State-of-the-Art Genetic Modalities to Engineer Cyanobacteria for Sustainable Biosynthesis of Biofuel and Fine-Chemicals to Meet Bio–Economy Challenges
Abstract
:1. Introduction
2. Advanced Tools for Engineering Cyanobacteria
2.1. Genome-Scale Modeling
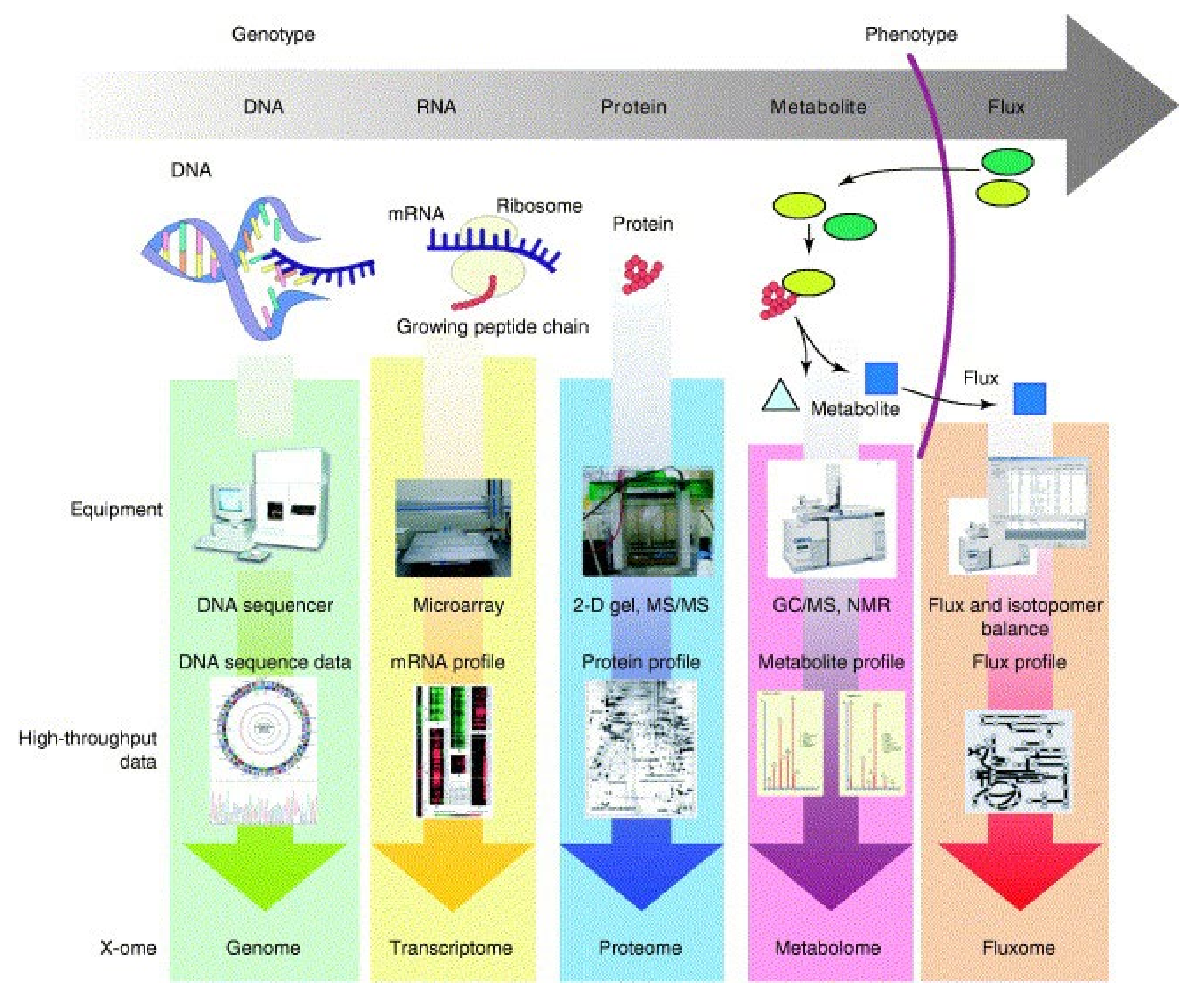
2.2. High Throughput Omics
2.3. Synthetic Biology Tools
2.4. CRISPR/Cas Technology
2.5. 13C-Based Metabolic Flux Analysis
3. Engineering Cyanobacteria for Environmental Stress Resistance
- (1)
- Seawater or industrial wastewater is utilized for larger-scale cyanobacterial culturing than that of sterilized fresh water because of economic and environmental costs, where the presence of heavy metals, salts, and many other potential toxins will interrupt the normal cellular growth and metabolism of cyanobacterial cell factories.
