Revolutionizing Epithelial Differentiability Analysis in Small Airway-on-a-Chip Models Using Label-Free Imaging and Computational Techniques
Abstract
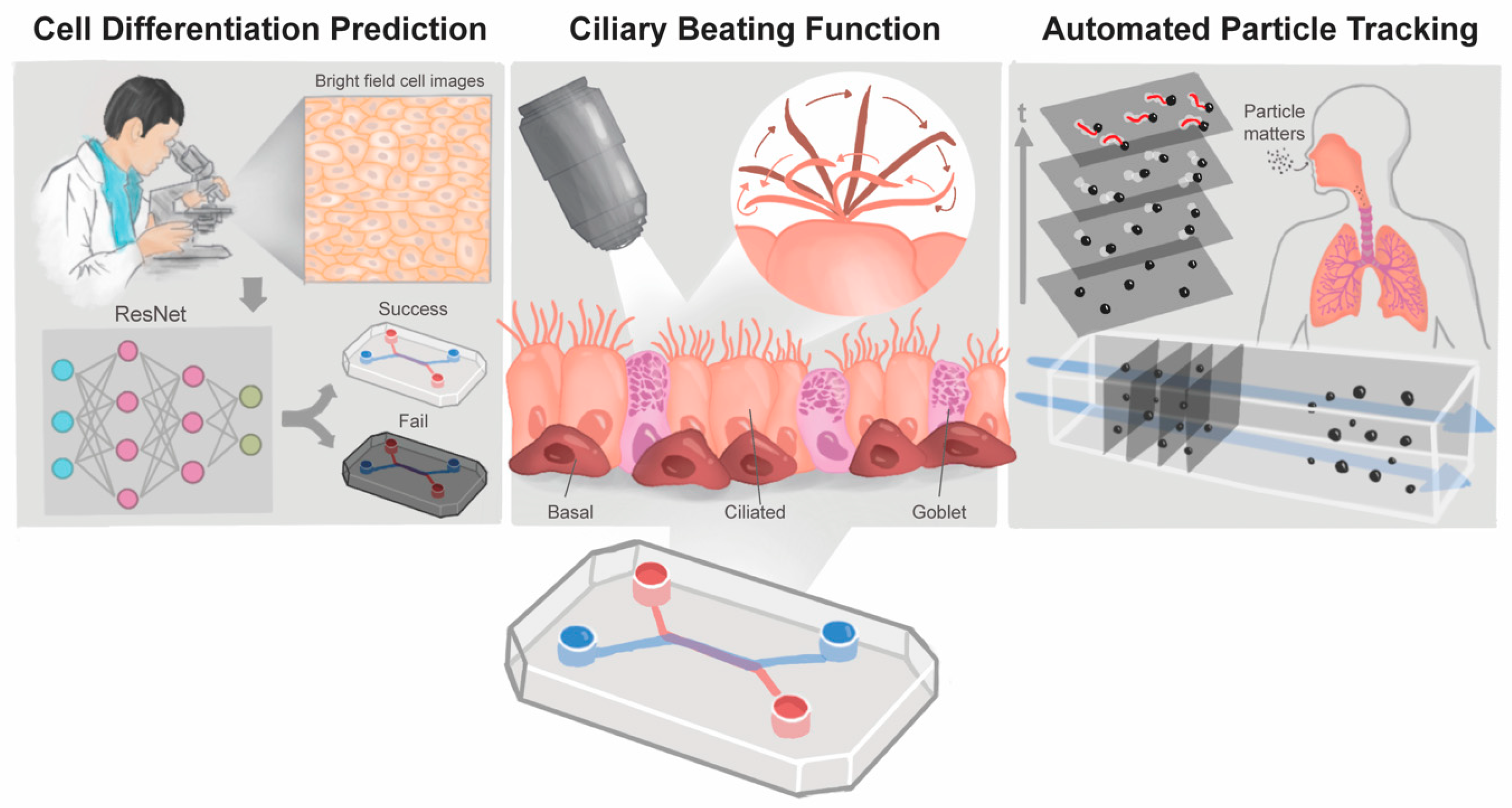
:1. Introduction
2. Materials and Methods
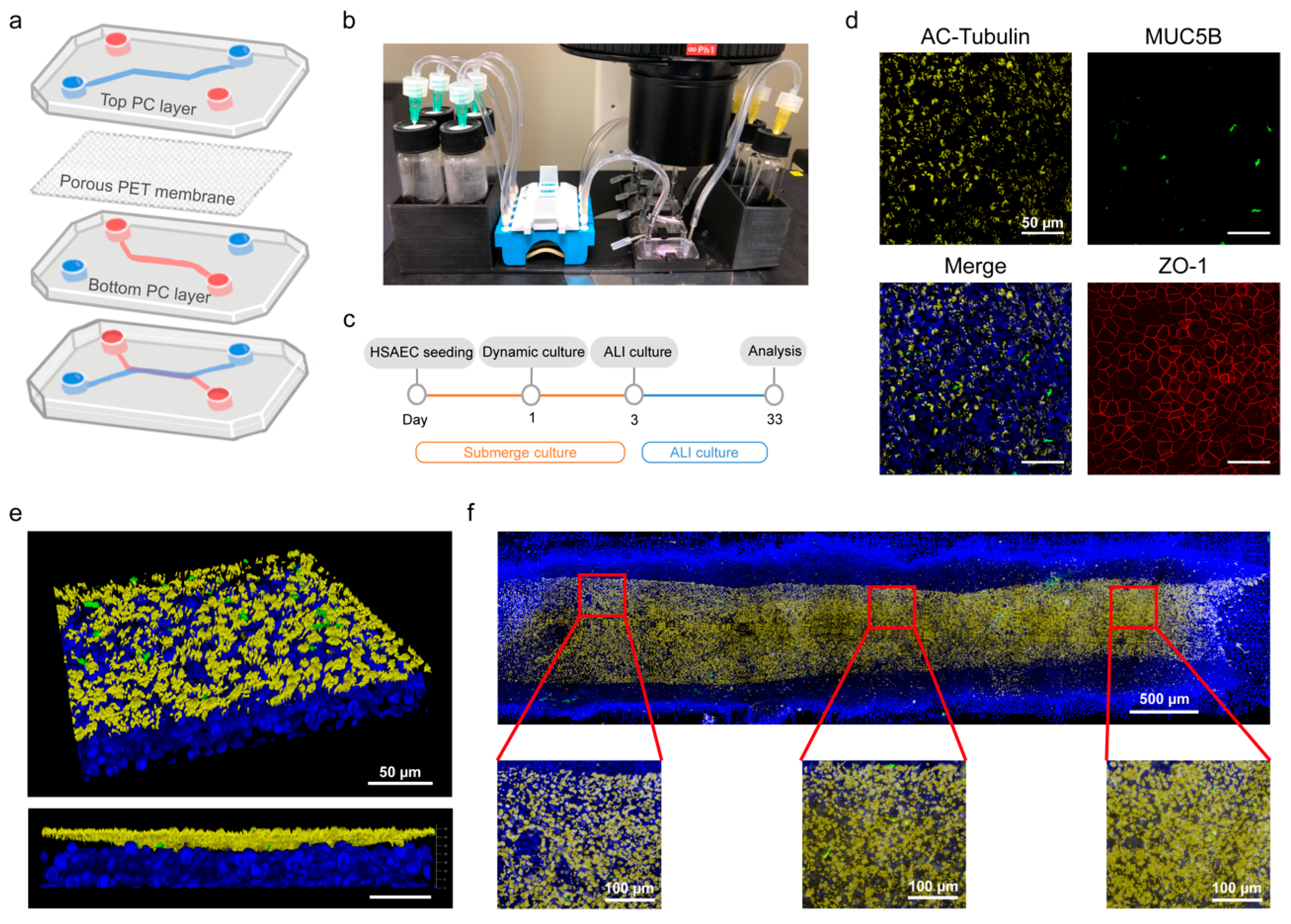
2.1. Chip Device Manufacturing
2.2. Small Airway-on-a-Chip Culture
2.3. Immunofluorescence Staining
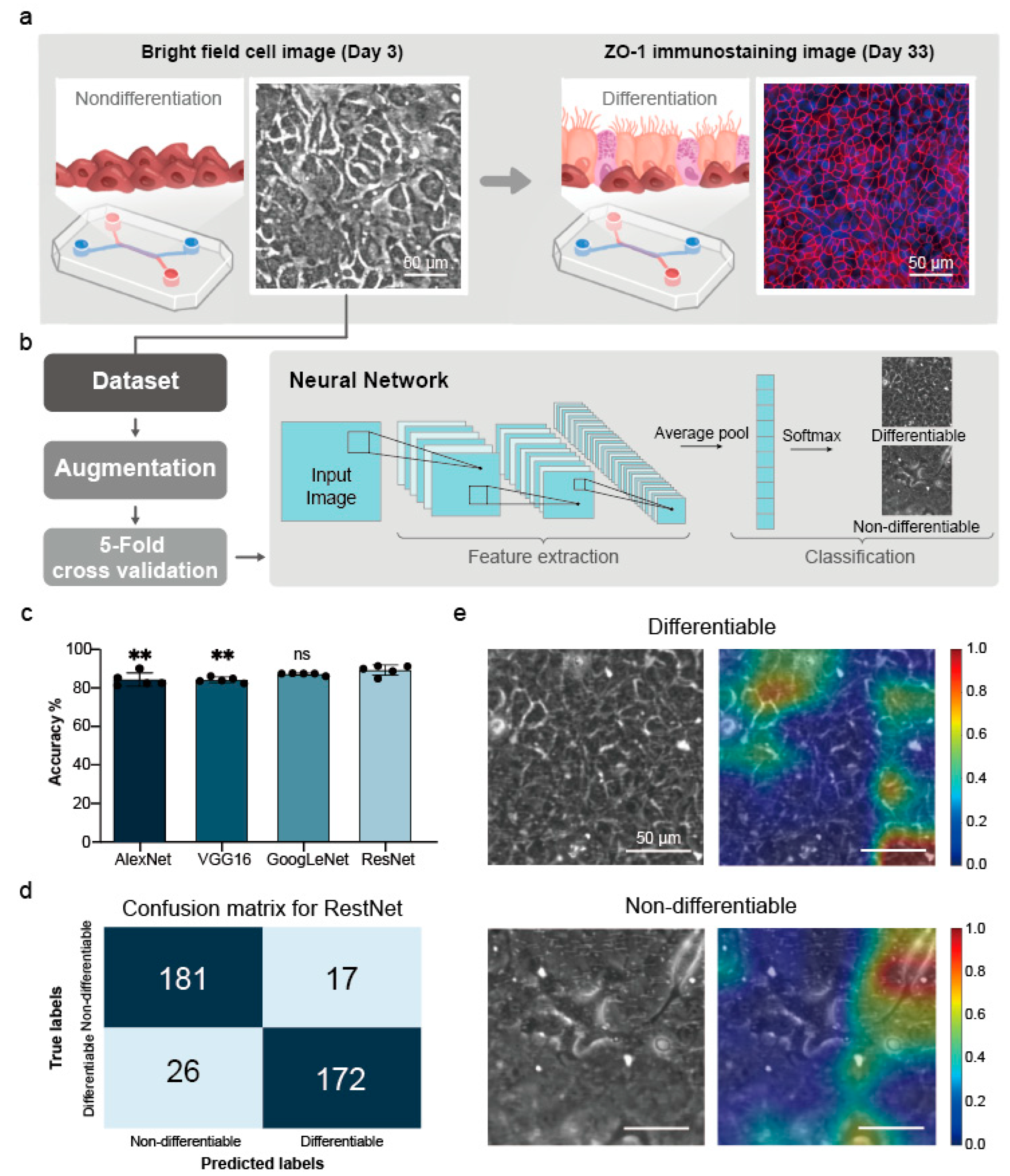
2.4. Image-Based Prediction of Cell Differentiation
2.4.1. HSAEC Dataset
2.4.2. Data Augmentation
2.4.3. Establishment and Validation of the Small Airway Survival Recognition Model
2.4.4. Feature Visualization
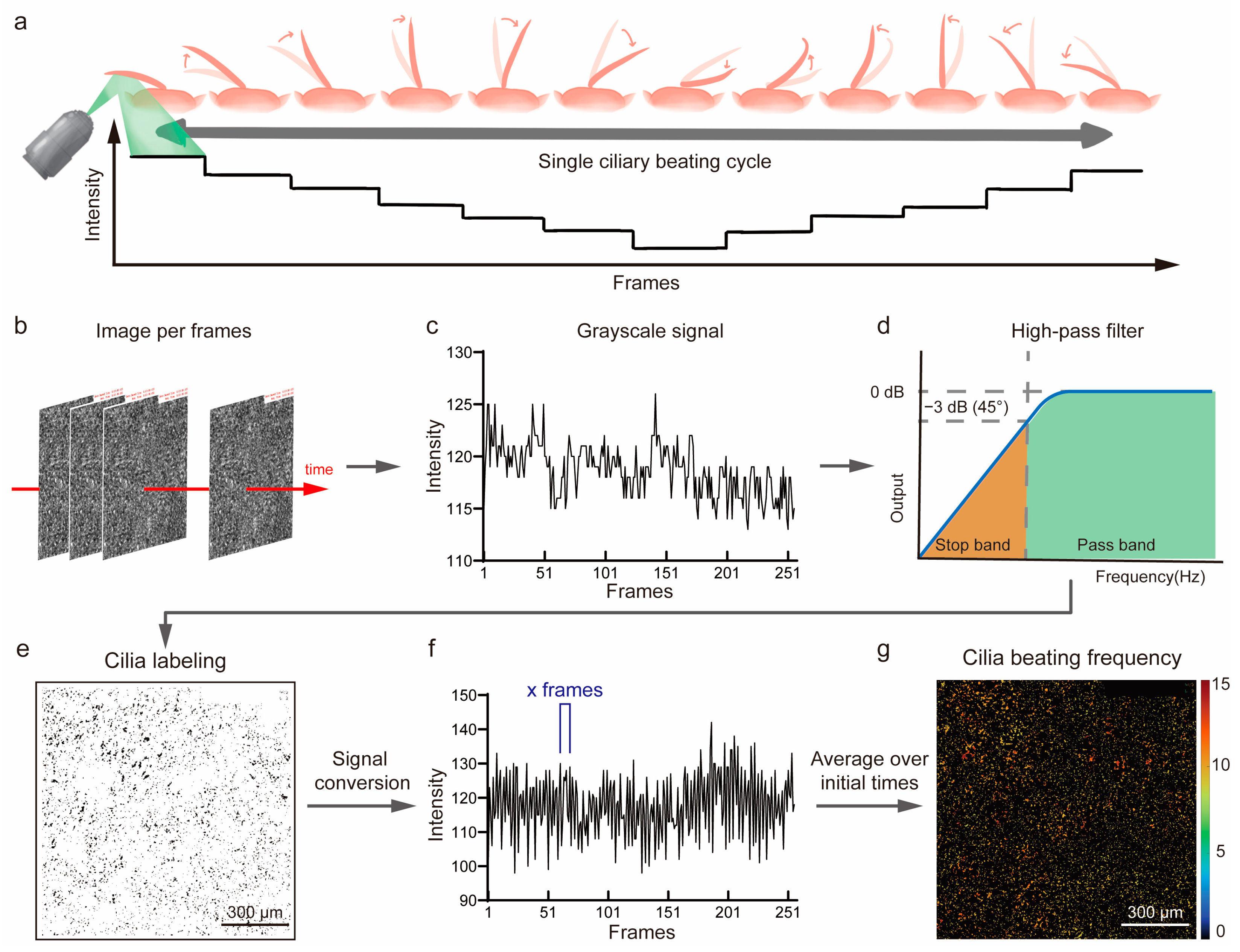
2.5. The Automated Ciliary Beating Frequency Calculating Model
2.6. Fluorescent Particles Tracking Analysis
2.7. Statistical Analysis
3. Results and Discussion
3.1. Mass Production of Polycarbonate(PC) Chips and Small Airway-on-a-Chip Differentiation
3.2. Predicting Differentiation Outcomes in Small Airway Epithelial Cells Using Deep Learning
3.2.1. Dataset Results
3.2.2. Data Augmentation Results
3.2.3. Five-Fold Cross-Validation Results
3.2.4. Feature Visualization Results
3.3. Analysis of Ciliary Beat Images
3.4. Automated Fluorescent Particle Tracking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Radisic, M.; Loskill, P. Beyond Pdms and Membranes: New Materials for Organ-on-a-Chip Devices. ACS Biomater. Sci. Eng. 2021, 7, 2861–2863. [Google Scholar] [CrossRef] [PubMed]
