G-Quadruplexes as An Alternative Recognition Element in Disease-Related Target Sensing
Abstract
:1. Introduction
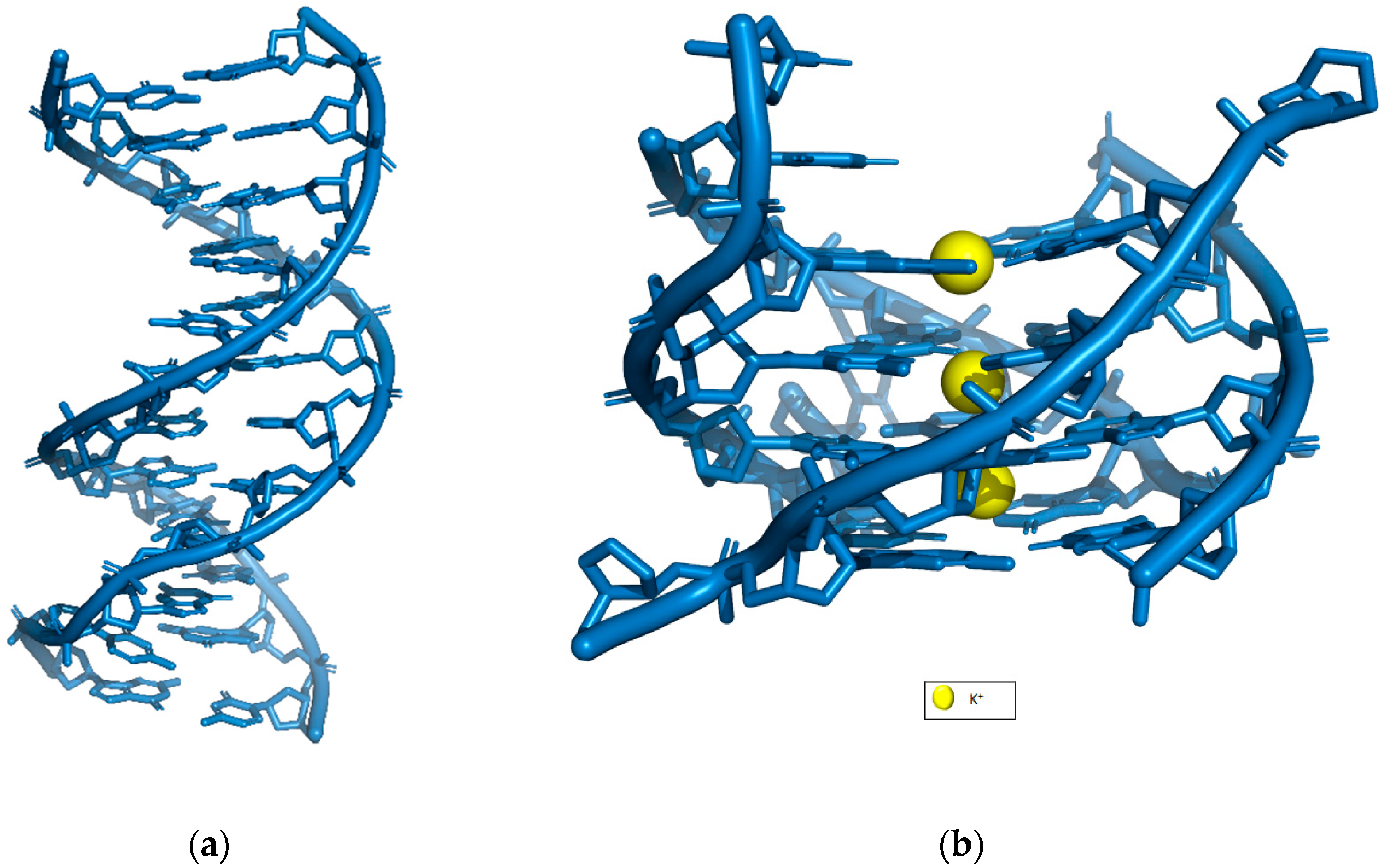
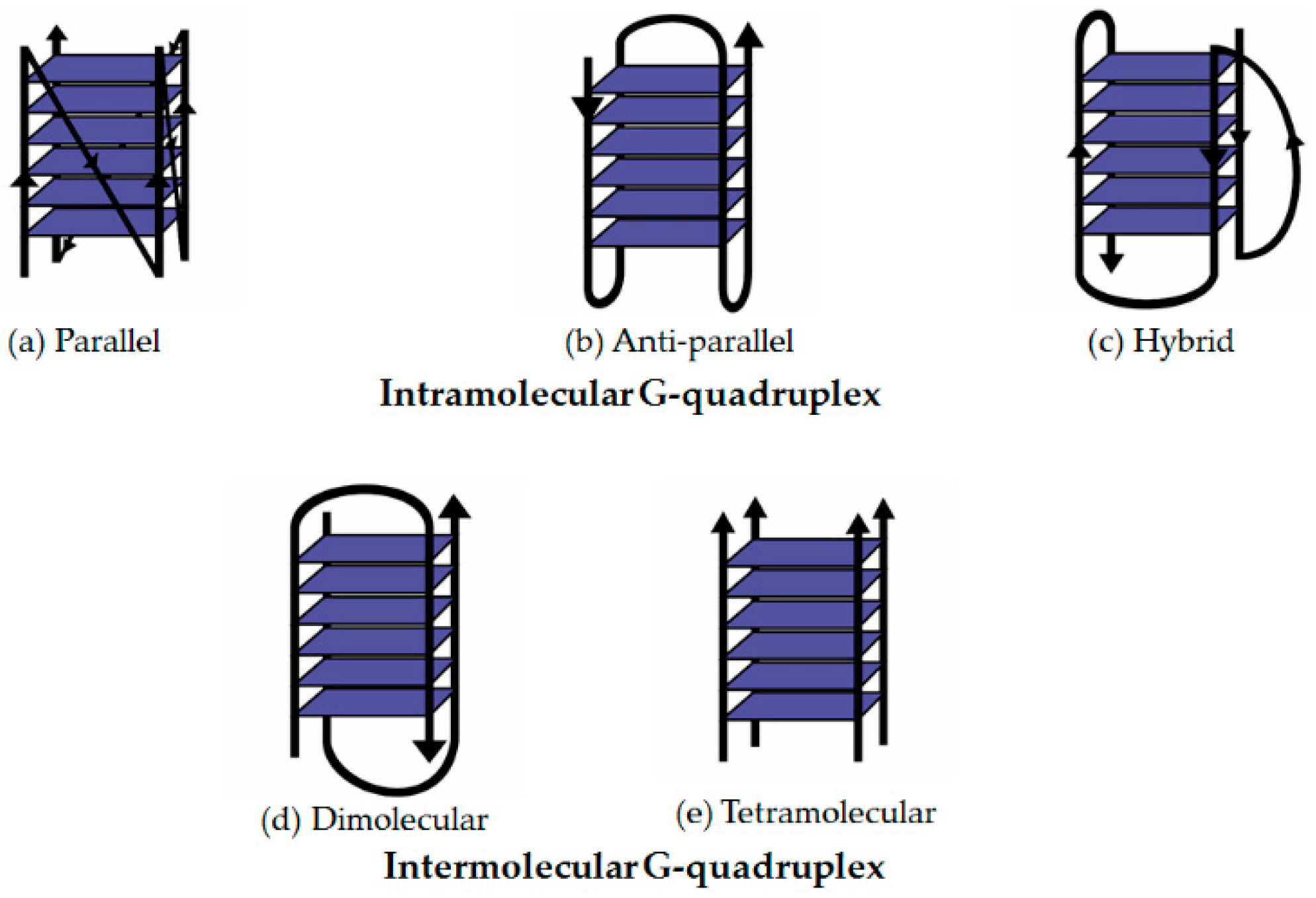
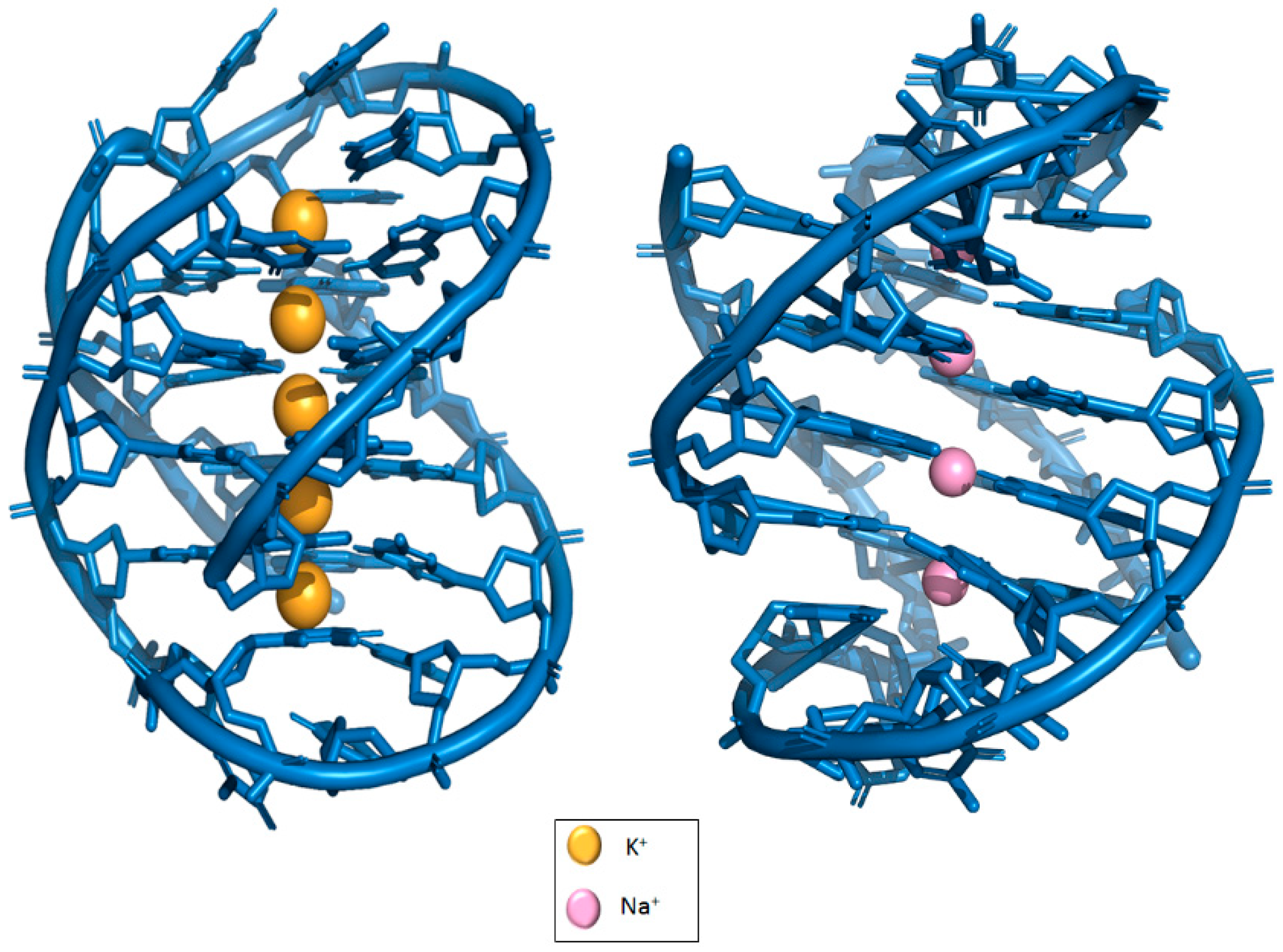
2. The Basic Structure of G-Quadruplex
3. G-Quadruplex-Based Detection System
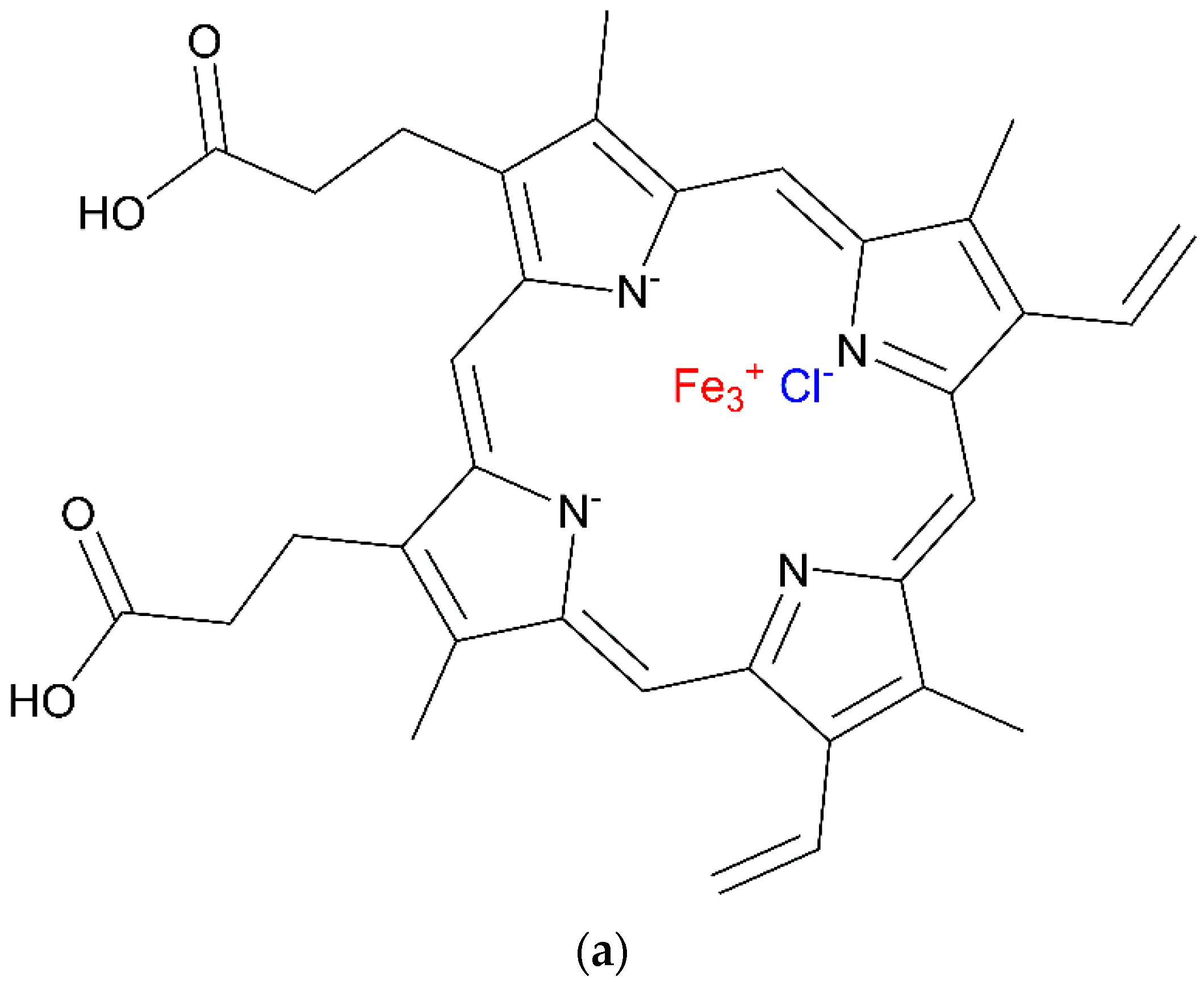
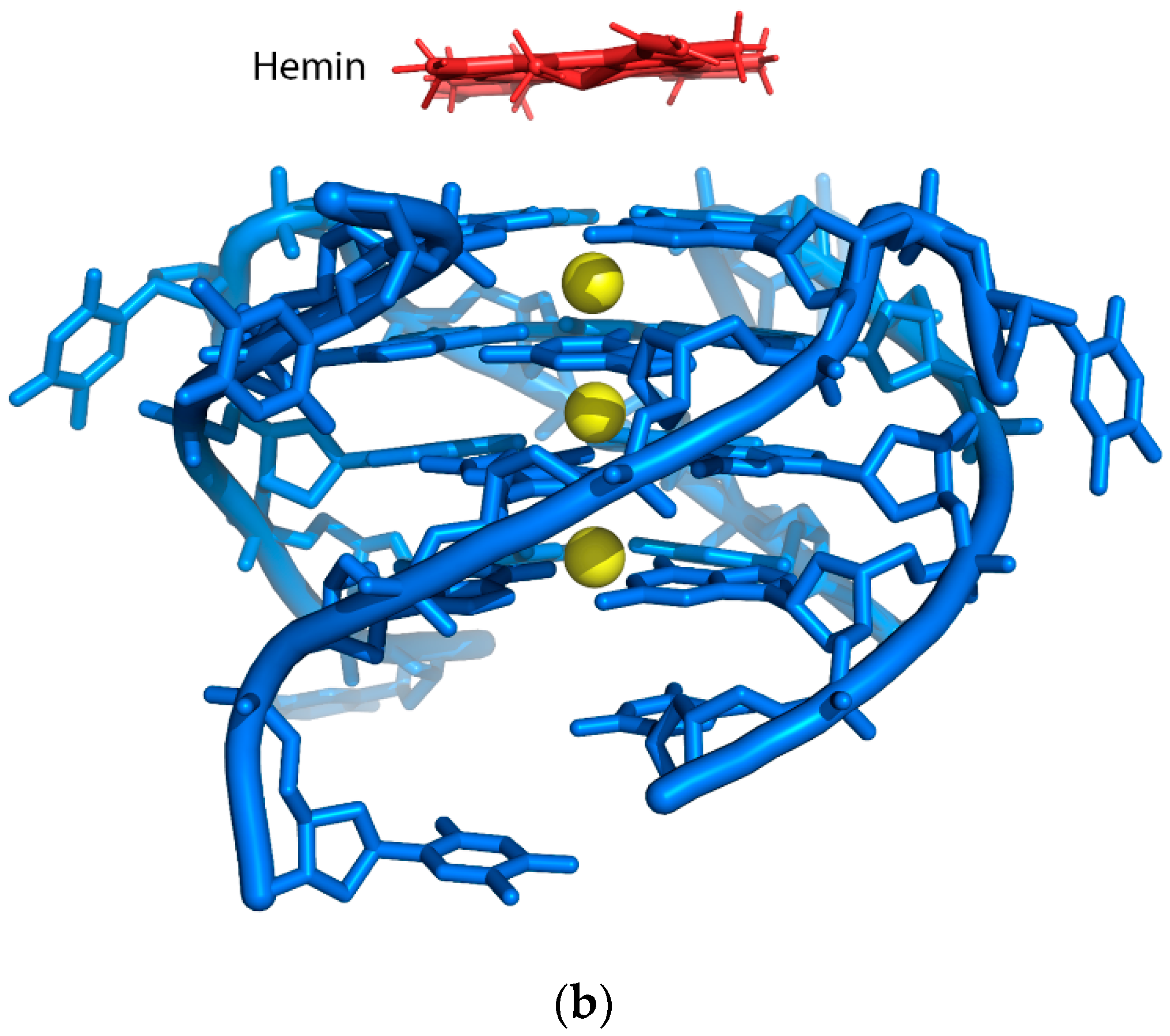
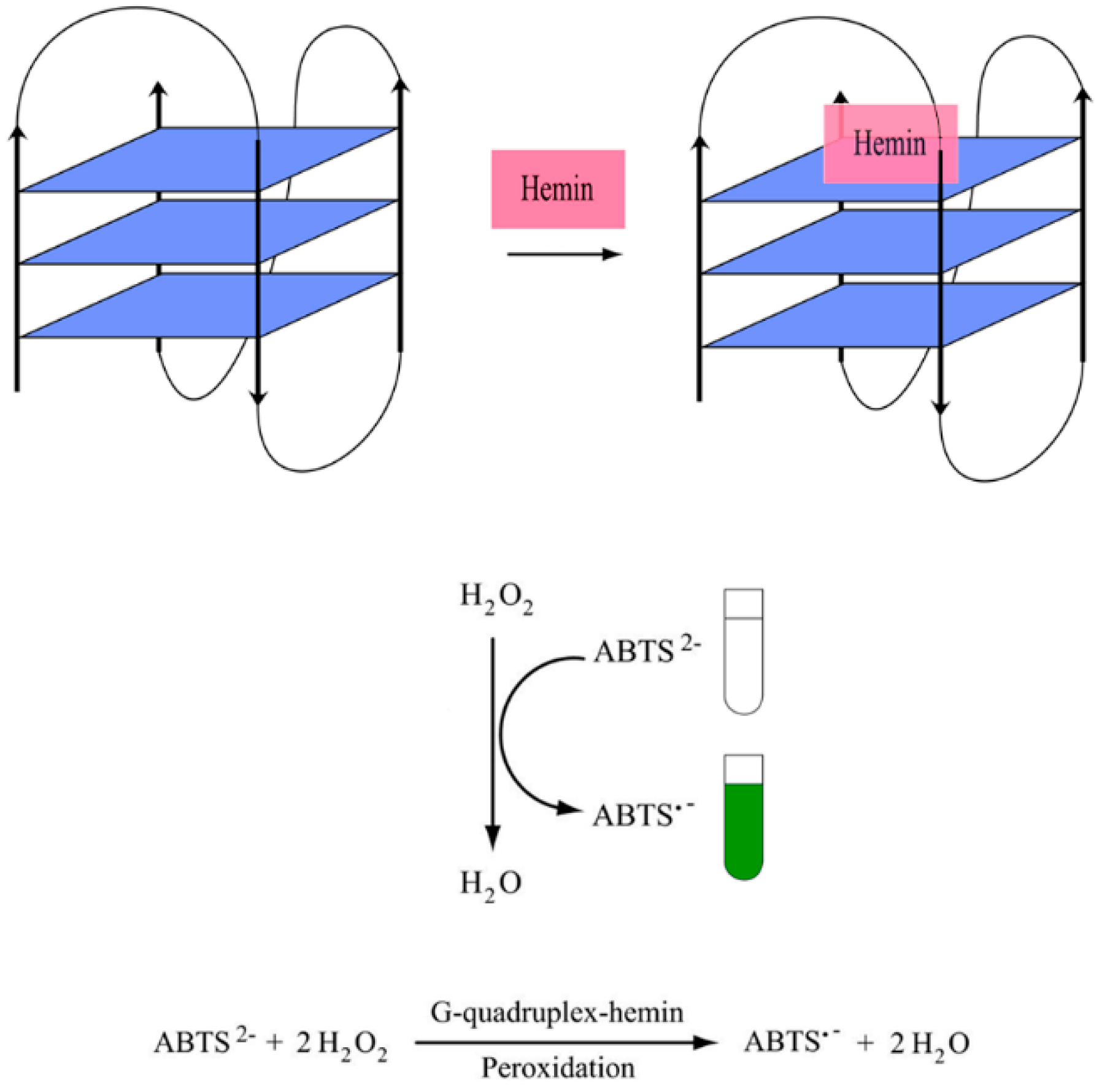
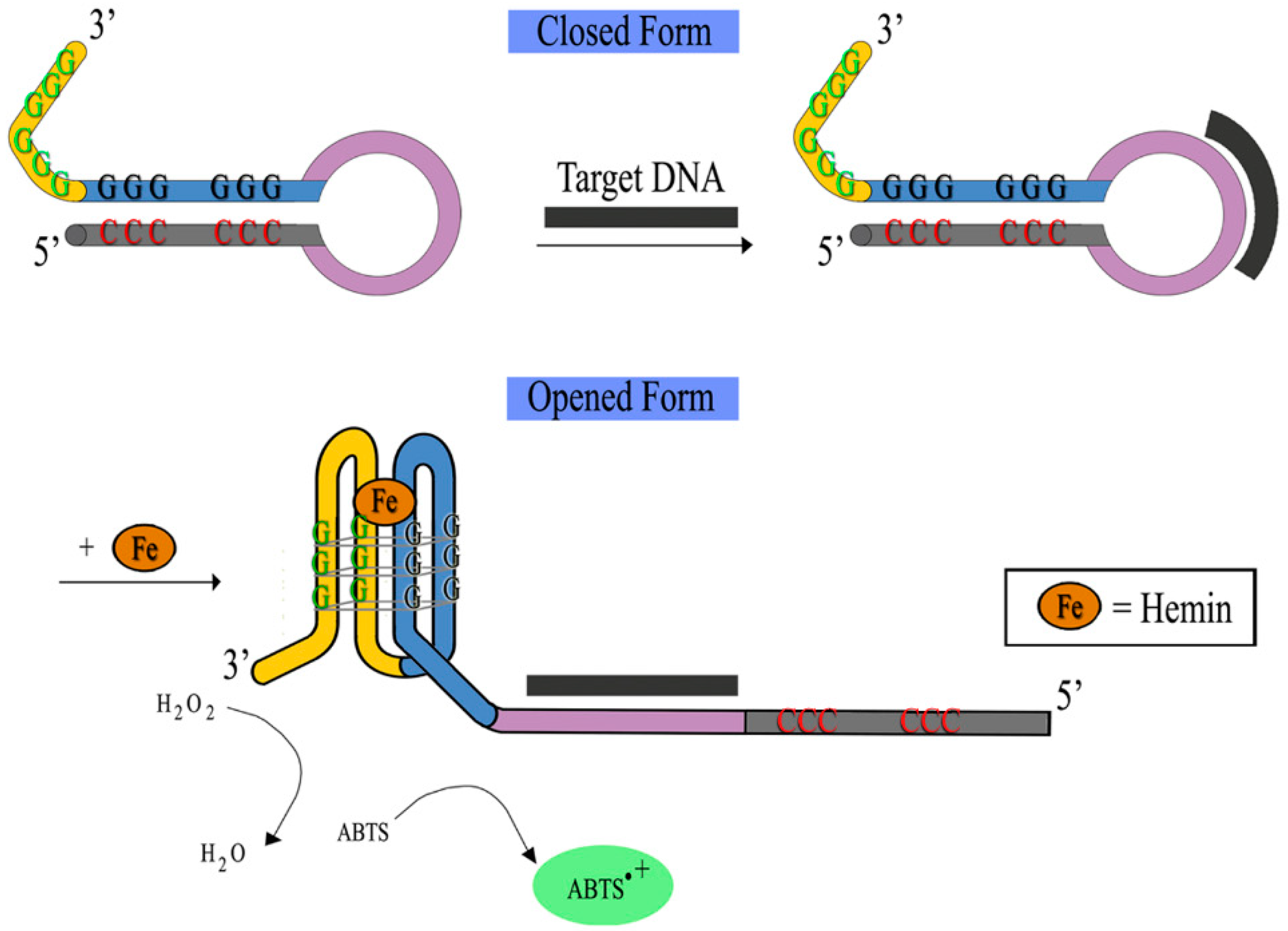
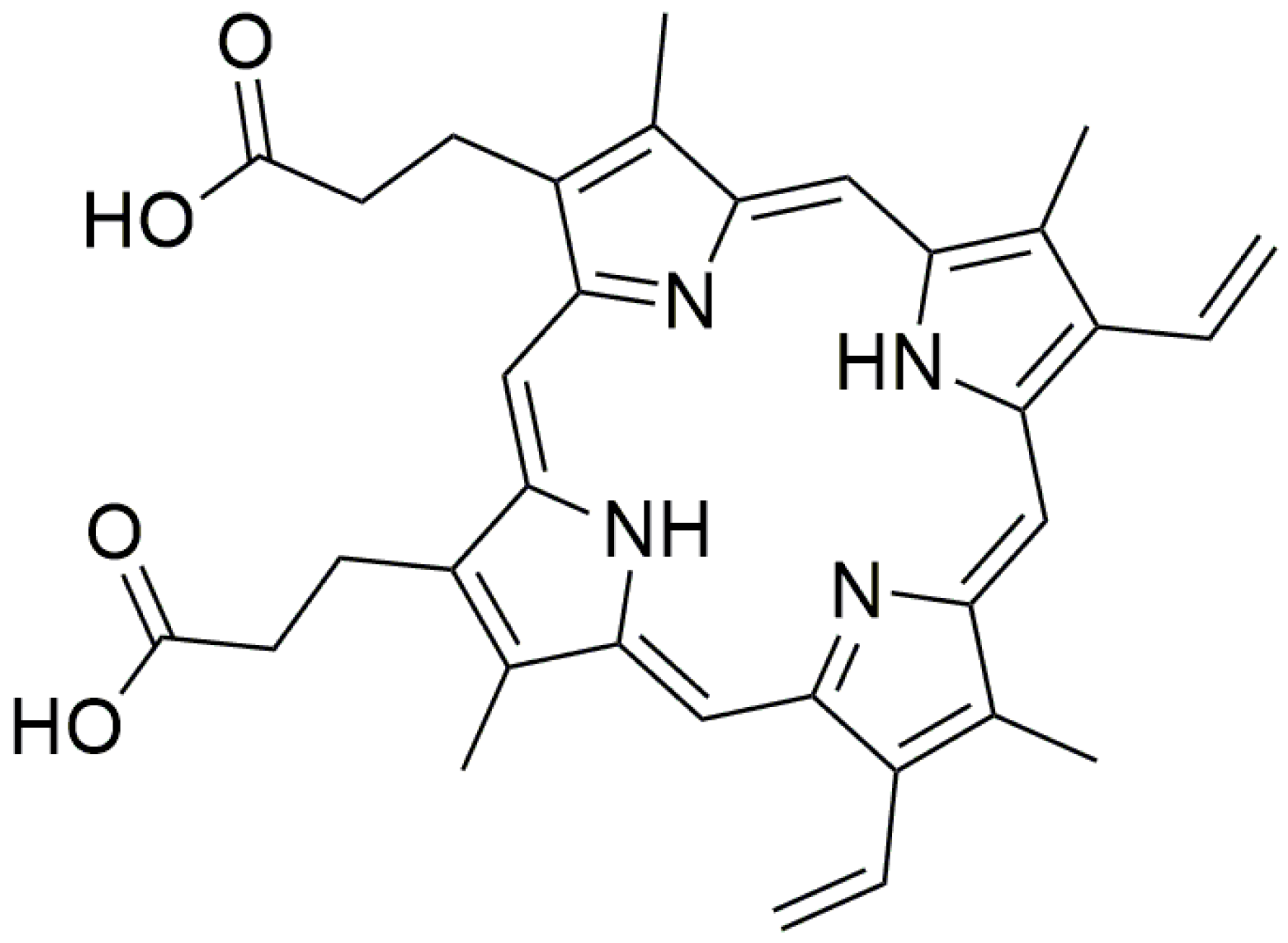
3.1. DNAzyme-Based Colorimetric G-Quadruplex/Hemin System
3.1.1. Conventional DNAzyme-Based G-Quadruplex/Hemin System
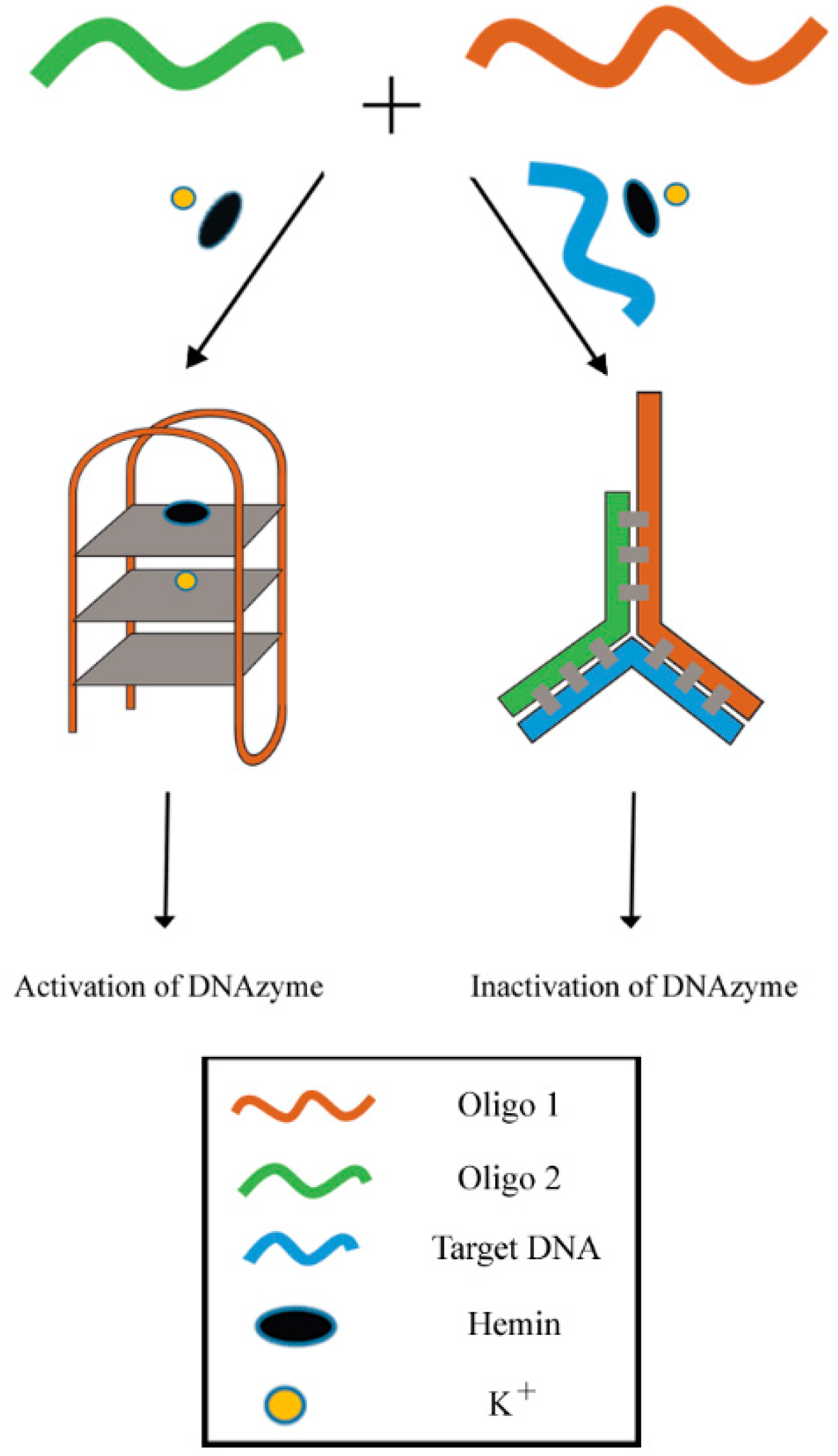
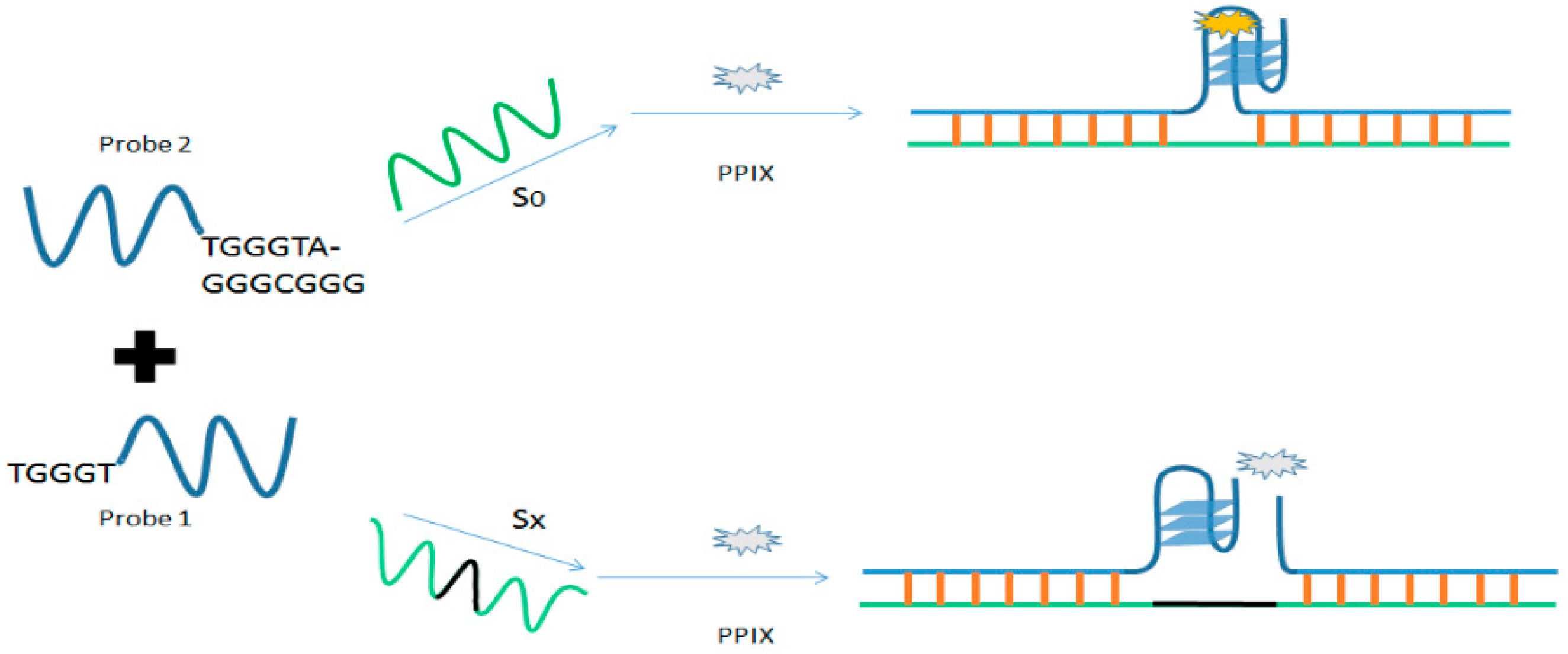
3.1.2. DNAzyme-Based Colorimetric Split G-Quadruplex System
3.1.3. DNAzyme-Based Colorimetric G-Quadruplex/Hemin Molecular Beacon System
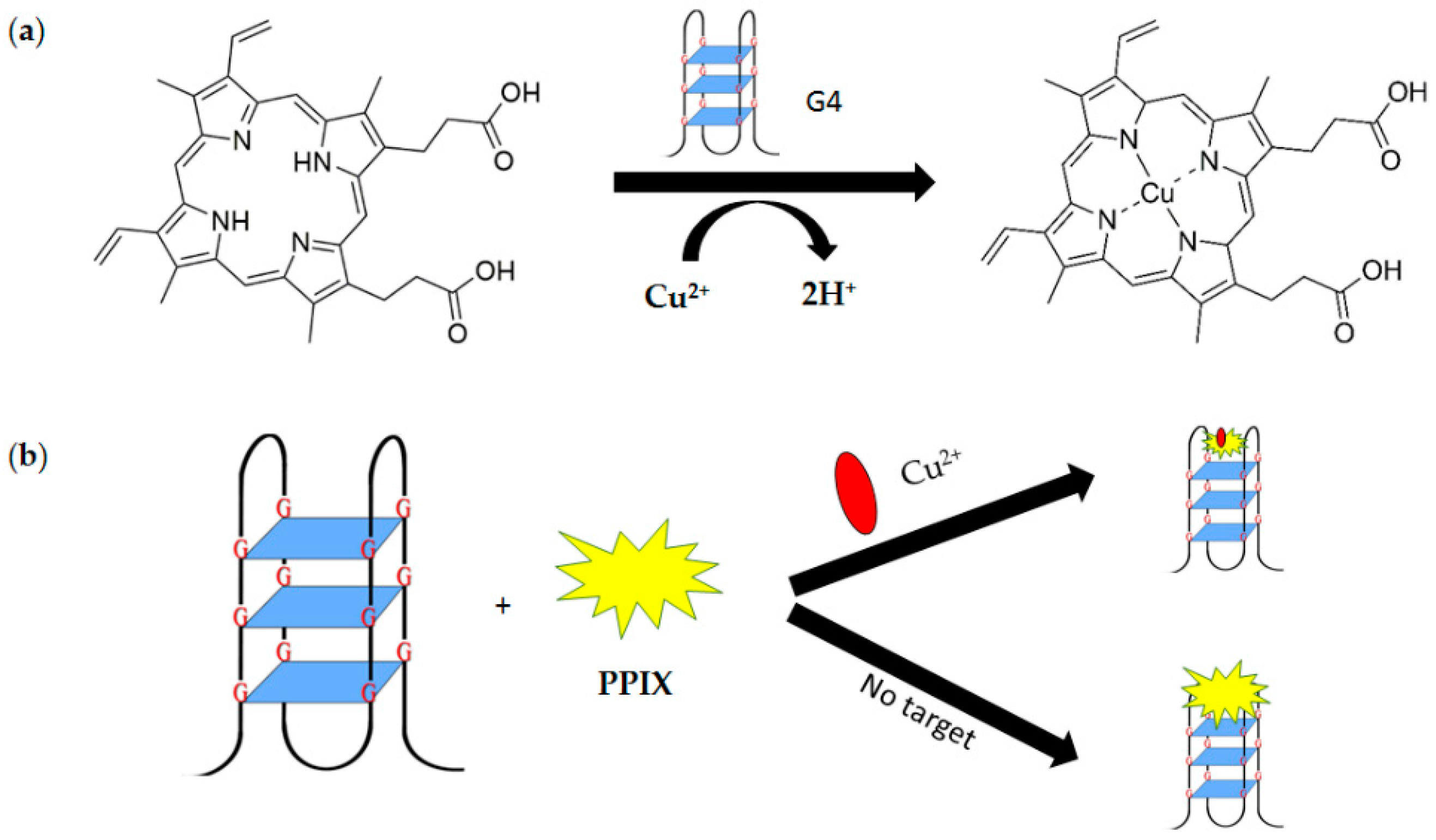
3.2. Non-Covalent Fluorescence-Based G-Quadruplex System
3.2.1. Conventional Non-Covalent Fluorescence-Based G-Quadruplex Sensing System
3.2.2. Non-Covalent Fluorescence Aptamer-Based G-Quadruplex Sensing System
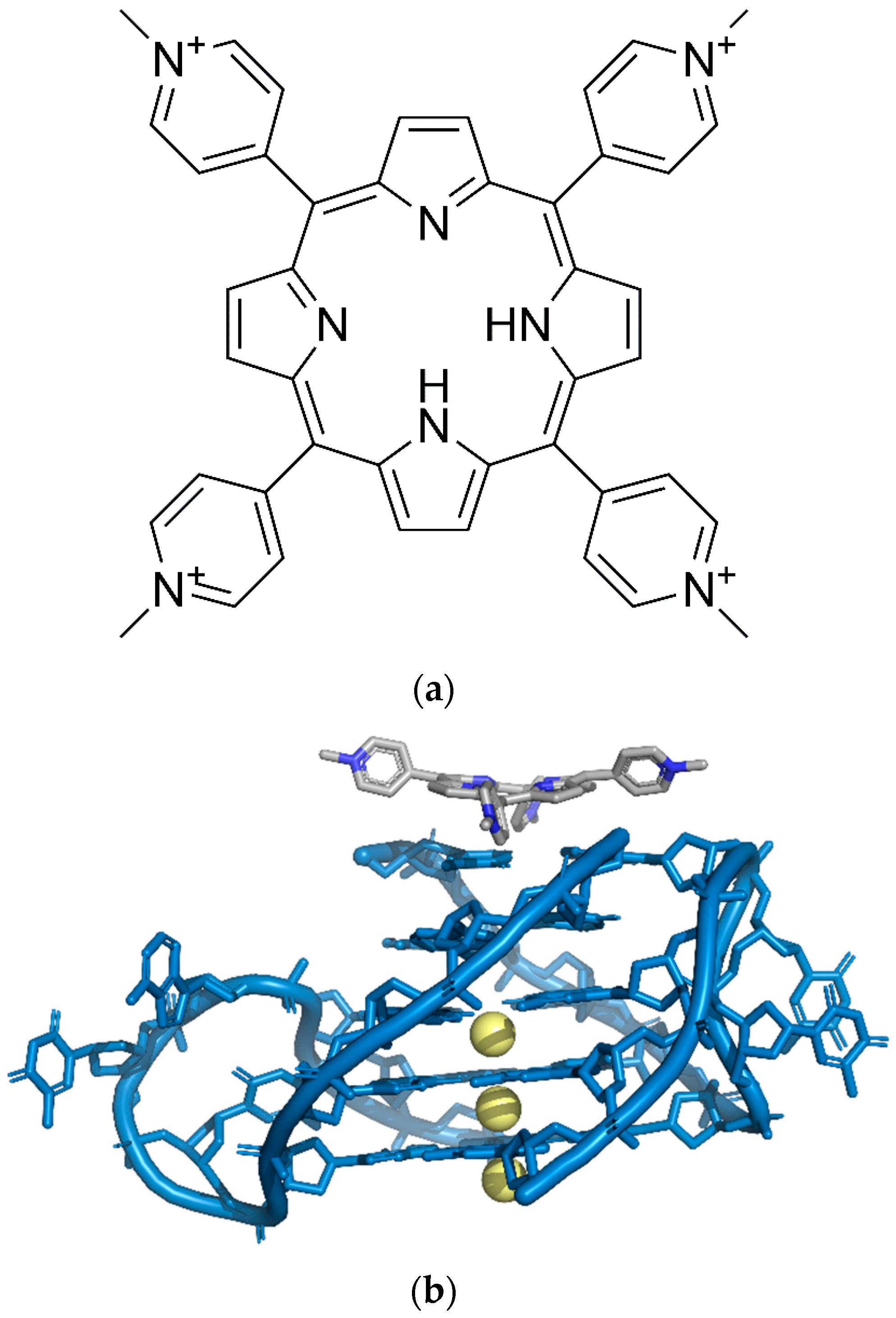
3.3. Luminescence-Based G-Quadruplex Sensing System
3.3.1. Conventional Luminescence-Based G-Quadruplex Sensing System
3.3.2. Luminescence Aptamer-Based G-Quadruplex Sensing System
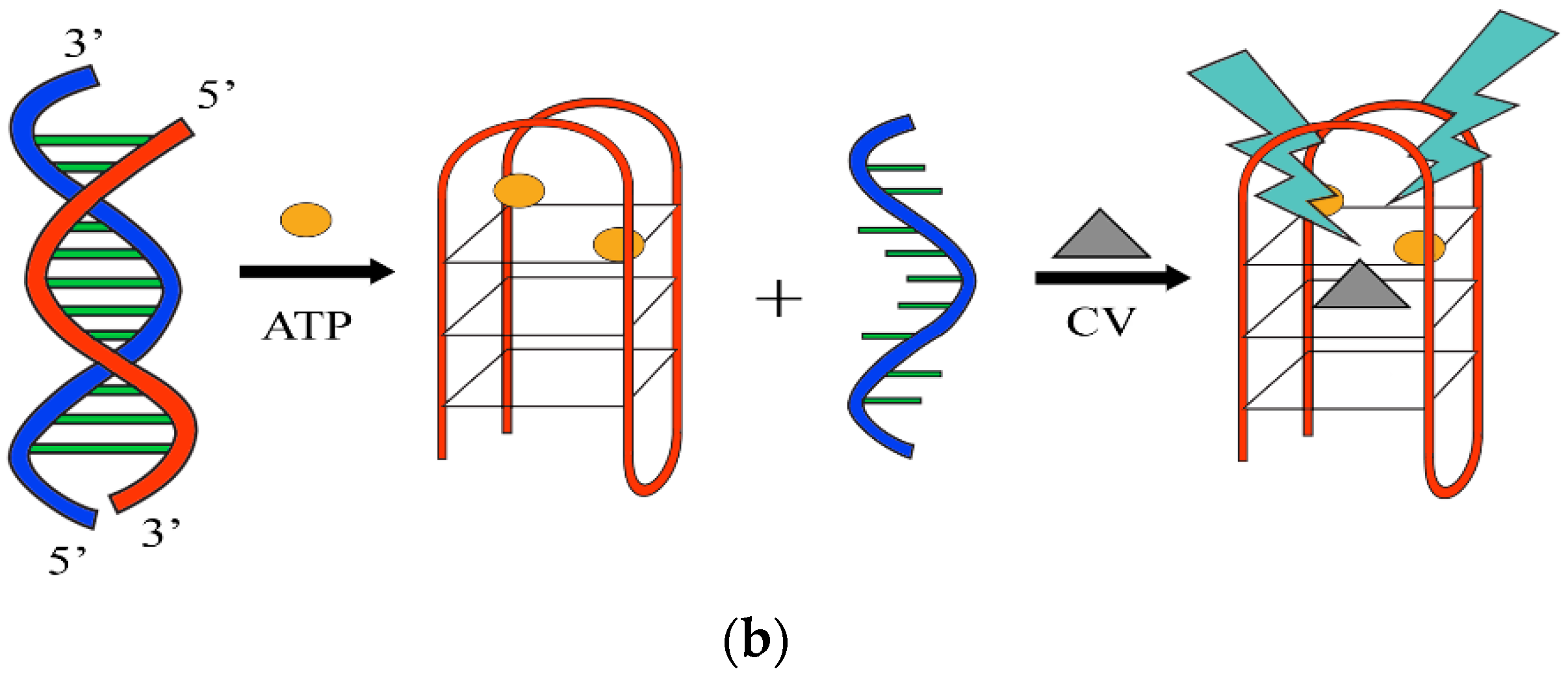
3.4. G-Quadruplex-Based Higher Order Sensor and Actuator Approaches
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Wang, M.; He, B.; Lu, L.; Leung, C.-H.; Mergny, J.-L.; Ma, D.-L. Label-free luminescent detection of LMP1 gene deletion using an intermolecular G-quadruplex-based switch-on probe. Biosens. Bioelectron. 2015, 70, 338–344. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Che, T.; Wang, Y.-Q.; Huang, Z.-L.; Tan, J.-H.; Huang, Z.-S.; Chen, S.-B. Natural Alkaloids and Heterocycles as G-Quadruplex Ligands and Potential Anticancer Agents. Molecules 2018, 23, 493. [Google Scholar] [Green Version]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Plückthun, A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef] [Green Version]
- Alba, J.J.; Sadurní, A.; Gargallo, R. Nucleic acid i-motif structures in analytical chemistry. Crit. Rev. Anal. Chem. 2016, 46, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, W.J.; Heddi, B.; Schmitt, E.; Lim, K.W.; Mechulam, Y.; Phan, A.T. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad. Sci. USA 2015, 112, 2729–2733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yang, D. Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. Curr. Protoc. Nucleic Acid Chem. 2012, 50, 17.15.11–17.15.17. [Google Scholar]
- Collie, G.W.; Promontorio, R.; Hampel, S.M.; Micco, M.; Neidle, S.; Parkinson, G.N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. [Google Scholar] [CrossRef]
- Hazel, P.; Parkinson, G.N.; Neidle, S. Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex. J. Am. Chem. Soc. 2006, 128, 5480–5487. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Volumetric contributions of loop regions of G-quadruplex DNA to the formation of the tertiary structure. Biophys. Chem. 2017, 231, 146–154. [Google Scholar] [CrossRef]