- (2)
- Application of selective conditions for bio-contamination control such as extreme low/high pH and elevated NaCl concentration can also impede the normal growth and metabolism.
- (3)
- The accumulation of toxic intermediates products and metabolites in the cultivation system.
- (4)
- Temperature or light intensities in the real environmental situations will be controlled in a rhythm, with peak levels that are too extreme for cyanobacterial strains to acclimate.
3.1. Introducing Heterologous Stress Tolerance Proteins
3.2. Enhancing Cyanobacterial Robustness by Overexpressing Heat Shock Proteins
4. Engineering Cyanobacteria for Biofuel and Fine Chemicals Production
4.1. Cyanobacteria—Biofuel
4.2. Cyanobacteria—Isobutanol and 1-Butanol
4.3. Cyanobacteria—Hydrogen
4.4. Cyanobacteria—1,3-Propanediol
4.5. Cyanobacteria—Fatty Acid and Hydrocarbons Biosynthesis
4.6. Engineering Cynobacteria for Organic Acids Biosynthesis
4.6.1. Cyanobacteria—Lactic Acid
4.6.2. Cyanobacteria—3-Hydroxpropionate
4.7. Engineering Cyanobacteria for Carbohydrates/Sugars Biosynthesis
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheng, H.; Wang, S.; Bilal, M.; Ge, X.; Zhang, C.; Fickers, P.; Cheng, H. Identification, characterization of two NADPH-dependent erythrose reductases in the yeast Yarrowia lipolytica and improvement of erythritol productivity using metabolic engineering. Microb. Cell Fact. 2018, 17, 133. [Google Scholar] [CrossRef] [PubMed]
- Knoot, C.J.; Ungerer, J.; Wangikar, P.P.; Pakrasi, H.B. Cyanobacteria: Promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018, 293, 5044–5052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. 4-Hydroxybenzoic acid—A versatile platform intermediate for value-added compounds. Appl. Microbiol. Biotechnol. 2018, 102, 3561–3571. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Lan, E.I. Advances in metabolic engineering of cyanobacteria for photosynthetic biochemical production. Metabolites 2015, 5, 636–658. [Google Scholar] [CrossRef] [PubMed]
- Luan, G.; Lu, X. Tailoring cyanobacterial cell factory for improved industrial properties. Biotechnol. Adv. 2018, 36, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, S.; Song, X.; Diao, J.; Chen, L.; Zhang, W. Toolboxes for cyanobacteria: Recent advances and future direction. Biotechnol. Adv. 2018, 36, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Whitton, B.A.; Potts, M. (Eds.) The Ecology of Cyanobacteria: Their Diversity in Time and Space; Springer: Berlin, Germany, 2007. [Google Scholar]
- Hamilton, T.L.; Bryant, D.A.; Macalady, J.L. The role of biology in planetary evolution: Cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ. Microbiol. 2016, 18, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Dismukes, G.C.; Carrieri, D.; Bennette, N.; Ananyev, G.M.; Posewitz, M.C. Aquatic phototrophs: Efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 2008, 19, 235–240. [Google Scholar] [CrossRef]
- Branco dos Santos, F.; Du, W.; Hellingwerf, K.J. Synechocystis: Not just a plug-bug for CO2, but a green E. coli. Front. Bioeng. Biotechnol. 2014, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Pate, R.; Klise, G.; Wu, B. Resource demand implications for US algae biofuels production scale-up. Appl. Energy 2011, 88, 3377–3388. [Google Scholar] [CrossRef]
- Sharma, N.K.; Rai, A.K.; Stal, L.J. Cyanobacteria: An Economic Perspective; John Wiley & Sons. Ltd.: Chichester, UK, 2013. [Google Scholar]
- Markou, G.; Georgakakis, D. Cultivation of filamentous cyanobacteria (blue-green algae) in agro-industrial wastes and wastewaters: A review. Appl. Energy 2011, 88, 3389–3401. [Google Scholar] [CrossRef]
- Deng, M.D.; Coleman, J.R. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 1999, 65, 523–528. [Google Scholar] [PubMed]
- Song, K.; Tan, X.; Liang, Y.; Lu, X. The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl. Microbiol. Biotechnol. 2016, 100, 7865–7875. [Google Scholar] [CrossRef] [PubMed]
