-thermal Processing of Liquid Foods. Food Bioprocess Technol. 2009, 2, 138–155. [Google Scholar] [CrossRef]
- Moore, G.; Ali, S.; Cloutman-Green, E.A.; Bradley, C.R.; Wilkinson, M.A.; Hartley, J.C.; Fraise, A.P.; Wilson, A.P. Use of UV-C radiation to disinfect non-critical patient care items: A laboratory assessment of the Nanoclave Cabinet. BMC Infect. Dis. 2012, 12, 174. [Google Scholar] [CrossRef]
- Veerachandra, Y.; Achyut, A.; Juan, M. Effect of ultraviolet light treatment on microbiological safety and quality of fresh produce: An overview. Front. Nutr. 2022, 9, 871243. [Google Scholar]
- Khan, M.; McDonald, M.; Mundada, K.; Willcox, M. Efficacy of Ultraviolet Radiations against Coronavirus, Bacteria, Fungi, Fungal Spores and Biofilm. Hygiene 2022, 2, 120–131. [Google Scholar] [CrossRef]
- Kim, H.J.; Yoon, H.W.; Lee, M.A.; Kim, Y.H.; Lee, C.J. Impact of UV-C Irradiation on Bacterial Disinfection in a Drinking Water Purification System. J. Microbiol. Biotechnol. 2023, 33, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Sasani, G.M.; Alamdari, S.; Han, W.; Park, H. Impact of nanostructured thin ZnO film in ultraviolet protection. Int. J. Nanomed. 2017, 12, 207–216. [Google Scholar] [CrossRef] [PubMed]





| Strain | Setting for UVA | Setting for UVC | Time (h) | ATP (RLU) | ||
|---|---|---|---|---|---|---|
| Dark | UV Light | Natural Light | ||||
| B. subtilis | 65 mW | 65 mW | 12 | 2431 | 0 | 3893 |
| 117 mW | 117 mW | 12 | 7580 | 4 | 7937 | |
| 42 mW | 65 mW | 12 | 95 | 5 | 1062 | |
| E. coli K12 | 117 mW | 117 mW | 12 | 7004 | 0 | 6031 |
| 65 mW | 65 mW | 12 | 5389 | 0 | 7496 | |
| S. epidermidis | 117 mW | 117 mW | 12 | 7701 | 0 | 8811 |
| 65 mW | 65 mW | 12 | 8272 | 0 | 8899 | |
| 42 mW | 65 mW | 12 | 8297 | 0 | 3561 | |
| Strain | Conditions | Reduction in CFU |
|---|---|---|
| E. coli K12 | Natural light | 32 ± 3% |
| Dark | 35 ± 8% | |
| S. epidermidis | Natural light | 34 ± 5% |
| Dark | 36 ± 2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hunter, E.; Percival, B.; Eberl, D.T.; White, S.J. A Pilot Study to Investigate the Antimicrobial Activity of Pulsed UVA and UVC. Aerobiology 2023, 1, 82-97. https://doi.org/10.3390/aerobiology1020007
Hunter E, Percival B, Eberl DT, White SJ. A Pilot Study to Investigate the Antimicrobial Activity of Pulsed UVA and UVC. Aerobiology. 2023; 1(2):82-97. https://doi.org/10.3390/aerobiology1020007
Chicago/Turabian StyleHunter, Elena, Benita Percival, Daniela T. Eberl, and Samuel J. White. 2023. "A Pilot Study to Investigate the Antimicrobial Activity of Pulsed UVA and UVC" Aerobiology 1, no. 2: 82-97. https://doi.org/10.3390/aerobiology1020007
APA StyleHunter, E., Percival, B., Eberl, D. T., & White, S. J. (2023). A Pilot Study to Investigate the Antimicrobial Activity of Pulsed UVA and UVC. Aerobiology, 1(2), 82-97. https://doi.org/10.3390/aerobiology1020007