- Bennet, T.J.; Randhawa, A.; Hua, J.; Cheung, K.C. Airway-on-a-chip: Designs and Applications for Lung Repair and Disease. Cells 2021, 10, 1602. [Google Scholar] [CrossRef]
- Sakolish, C.; Georgescu, A.; Huh, D.D.; Rusyn, I. A Model of Human Small Airway on a Chip for Studies of Subacute Effects of Inhalation Toxicants. Toxicol. Sci. 2022, 187, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Bai, H.; Rodas, M.; Cao, W.; Oh, C.Y.; Jiang, A.; Moller, R.; Hoagland, D.; Oishi, K.; Horiuchi, S. A Human-Airway-on-a-Chip for the Rapid Identification of Candidate Antiviral Therapeutics and Prophylactics. Nat. Biomed. Eng. 2021, 5, 815–829. [Google Scholar] [CrossRef]
- Fuchs, S.; Johansson, S.; Tjell, A.; Werr, G.; Mayr, T.; Tenje, M. In-Line Analysis of Organ-on-Chip Systems with Sensors: Integration, Fabrication, Challenges, and Potential. ACS Biomater. Sci. Eng. 2021, 7, 2926–2948. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.; Bai, H.; Wang, H.; Hao, S.; Ding, Y.; Peng, B.; Zhang, J.; Li, L.; Huang, W. An Overview of Organs-on-Chips Based on Deep Learning. Research 2022, 2022, 9869518. [Google Scholar] [CrossRef] [PubMed]
- Renò, V.; Sciancalepore, M.; Dimauro, G.; Maglietta, R.; Cassano, M.; Gelardi, M. A Novel Approach for the Automatic Estimation of the Ciliated Cell Beating Frequency. Electronics 2020, 9, 1002. [Google Scholar] [CrossRef]