- Yang, D.; Okamoto, K. Structural insights into G-quadruplexes: Towards new anticancer drugs. Future Med. Chem. 2010, 2, 619–646. [Google Scholar] [CrossRef]
- Keniry, M.A. Quadruplex structures in nucleic acids. Biopolym. Orig. Res. Biomol. 2000, 56, 123–146. [Google Scholar] [CrossRef]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Ju, H.; Zhou, J. Thermal denaturation profile: A straightforward signature to characterize parallel G-quadruplexes. Biochimie 2019, 157, 22–25. [Google Scholar] [CrossRef]
- Esposito, V.; Galeone, A.; Mayol, L.; Oliviero, G.; Virgilio, A.; Randazzo, L. A topological classification of G-quadruplex structures. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1155–1159. [Google Scholar] [CrossRef]
- Campbell, N.; Collie, G.W.; Neidle, S. Crystallography of DNA and RNA G-Quadruplex Nucleic Acids and Their Ligand Complexes. Curr. Protoc. Nucleic Acid Chem. 2012, 50, 17.16.11–17.16.22. [Google Scholar]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef]
- Nagesh, N.; Chatterji, D. Ammonium ion at low concentration stabilizes the G-quadruplex formation by telomeric sequence. J. Biochem. Biophys. Methods 1995, 30, 1–8. [Google Scholar] [CrossRef]
- Phillips, K.; Dauter, Z.; Murchie, A.I.; Lilley, D.M.; Luisi, B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol. 1997, 273, 171–182. [Google Scholar] [CrossRef]
- Hud, N.V.; Plavec, J. The role of cations in determining quadruplex structure and stability. Quadruplex Nucleic Acids 2006, 100–130. [Google Scholar] [CrossRef]
- Nasab, M.G.; Hassani, L.; Nejad, S.M.; Norouzi, D. Interaction of hemin with quadruplex DNA. J. Biol. Phys. 2017, 43, 5–14. [Google Scholar] [CrossRef]
- Hardin, C.C.; Watson, T.; Corregan, M.; Bailey, C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d (CGCG3GCG). Biochemistry 1992, 31, 833–841. [Google Scholar] [CrossRef]
- Venczel, E.A.; Sen, D. Parallel and antiparallel G-DNA structures from a complex telomeric sequence. Biochemistry 1993, 32, 6220–6228. [Google Scholar] [CrossRef]
- Izatt, R.M.; Christensen, J.J.; Rytting, J.H. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and and nucleotides. Chem. Rev. 1971, 71, 439–481. [Google Scholar] [CrossRef]
- Zhou, X.-H.; Kong, D.-M.; Shen, H.-X. Ag+ and cysteine quantitation based on G-quadruplex-hemin DNAzymes disruption by Ag+. Anal. Chem. 2009, 82, 789–793. [Google Scholar] [CrossRef]
- Tang, G.; Zhao, C.; Gao, J.; Tan, H. Colorimetric detection of hydrogen sulfide based on terbium-G-quadruplex-hemin DNAzyme. Sens. Actuators B Chem. 2016, 237, 795–801. [Google Scholar] [CrossRef]
- Hardin, C.C.; Perry, A.G.; White, K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolym. Orig. Res. Biomol. 2000, 56, 147–194. [Google Scholar] [CrossRef]
- Blume, S.W.; Guarcello, V.; Zacharias, W.; Miller, D.M. Divalent transition metal cations counteract potassium-induced quadruplex assembly of oligo (dG) sequences. Nucleic Acids Res. 1997, 25, 617–625. [Google Scholar] [CrossRef]