- Ducat, D.C.; Silver, P.A. Improving carbon fixation pathways. Curr. Opin. Chem. Biol. 2012, 16, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, T.L.; Young, E.J.; Ducat, D.C. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab. Eng. 2017, 44, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhao, H.; Li, Z.; Tan, X.; Lu, X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 2012, 5, 9857–9865. [Google Scholar] [CrossRef]
- Choi, S.Y.; Wang, J.Y.; Kwak, H.S.; Lee, S.M.; Um, Y.; Kim, Y.; Sim, S.J.; Choi, J.I.; Woo, H.M. Improvement of squalene production from CO2 in Synechococcus elongatus PCC 7942 by metabolic engineering and scalable production in a photobioreactor. ACS Synth. Biol. 2017, 6, 1289–1295. [Google Scholar] [CrossRef]
- Aikawa, S.; Nishida, A.; Ho, S.H.; Chang, J.S.; Hasunuma, T.; Kondo, A. Glycogen production for biofuels by the euryhaline cyanobacteria Synechococcus sp. strain PCC 7002 from an oceanic environment. Biotechnol. Biofuels 2014, 7, 88. [Google Scholar] [CrossRef]
- Lin, P.C.; Saha, R.; Zhang, F.; Pakrasi, H.B. Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2017, 7, 17503. [Google Scholar] [CrossRef]
- Wang, B.; Eckert, C.; Maness, P.C.; Yu, J. A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth. Biol. 2017, 7, 276–286. [Google Scholar] [CrossRef]
- Davies, F.K.; Work, V.H.; Beliaev, A.S.; Posewitz, M.C. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014, 2, 21. [Google Scholar] [CrossRef]
- Korosh, T.C.; Markley, A.L.; Clark, R.L.; McGinley, L.L.; McMahon, K.D.; Pfleger, B.F. Engineering photosynthetic production of L-lysine. Metab. Eng. 2017, 44, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Liu, X.; Englund, E.; Lindberg, P.; Lindblad, P. Isobutanol production in Synechocystis PCC 6803 using heterologous and endogenous alcohol dehydrogenases. Metab. Eng. Commun. 2017, 5, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Kanno, M.; Carroll, A.L.; Atsumi, S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Nat. Commun. 2017, 8, 14724. [Google Scholar] [CrossRef] [PubMed]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Burgess, J.G.; Wright, P.C. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Chwa, J.W.; Kim, W.J.; Sim, S.J.; Um, Y.; Woo, H.M. Engineering of a modular and synthetic phosphoketolase pathway for photosynthetic production of acetone from CO2 in Synechococcus elongatus PCC 7942 under light and aerobic condition. Plant Biotechnol. J. 2016, 14, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhu, T.; Cai, Z.; Li, Y. From cyanochemicals to cyanofactories: A review and perspective. Microb. Cell Fact. 2016, 15, 2. [Google Scholar] [CrossRef]
- Gao, X.; Gao, F.; Liu, D.; Zhang, H.; Nie, X.; Yang, C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 2016, 9, 1400–1411. [Google Scholar] [CrossRef]
- Zhang, A.; Carroll, A.L.; Atsumi, S. Carbon recycling by cyanobacteria: Improving CO2 fixation through chemical production. FEMS Microbiol. Lett. 2017, 364, fnx165. [Google Scholar] [CrossRef]
- Whitney, S.M.; Houtz, R.L.; Alonso, H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011, 155, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Borirak, O.; de Koning, L.J.; van der Woude, A.D.; Hoefsloot, H.C.; Dekker, H.L.; Roseboom, W.; de Koster, C.G.; Hellingwerf, K.J. Quantitative proteomics analysis of an ethanol-and a lactate-producing mutant strain of Synechocystis sp. PCC6803. Biotechnol. Biofuels 2015, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, J.; Pakrasi, H.B. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci. Rep. 2016, 6, 39681. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Shen, C.R.; Li, H.; Sung, L.Y.; Wu, M.Y.; Hu, Y.C. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb. Cell Fact. 2016, 15, 196. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Yoshikawa, K.; Toya, Y.; Matsuda, F.; Shimizu, H. Metabolic flux analysis of the Synechocystis sp. PCC 6803 Δ nrtABCD mutant reveals a mechanism for metabolic adaptation to nitrogen-limited conditions. Plant Cell Physiol. 2017, 58, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Chou, H.; Ham, T.S.; Lee, T.S.; Keasling, J.D. Metabolic engineering of microorganisms for biofuels production: From bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 2008, 19, 556–563. [Google Scholar] [CrossRef]