- Tratnjek, L.; Kreft, M.; Kristan, K.; Kreft, M.E. Ciliary Beat Frequency of in Vitro Human Nasal Epithelium Measured with the Simple High-Speed Microscopy Is Applicable for Safety Studies of Nasal Drug Formulations. Toxicol Vitr. 2020, 66, 104865. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sarwinda, D.; Paradisa, R.H.; Bustamam, A.; Anggia, P. Deep Learning in Image Classification Using Residual Network (Resnet) Variants for Detection of Colorectal Cancer. Procedia Comput. Sci. 2021, 179, 423–431. [Google Scholar] [CrossRef]
- Tian, Y. Artificial Intelligence Image Recognition Method Based on Convolutional Neural Network Algorithm. IEEE Access 2020, 8, 125731–125744. [Google Scholar] [CrossRef]
- Lin, K.C.; Yang, J.W.; Ho, P.Y.; Yen, C.Z.; Huang, H.W.; Lin, H.Y.; Chung, J.; Chen, G.Y. Development of an Alveolar Chip Model to Mimic Respiratory Conditions due to Fine Particulate Matter Exposure. Appl. Mater. Today 2022, 26, 101281. [Google Scholar] [CrossRef]
- Baldassi, D.; Gabold, B.; Merkel, O.M. Air− Liquid Interface Cultures of the Healthy and Diseased Human Respiratory Tract: Promises, Challenges, and Future Directions. Adv. Nanobiomed Res. 2021, 1, 2000111. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-Weighted Visual Explanations for Convolutional Neural Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 24–25. [Google Scholar]