- Lu, H.; Li, S.; Chen, J.; Xia, J.; Zhang, J.; Huang, Y.; Liu, X.; Wu, H.-C.; Zhao, Y.; Chai, Z. Metal ions modulate the conformation and stability of a G-quadruplex with or without a small-molecule ligand. Metallomics 2015, 7, 1508–1514. [Google Scholar] [CrossRef]
- Kwan, I.C.; She, Y.-M.; Wu, G. Trivalent lanthanide metal ions promote formation of stacking G-quartets. Chem. Commun. 2007, 41, 4286–4288. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Vankayalapati, H.; Shin-Ya, K.; Wierzba, K.; Hurley, L.H. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002, 124, 2098–2099. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Gleason-Guzman, M.; Izbicka, E.; Nishioka, D.; Hurley, L.H. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003, 63, 3247–3256. [Google Scholar]
- Morris, M.J.; Wingate, K.L.; Silwal, J.; Leeper, T.C.; Basu, S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012, 40, 4137–4145. [Google Scholar] [CrossRef] [Green Version]
- Moruno-Manchon, J.F.; Koellhoffer, E.C.; Gopakumar, J.; Hambarde, S.; Kim, N.; McCullough, L.D.; Tsvetkov, A.S. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany) 2017, 9, 1957. [Google Scholar] [CrossRef]
- Kawauchi, K.; Sugimoto, W.; Yasui, T.; Murata, K.; Itoh, K.; Takagi, K.; Tsuruoka, T.; Akamatsu, K.; Tateishi-Karimata, H.; Sugimoto, N. An anionic phthalocyanine decreases NRAS expression by breaking down its RNA G-quadruplex. Nat. Commun. 2018, 9, 2271. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Effect of pressure on thermal stability of G-quadruplex DNA and double-stranded DNA structures. Molecules 2013, 18, 13297–13319. [Google Scholar] [CrossRef]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef] [Green Version]
- Kankia, B.I. Self-dissociative primers for nucleic acid amplification and detection based on DNA quadruplexes with intrinsic fluorescence. Anal. Biochem. 2011, 409, 59–65. [Google Scholar] [CrossRef]
- Dutta, K.; Fujimoto, T.; Inoue, M.; Miyoshi, D.; Sugimoto, N. Development of new functional nanostructures consisting of both DNA duplex and quadruplex. Chem. Commun. 2010, 46, 7772–7774. [Google Scholar] [CrossRef]
- Travascio, P.; Bennet, A.J.; Wang, D.Y.; Sen, D. A ribozyme and a catalytic DNA with peroxidase activity: Active sites versus cofactor-binding sites. Chem. Biol. 1999, 6, 779–787. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef] [Green Version]
- Travascio, P.; Witting, P.K.; Mauk, A.G.; Sen, D. The peroxidase activity of a hemin—DNA oligonucleotide complex: Free radical damage to specific guanine bases of the DNA. J. Am. Chem. Soc. 2001, 123, 1337–1348. [Google Scholar] [CrossRef]
- Fan, D.; Zhu, J.; Zhai, Q.; Wang, E.; Dong, S. Cascade DNA logic device programmed ratiometric DNA analysis and logic devices based on a fluorescent dual-signal probe of a G-quadruplex DNAzyme. Chem. Commun. 2016, 52, 3766–3769. [Google Scholar] [CrossRef]