- Xu, C.; Liu, L.; Zhang, Z.; Jin, D.; Qiu, J.; Chen, M. Genome-scale metabolic model in guiding metabolic engineering of microbial improvement. Appl. Microbiol. Biotechnol. 2013, 97, 519–539. [Google Scholar] [CrossRef]
- Triana, J.; Montagud, A.; Siurana, M.; Urchueguía, A.; Gamermann, D.; Torres, J.; Tena, J.; de Córdoba, P.; Urchueguía, J. Generation and evaluation of a genome-scale metabolic network model of Synechococcus elongatus PCC7942. Metabolites 2014, 4, 680–698. [Google Scholar] [CrossRef]
- Broddrick, J.T.; Rubin, B.E.; Welkie, D.G.; Du, N.; Mih, N.; Diamond, S.; Lee, J.J.; Golden, S.S.; Palsson, B.O. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. USA 2016, 113, E8344–E8353. [Google Scholar] [CrossRef]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO 2. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef]
- Mueller, T.J.; Ungerer, J.L.; Pakrasi, H.B.; Maranas, C.D. Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973. Sci. Rep. 2017, 7, 41569. [Google Scholar] [CrossRef] [PubMed]
- Hendry, J.I.; Gopalakrishnan, S.; Ungerer, J.; Pakrasi, H.B.; Tang, Y.J.; Maranas, C.D. Genome-scale fluxome of Synechococcus elongatus UTEX 2973 using transient 13C-labeling data. Plant Physiol. 2019, 179, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Lee, D.Y.; Kim, T.Y. Systems biotechnology for strain improvement. Trends Biotechnol. 2005, 23, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, K.A.; Gaudet, A.D.; Amici, S.A.; Popovich, P.G.; Guerau-de-Arellano, M. Control of the inflammatory macrophage transcriptional signature by miR-155. PLoS ONE 2016, 11, e0159724. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Hess, W.R. Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev. 2015, 39, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Berla, B.M.; Saha, R.; Immethun, C.M.; Maranas, C.D.; Moon, T.S.; Pakrasi, H. Synthetic biology of cyanobacteria: Unique challenges and opportunities. Front. Microbiol. 2013, 4, 246. [Google Scholar] [CrossRef]
- Markley, A.L.; Begemann, M.B.; Clarke, R.E.; Gordon, G.C.; Pfleger, B.F. Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synth. Biol. 2014, 4, 595–603. [Google Scholar] [CrossRef]
- Xu, Y.; Alvey, R.M.; Byrne, P.O.; Graham, J.E.; Shen, G.; Bryant, D.A. Expression of genes in cyanobacteria: Adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. In Photosynthesis Research Protocols; Humana Press: Totowa, NJ, USA, 2011; pp. 273–293. [Google Scholar]
- Nozzi, N.E.; Case, A.E.; Carroll, A.L.; Atsumi, S. Systematic approaches to efficiently produce 2, 3-butanediol in a marine cyanobacterium. ACS Synth. Biol. 2017, 6, 2136–2144. [Google Scholar] [CrossRef]
- Zess, E.K.; Begemann, M.B.; Pfleger, B.F. Construction of new synthetic biology tools for the control of gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. Biotechnol. Bioeng. 2016, 113, 424–432. [Google Scholar] [CrossRef]
- Ohbayashi, R.; Akai, H.; Yoshikawa, H.; Hess, W.R.; Watanabe, S. A tightly inducible riboswitch system in Synechocystis sp. PCC 6803. J. Gen. Appl. Microbiol. 2016, 62, 154–159. [Google Scholar] [CrossRef]