- Zhang, B.; Radisic, M. Organ-on-a-Chip Devices Advance to Market. Lab A Chip 2017, 17, 2395–2420. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Chou, H.-C.; Lin, K.-C.; Yang, J.-W.; Xie, R.-H.; Chen, C.-Y.; Liu, X.-Y.; Chung, J.H.Y.; Chen, G.-Y. Investigation of the Role of the Autophagic Protein Lc3b in the Regulation of Human Airway Epithelium Cell Differentiation in Copd Using a Biomimetic Model. Mater. Today Bio 2022, 13, 100182. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, R.; Laurent, V.; Roquefort, P.; Haute, T.; Ramel, S.; Le Gall, T.; Aubry, T.; Montier, T. Optimizations of In Vitro Mucus and Cell Culture Models to Better Predict In Vivo Gene Transfer in Pathological Lung Respiratory Airways: Cystic Fibrosis as an Example. Pharmaceutics 2020, 13, 47. [Google Scholar] [CrossRef]
- Hawkins, F.J.; Suzuki, S.; Beermann, M.L.; Barillà, C.; Wang, R.; Villacorta-Martin, C.; Berical, A.; Jean, J.; Le Suer, J.; Matte, T. Derivation of Airway Basal Stem Cells from Human Pluripotent Stem Cells. Cell Stem Cell 2021, 28, 79–95.e8. [Google Scholar] [CrossRef]