- Zhang, K.; Lv, S.; Lin, Z.; Tang, D. CdS: Mn quantum dot-functionalized gC 3 N 4 nanohybrids as signal-generation tags for photoelectrochemical immunoassay of prostate specific antigen coupling DNAzyme concatamer with enzymatic biocatalytic precipitation. Biosens. Bioelectron. 2017, 95, 34–40. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, J.; Cheng, M.; Monchaud, D.; Zhou, J.; Ju, H. A Thermophilic Tetramolecular G-Quadruplex/Hemin DNAzyme. Angew. Chem. 2017, 129, 16863–16867. [Google Scholar] [CrossRef]
- Omar, N.; Loh, Q.; Tye, G.J.; Choong, Y.S.; Noordin, R.; Glökler, J.; Lim, T.S. Development of an antigen-DNAzyme based probe for a direct antibody-antigen assay using the intrinsic DNAzyme activity of a daunomycin aptamer. Sensors 2013, 14, 346–355. [Google Scholar] [CrossRef]
- Loh, Q.; Omar, N.; Glökler, J.; Lim, T.S. IQPA: Isothermal nucleic acid amplification-based immunoassay using DNAzyme as the reporter system. Anal. Biochem. 2014, 463, 67–69. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, Q.; Chen, P.; Chen, J.; Chen, G.; Fu, F. A novel Tb3+-promoted G-quadruplex-hemin DNAzyme for the development of label-free visual biosensors. Biosens. Bioelectron. 2011, 26, 4053–4057. [Google Scholar] [CrossRef]
- Ye, M.; Zhang, Y.; Li, H.; Zhang, Y.; Tan, P.; Tang, H.; Yao, S. A novel method for the detection of point mutation in DNA using single-base-coded CdS nanoprobes. Biosens. Bioelectron. 2009, 24, 2339–2345. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Y.; Quirke, P.; Zhou, D. Ultrasensitive single-nucleotide polymorphism detection using target-recycled ligation, strand displacement and enzymatic amplification. Nanoscale 2013, 5, 5027–5035. [Google Scholar] [CrossRef]
- Shen, W.; Le Lim, C.; Gao, Z. A ferrofluid-based homogeneous assay for highly sensitive and selective detection of single-nucleotide polymorphisms. Chem. Commun. 2013, 49, 8114–8116. [Google Scholar] [CrossRef] [Green Version]
- Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; He, F.; Zhao, M.; Ling, L. Turn-off colorimetric sensor for sequence-specific recognition of single-stranded DNA based upon Y-shaped DNA structure. Sci. Rep. 2018, 8, 12021. [Google Scholar] [CrossRef]
- Sun, H.; Xiang, J.; Gai, W.; Liu, Y.; Guan, A.; Yang, Q.; Li, Q.; Shang, Q.; Su, H.; Tang, Y. Quantification of the Na+/K+ ratio based on the different response of a newly identified G-quadruplex to Na+ and K+. Chem. Commun. 2013, 49, 4510–4512. [Google Scholar] [CrossRef]
- Sun, H.; Chen, H.; Zhang, X.; Liu, Y.; Guan, A.; Li, Q.; Yang, Q.; Shi, Y.; Xu, S.; Tang, Y. Colorimetric detection of sodium ion in serum based on the G-quadruplex conformation related DNAzyme activity. Anal. Chim. Acta 2016, 912, 133–138. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Y. Responsive lanthanide coordination polymer for hydrogen sulfide. Anal. Chem. 2013, 85, 11020–11025. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M. A binary DNA probe for highly specific nucleic acid recognition. J. Am. Chem. Soc. 2006, 128, 10625–10628. [Google Scholar] [CrossRef]