- Abe, K.; Sakai, Y.; Nakashima, S.; Araki, M.; Yoshida, W.; Sode, K.; Ikebukuro, K. Design of riboregulators for control of cyanobacterial (Synechocystis) protein expression. Biotechnol. Lett. 2014, 36, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Albers, S.C.; Gallegos, V.A.; Peebles, C.A. Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J. Biotechnol. 2015, 216, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Armshaw, P.; Carey, D.; Sheahan, C.; Pembroke, J.T. Utilising the native plasmid, pCA2. 4, from the cyanobacterium Synechocystis sp. strain PCC6803 as a cloning site for enhanced product production. Biotechnol. Biofuels 2015, 8, 201. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.H.; Berla, B.M.; Pakrasi, H.B. Fine tuning of photoautotrophic protein production by combining promoters and neutral sites in Synechocystis 6803, a cyanobacterium. Appl. Environ. Microbiol. 2015, 81, 6857–6863. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Liang, F.; Lindberg, P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6, 36640. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.D.; Metcalf, W.W. Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc. Natl. Acad. Sci. USA 2017, 114, 2976–2981. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, C.R.; Huang, C.H.; Sung, L.Y.; Wu, M.Y.; Hu, Y.C. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 2016, 38, 293–302. [Google Scholar] [CrossRef]
- Behler, J.; Vijay, D.; Hess, W.R.; Akhtar, M.K. CRISPR-based technologies for metabolic engineering in cyanobacteria. Trends Biotechnol. 2018, 36, 996–1010. [Google Scholar] [CrossRef]
- Ramey, C.J.; Barón-Sola, A.; Aucoin, H.R.; Boyle, N.R. Genome engineering in cyanobacteria: Where we are and where we need to go. ACS Synth. Biol. 2015, 4, 1186–1196. [Google Scholar] [CrossRef]
- Wendt, K.E.; Ungerer, J.; Cobb, R.E.; Zhao, H.; Pakrasi, H.B. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact. 2016, 15, 115. [Google Scholar] [CrossRef]
- Young, J.D.; Shastri, A.A.; Stephanopoulos, G.; Morgan, J.A. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng. 2011, 13, 656–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jazmin, L.J.; O’Grady, J.P.; Ma, F.; Allen, D.K.; Morgan, J.A.; Young, J.D. Isotopically nonstationary MFA (INST-MFA) of autotrophic metabolism. In Plant Metabolic Flux Analysis; Humana Press: Totowa, NJ, USA, 2014; pp. 181–210. [Google Scholar]
- Jazmin, L.J.; Xu, Y.; Cheah, Y.E.; Adebiyi, A.O.; Johnson, C.H.; Young, J.D. Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab. Eng. 2017, 42, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, E.I.; Wei, C.T. Metabolic engineering of cyanobacteria for the photosynthetic production of succinate. Metab. Eng. 2016, 38, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Laloknam, S.; Tanaka, K.; Buaboocha, T.; Waditee, R.; Incharoensakdi, A.; Hibino, T.; Tanaka, Y.; Takabe, T. Halotolerant cyanobacterium Aphanothece halophytica contains a betaine transporter active at alkaline pH and high salinity. Appl. Environ. Microbiol. 2006, 72, 6018–6026. [Google Scholar] [CrossRef] [PubMed]
- Waditee, R.; Hibino, T.; Nakamura, T.; Incharoensakdi, A.; Takabe, T. Overexpression of a Na+/H+ antiporter confers salt tolerance on a freshwater cyanobacterium, making it capable of growth in sea water. Proc. Natl. Acad. Sci. USA 2002, 99, 4109–4114. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Sharma, N.K.; Prasad, S.B.; Yadav, S.S.; Narayan, G.; Rai, A.K. The freshwater cyanobacterium Anabaena doliolum transformed with ApGSMT-DMT exhibited enhanced salt tolerance and protection to nitrogenase activity, but became halophilic. Microbiology 2013, 159, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Chaurasia, A.K.; Apte, S.K. Overexpression of the groESL operon enhances the heat and salinity stress tolerance of the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC7120. Appl. Environ. Microbiol. 2009, 75, 6008–6012. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Esquer, C.R.; Vermaas, W.F. ClpB1 overproduction in Synechocystis sp. strain PCC 6803 increases tolerance to rapid heat shock. Appl. Environ. Microbiol. 2013, 79, 6220–6227. [Google Scholar] [CrossRef]