- Leung, C.; Wadsworth, S.J.; Yang, S.J.; Dorscheid, D.R. Structural and Functional Variations in Human Bronchial Epithelial Cells Cultured in Air-Liquid Interface Using Different Growth Media. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 318, L1063–L1073. [Google Scholar] [CrossRef] [PubMed]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Abdollahi, B.; Tomita, N.; Hassanpour, S. Data Augmentation in Training Deep Learning Models for Medical Image Analysis. In Deep Learners and Deep Learner Descriptors for Medical Applications; Springer: Berlin/Heidelberg, Germany, 2020; pp. 167–180. [Google Scholar]
- Waisman, A.; La Greca, A.; Möbbs, A.M.; Scarafía, M.A.; Velazque, N.L.S.; Neiman, G.; Moro, L.N.; Luzzani, C.; Sevlever, G.E.; Guberman, A.S. Deep Learning Neural Networks Highly Predict very Early Onset of Pluripotent Stem Cell Differentiation. Stem Cell Rep. 2019, 12, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, R.; Wu, Z.; Song, S.; Cheng, L.; Zhu, R. Deep Learning-Based Predictive Identification of Neural Stem Cell Differentiation. Nat. Commun. 2021, 12, 2614. [Google Scholar] [CrossRef]
- Kojima, T.; Go, M.; Takano, K.; Kurose, M.; Ohkuni, T.; Koizumi, J.; Kamekura, R.; Ogasawara, N.; Masaki, T.; Fuchimoto, J.; et al. Regulation of Tight Junctions in Upper Airway Epithelium. Biomed Res. Int. 2013, 2013, 947072. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, R.; Lloyd, C.M.; Molyneaux, P.L. Respiratory Microbiome and Epithelial Interactions Shape Immunity in the Lungs. Immunology 2020, 160, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Nawroth, J.C.; van der Does, A.M.; Ryan, A.; Kanso, E. Multiscale Mechanics of Mucociliary Clearance in the Lung. Philos. Trans. R. Soc. B 2020, 375, 20190160. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.J.; Yoon, J.-K.; Paulson, B.; Lee, C.-H.; Yim, J.-J.; Kim, J.-I.; Kim, J.K. Image Correlation-Based Method to Assess Ciliary Beat Frequency in Human Airway Organoids. IEEE Trans. Med. Imaging 2021, 41, 374–382. [Google Scholar] [CrossRef]
- Huck, B.; Murgia, X.; Frisch, S.; Hittinger, M.; Hidalgo, A.; Loretz, B.; Lehr, C.-M. Models Using Native Tracheobronchial Mucus in the Context of Pulmonary Drug Delivery Research: Composition, Structure and Barrier Properties. Adv. Drug Deliv. Rev. 2022, 183, 114141. [Google Scholar] [CrossRef]
- Khelloufi, M.-K.; Loiseau, E.; Jaeger, M.; Molinari, N.; Chanez, P.; Gras, D.; Viallat, A. Spatiotemporal Organization of Cilia Drives Multiscale Mucus Swirls in Model Human Bronchial Epithelium. Sci. Rep. 2018, 8, 2447. [Google Scholar] [CrossRef]
- Robinot, R.; Hubert, M.; de Melo, G.D.; Lazarini, F.; Bruel, T.; Smith, N.; Levallois, S.; Larrous, F.; Fernandes, J.; Gellenoncourt, S. SARS-CoV-2 Infection Induces the Dedifferentiation of Multiciliated Cells and Impairs Mucociliary Clearance. Nat. Commun. 2021, 12, 4354. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.-L.; Xie, R.