- Ma, D.-L.; Wang, M.; He, B.; Yang, C.; Wang, W.; Leung, C.-H. A luminescent cocaine detection platform using a split G-Quadruplex-selective iridium (III) complex and a three-Way DNA junction architecture. ACS Appl. Mater. Interfaces 2015, 7, 19060–19067. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M. Split DNA enzyme for visual single nucleotide polymorphism typing. J. Am. Chem. Soc. 2008, 130, 2934–2935. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, D.; Zhou, Y.; Zhou, X. Highly effective colorimetric and visual detection of nucleic acids using an asymmetrically split peroxidase DNAzyme. J. Am. Chem. Soc. 2008, 130, 13095–13102. [Google Scholar] [CrossRef]
- Deng, M.; Feng, S.; Luo, F.; Wang, S.; Sun, X.; Zhou, X.; Zhang, X.-L. Visual detection of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis strains by use of an asymmetrically split peroxidase DNAzyme. J. Clin. Microbiol. 2012, 50, 3443–3450. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Dong, S.; Wang, E. How to split a G-quadruplex for DNA detection: New insight into the formation of DNA split G-quadruplex. Chem. Sci. 2015, 6, 4822–4827. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303. [Google Scholar] [CrossRef]
- Dong, H.; Ma, J.; Wang, J.; Wu, Z.-S.; Sinko, P.J.; Jia, L. A biofunctional molecular beacon for detecting single base mutations in cancer cells. Mol. Ther. Nucleic Acids 2016, 5, e302. [Google Scholar] [CrossRef]
- Zhang, J.-Q.; Wang, Y.-S.; Xue, J.-H.; He, Y.; Yang, H.-X.; Liang, J.; Shi, L.-F.; Xiao, X.-L. A gold nanoparticles-modified aptamer beacon for urinary adenosine detection based on structure-switching/fluorescence-“turning on” mechanism. J. Pharm. Biomed. Anal. 2012, 70, 362–368. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, J.; Li, T.; Wang, E. Bifunctional colorimetric oligonucleotide probe based on a G-quadruplex DNAzyme molecular beacon. Anal. Chem. 2011, 83, 8871–8876. [Google Scholar] [CrossRef]
- Zheng, Z.; Han, J.; Pang, W.; Hu, J. G-quadruplex DNAzyme molecular beacon for amplified colorimetric biosensing of Pseudostellaria heterophylla. Sensors 2013, 13, 1064–1075. [Google Scholar] [CrossRef]
- Li, H.; Wu, Z.; Qiu, L.; Liu, J.; Wang, C.; Shen, G.; Yu, R. Ultrasensitive label-free amplified colorimetric detection of p53 based on G-quadruplex MBzymes. Biosens. Bioelectron. 2013, 50, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Vummidi, B.R.; Alzeer, J.; Luedtke, N.W. Fluorescent probes for G-quadruplex structures. ChemBioChem 2013, 14, 540–558. [Google Scholar] [CrossRef]
- He, H.-Z.; Chan, D.S.-H.; Leung, C.-H.; Ma, D.-L. G-quadruplexes for luminescent sensing and logic gates. Nucleic Acids Res. 2013, 41, 4345–4359. [Google Scholar] [CrossRef] [Green Version]
- Bhasikuttan, A.C.; Mohanty, J. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: Promising fluorescence sensors. Chem. Commun. 2015, 51, 7581–7597. [Google Scholar] [CrossRef]
- Wochner, A.; Menger, M.; Orgel, D.; Cech, B.; Rimmele, M.; Erdmann, V.A.; Glökler, J. A DNA aptamer with high affinity and specificity for therapeutic anthracyclines. Anal. Biochem. 2008, 373, 34–42. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. Parallel G-quadruplex-specific fluorescent probe for monitoring DNA structural changes and label-free detection of potassium ion. Anal. Chem. 2010, 82, 7576–7580. [Google Scholar] [CrossRef]
- Stevens, A.J.; Kennedy, H.L.; Kennedy, M.A. Fluorescence methods for probing G-quadruplex structure in single-and double-stranded DNA. Biochemistry 2016, 55, 3714–3725. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Li, T.; Dong, S.; Wang, E. Enzyme-Free Unlabeled DNA Logic Circuits Based on Toehold-Mediated Strand Displacement and Split G-Quadruplex Enhanced Fluorescence. Adv. Mater. 2013, 25, 2440–2444. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Wang, E. Measurement of the base number of DNA using a special calliper made of a split G-quadruplex. Chem. Commun. 2012, 48, 11990–11992. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, J.; Ai, J.; Zhou, Z.; Jia, X.; Wang, E. Label-free G-quadruplex-specific fluorescent probe for sensitive detection of copper (II) ion. Biosens. Bioelectron. 2013, 39, 268–273. [Google Scholar] [CrossRef]
- Barnham, K.J.; Masters, C.L.; Bush, A.I. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004, 3, 205. [Google Scholar] [CrossRef]
- Kim, H.R.; Lee, I.J.; Kim, D.-E. Label/Quencher-Free Detection of Exon Deletion Mutation in Epidermal Growth Factor Receptor Gene Using G-Quadruplex-Inducing DNA Probe. J. Microbiol. Biotechnol. 2017, 27, 72–76. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Deng, Q.; Hou, J.-Q.; Hu, D.-P.; Wang, Z.-Y.; Zhang, K.; Luyt, L.G.; Wong, W.-L.; Chow, C.-F. Molecular engineering of thiazole orange dye: Change of fluorescent signaling from universal to specific upon binding with nucleic acids in bioassay. Acs Chem. Biol. 2016, 11, 1019–1029. [Google Scholar] [CrossRef]
- Guo, R.-J.; Yan, J.-W.; Chen, S.-B.; Gu, L.-Q.; Huang, Z.-S.; Tan, J.-H. A simple structural modification to thiazole orange to improve the selective detection of G-quadruplexes. Dye. Pigment. 2016, 126, 76–85. [Google Scholar] [CrossRef]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Et Biophys. Acta (Bba)-Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef]
- González, V.; Martín, M.; Fernández, G.; García-Sacristán, A. Use of aptamers as diagnostics tools and antiviral agents for human viruses. Pharmaceuticals 2016, 9, 78. [Google Scholar] [CrossRef]
- Ma, D.L.; Wang, W.; Mao, Z.; Kang, T.S.; Han, Q.B.; Chan, P.W.H.; Leung, C.H. Utilization of G-Quadruplex-Forming Aptamers for the Construction of Luminescence Sensing Platforms. Chempluschem 2017, 82, 8–17. [Google Scholar] [CrossRef]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. Trac Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar]
- Callan, J.F.; Mulrooney, R.C.; Kamila, S. Luminescent detection of ATP in aqueous solution using positively charged CdSe–ZnS quantum dots. J. Fluoresc. 2008, 18, 1157–1161. [Google Scholar] [CrossRef]