-H.; Chen, C.-Y.; Yang, J.-W.; Hsieh, K.-Y.; Liu, X.-Y.; Xin, J.-Y.; Kung, C.-K.; Chung, J.H.Y.; Chen, G.-Y. Revolutionizing Epithelial Differentiability Analysis in Small Airway-on-a-Chip Models Using Label-Free Imaging and Computational Techniques. Biosensors 2024, 14, 581. https://doi.org/10.3390/bios14120581
Chen S-L, Xie R-H, Chen C-Y, Yang J-W, Hsieh K-Y, Liu X-Y, Xin J-Y, Kung C-K, Chung JHY, Chen G-Y. Revolutionizing Epithelial Differentiability Analysis in Small Airway-on-a-Chip Models Using Label-Free Imaging and Computational Techniques. Biosensors. 2024; 14(12):581. https://doi.org/10.3390/bios14120581
Chicago/Turabian StyleChen, Shiue-Luen, Ren-Hao Xie, Chong-You Chen, Jia-Wei Yang, Kuan-Yu Hsieh, Xin-Yi Liu, Jia-Yi Xin, Ching-Kai Kung, Johnson H. Y. Chung, and Guan-Yu Chen. 2024. "Revolutionizing Epithelial Differentiability Analysis in Small Airway-on-a-Chip Models Using Label-Free Imaging and Computational Techniques" Biosensors 14, no. 12: 581. https://doi.org/10.3390/bios14120581
APA StyleChen, S.-L., Xie, R.-H., Chen, C.-Y., Yang, J.-W., Hsieh, K.-Y., Liu, X.-Y., Xin, J.-Y., Kung, C.-K., Chung, J. H. Y., & Chen, G.-Y. (2024). Revolutionizing Epithelial Differentiability Analysis in Small Airway-on-a-Chip Models Using Label-Free Imaging and Computational Techniques. Biosensors, 14(12), 581. https://doi.org/10.3390/bios14120581