- He, H.-Z.; Ma, V.P.-Y.; Leung, K.-H.; Chan, D.S.-H.; Yang, H.; Cheng, Z.; Leung, C.-H.; Ma, D.-L. A label-free G-quadruplex-based switch-on fluorescence assay for the selective detection of ATP. Analyst 2012, 137, 1538–1540. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, W.; Zhou, M.; Xiang, Y.; Yuan, R.; Chai, Y. Toehold strand displacement-driven assembly of G-quadruplex DNA for enzyme-free and non-label sensitive fluorescent detection of thrombin. Biosens. Bioelectron. 2015, 64, 306–310. [Google Scholar] [CrossRef]
- Leung, K.-H.; He, H.-Z.; Ma, V.P.-Y.; Chan, D.S.-H.; Leung, C.-H.; Ma, D.-L. A luminescent G-quadruplex switch-on probe for the highly selective and tunable detection of cysteine and glutathione. Chem. Commun. 2013, 49, 771–773. [Google Scholar] [CrossRef]
- Foldbjerg, R.; Dang, D.A.; Autrup, H. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch. Toxicol. 2011, 85, 743–750. [Google Scholar] [CrossRef]
- AshaRani, P.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 2008, 3, 279–290. [Google Scholar] [CrossRef]
- Man, B.Y.-W.; Chan, D.S.-H.; Yang, H.; Ang, S.-W.; Yang, F.; Yan, S.-C.; Ho, C.-M.; Wu, P.; Che, C.-M.; Leung, C.-H. A selective G-quadruplex-based luminescent switch-on probe for the detection of nanomolar silver (i) ions in aqueous solution. Chem. Commun. 2010, 46, 8534–8536. [Google Scholar] [CrossRef]
- Leung, K.-H.; He, H.-Z.; Ma, V.P.-Y.; Yang, H.; Chan, D.S.-H.; Leung, C.-H.; Ma, D.-L. A G-quadruplex-selective luminescent switch-on probe for the detection of sub-nanomolar human neutrophil elastase. RSC Adv. 2013, 3, 1656–1659. [Google Scholar] [CrossRef]
- He, J.-L.; Wu, Z.-S.; Zhang, S.-B.; Shen, G.-L.; Yu, R.-Q. Fluorescence aptasensor based on competitive-binding for human neutrophil elastase detection. Talanta 2010, 80, 1264–1268. [Google Scholar] [CrossRef]
- Cho, S.; Park, L.; Chong, R.; Kim, Y.T.; Lee, J.H. Rapid and simple G-quadruplex DNA aptasensor with guanine chemiluminescence detection. Biosens. Bioelectron. 2014, 52, 310–316. [Google Scholar] [CrossRef]
- Schlachter, C.; Lisdat, F.; Frohme, M.; Erdmann, V.A.; Konthur, Z.; Lehrach, H.; Glökler, J. Pushing the detection limits: The evanescent field in surface plasmon resonance and analyte-induced folding observation of long human telomeric repeats. Biosens. Bioelectron. 2012, 31, 571–574. [Google Scholar] [CrossRef]
- Dong, J.; Cui, X.; Deng, Y.; Tang, Z. Amplified detection of nucleic acid by G-quadruplex based hybridization chain reaction. Biosens. Bioelectron. 2012, 38, 258–263. [Google Scholar] [CrossRef] [Green Version]
- Hou, T.; Li, W.; Liu, X.; Li, F. Label-free and enzyme-free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: A facile, sensitive, and highly specific microRNA assay. Anal. Chem. 2015, 87, 11368–11374. [Google Scholar] [CrossRef]
- Olejko, L.; Cywiński, P.J.; Bald, I. An ion-controlled four-color fluorescent telomeric switch on DNA origami structures. Nanoscale 2016, 8, 10339–10347. [Google Scholar] [CrossRef]
- Wang, J.; Yue, L.; Wang, S.; Willner, I. Triggered Reversible Reconfiguration of G-Quadruplex-Bridged “Domino”-Type Origami Dimers: Application of the Systems for Programmed Catalysis. ACS Nano 2018, 8, 10339–10347. [Google Scholar] [CrossRef]
- Deng, M.; Feng, S.; Luo, F.; Wang, S.; Sun, X.; Zhou, X.; Zhang, X.-L. Visual detection of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis using an asymmetrically split peroxidase DNAzyme. J. Clin. Microbiol. 2012, 12, 12324–12336. [Google Scholar]




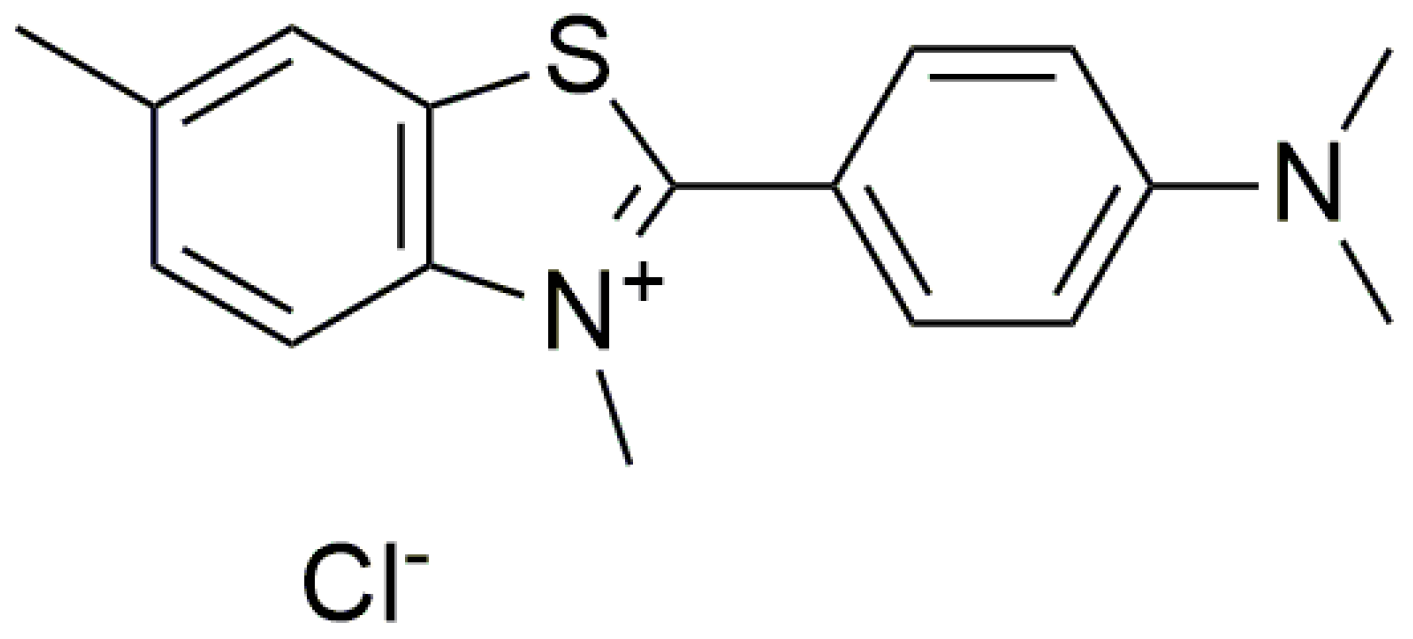





| Conformation | λ (nm) | |||
|---|---|---|---|---|
| 243 | 260 | 275 | 295 | |
| Parallel | + | − | − | − |
| Antiparallel | + | −/+ | + | − |
| Hybrid | + | + | + | − |
| Sensing System | Target Molecules | Facilitating Ligands | Advantages | Disadvantages | Limit of Detection | References |
|---|---|---|---|---|---|---|
| G4/Hemin-basedDNAzyme | ssDNA (SNP) | Hemin |
| - | 0.95 nM | [56] |
| Sodium ions | 0.6 µM | [58] | ||||
| Hydrogen sulfide (H2S) | Hemin, Ag+ and Tb3+ (as substitute for K+) |
|
| 13 nM | [29] | |
| DNAzyme-based colorimetric split G4 | ssDNA (rifampin-resistant M.tb) | Hemin |
|
| - | [106] |
| DNAzyme-based colorimetric G4/Hemin Molecular Beacon | nrDNA ITS |
| - | 3.1 × 10−10 mol·L−1 | [70] | |
| p53 DNA | - | 25 fM | [71] | |||
| Conventional non-covalent fluorescence G4 | DNA (base number of DNA) |
|
|
| - | [79] |
| Copper (II) ion | 3.0 nM | [80] | ||||
| DNA (EGFR exon 19 deletion mutant) |
| 2.3 nM | [82] | |||
| Non-covalent fluorescence aptamer-based G4 | ATP | Crystal violet (CV) |
| - | 5 µM | [92] |
| Thrombin | N-Methyl mesoporphyrin (IX) (NMM) |
| - | 5 pM | [93] | |
| Conventional luminescence-based G4 | Silver ion | Chloro(2-phenyl-1,10-phenanthroline)-platinum(II) |
| - | 20 nM | [97] |
| Luminescence aptamer-based G4 | Enzyme | Iridium(III) complex[Ir(ppy)2(dpp)]+ |
| - | [98] | |
| Thrombin | 3,4,5-trimethoxy-phenylglyoxal (TMPG) |
| - | 12.3 nM | [100] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ida, J.; Chan, S.K.; Glökler, J.; Lim, Y.Y.; Choong, Y.S.; Lim, T.S. G-Quadruplexes as An Alternative Recognition Element in Disease-Related Target Sensing. Molecules 2019, 24, 1079. https://doi.org/10.3390/molecules24061079
Ida J, Chan SK, Glökler J, Lim YY, Choong YS, Lim TS. G-Quadruplexes as An Alternative Recognition Element in Disease-Related Target Sensing. Molecules. 2019; 24(6):1079. https://doi.org/10.3390/molecules24061079
Chicago/Turabian StyleIda, Jeunice, Soo Khim Chan, Jörn Glökler, Yee Ying Lim, Yee Siew Choong, and Theam Soon Lim. 2019. "G-Quadruplexes as An Alternative Recognition Element in Disease-Related Target Sensing" Molecules 24, no. 6: 1079. https://doi.org/10.3390/molecules24061079
APA StyleIda, J., Chan, S. K., Glökler, J., Lim, Y. Y., Choong, Y. S., & Lim, T. S. (2019). G-Quadruplexes as An Alternative Recognition Element in Disease-Related Target Sensing. Molecules, 24(6), 1079. https://doi.org/10.3390/molecules24061079