- Su, H.Y.; Chou, H.H.; Chow, T.J.; Lee, T.M.; Chang, J.S.; Huang, W.L.; Chen, H.J. Improvement of outdoor culture efficiency of cyanobacteria by over-expression of stress tolerance genes and its implication as bio-refinery feedstock. Bioresour. Technol. 2017, 244, 1294–1303. [Google Scholar] [CrossRef]
- Jiang, L.; Pei, H.; Hu, W.; Ji, Y.; Han, L.; Ma, G. The feasibility of using complex wastewater from a monosodium glutamate factory to cultivate Spirulina subsalsa and accumulate biochemical composition. Bioresour. Technol. 2015, 180, 304–310. [Google Scholar] [CrossRef]
- Iijima, H.; Nakaya, Y.; Kuwahara, A.; Hirai, M.Y.; Osanai, T. Seawater cultivation of freshwater cyanobacterium Synechocystis sp. PCC 6803 drastically alters amino acid composition and glycogen metabolism. Front. Microbiol. 2015, 6, 326. [Google Scholar] [CrossRef] [PubMed]
- Touloupakis, E.; Cicchi, B.; Benavides, A.M.S.; Torzillo, G. Effect of high pH on growth of Synechocystis sp. PCC 6803 cultures and their contamination by golden algae (Poterioochromonas sp.). Appl. Microbiol. Biotechnol. 2016, 100, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Luan, G.; Tan, X.; Zhang, H.; Lu, X. Rescuing ethanol photosynthetic production of cyanobacteria in non-sterilized outdoor cultivations with a bicarbonate-based pH-rising strategy. Biotechnol. Biofuels 2017, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Stephanopoulos, G. Challenges in engineering microbes for biofuels production. Science 2007, 315, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.S.; Matsui, M.; Abdullah, A.A.A. Cyanobacteria: Photoautotrophic microbial factories for the sustainable synthesis of industrial products. BioMed. Res. Int. 2015, 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Englund, E.; Lindberg, P.; Lindblad, P. Engineered cyanobacteria with enhanced growth show increased ethanol production and higher biofuel to biomass ratio. Metab. Eng. 2018, 46, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Angermayr, S.A.; Van der Woude, A.D.; Correddu, D.; Vreugdenhil, A.; Verrone, V.; Hellingwerf, K.J. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 2014, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, G.M.; Tashiro, Y.; Atsumi, S. Expanding ester biosynthesis in Escherichia coli. Nat. Chem. Biol. 2014, 10, 259. [Google Scholar] [CrossRef]
- Lan, E.I.; Liao, J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 6018–6023. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Shen, C.R.; Liao, J.C. Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosynth. Res. 2014, 120, 301–310. [Google Scholar] [CrossRef]
- Miao, R.; Xie, H.; Ho, F.M.; Lindblad, P. Protein engineering of α-ketoisovalerate decarboxylase for improved isobutanol production in Synechocystis PCC 6803. Metab. Eng. 2018, 47, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Pei, G.; Wang, J.; Chen, L.; Zhang, W. A novel small RNA CoaR regulates coenzyme A biosynthesis and tolerance of Synechocystis sp. PCC6803 to 1-butanol possibly via promoter-directed transcriptional silencing. Biotechnol. Biofuels 2017, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.J.; Lan, E.I. Photoautotrophic synthesis of butyrate by metabolically engineered cyanobacteria. Biotechnol. Bioeng. 2019, 116, 893–903. [Google Scholar] [CrossRef]
- Higo, A.; Ehira, S. Anaerobic butanol production driven by oxygen-evolving photosynthesis using the heterocyst-forming multicellular cyanobacterium Anabaena sp. PCC 7120. Appl. Microbiol. Biotechnol. 2019, 103, 2441–2447. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; De, D.; Chaudhuri, S.; Bhattacharya, S.K. Hydrogen production by cyanobacteria. Microb. Cell Fact. 2005, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Pinto, F.A.L.; Troshina, O.; Lindblad, P. A brief look at three decades of research on cyanobacterial hydrogen evolution. Int. J. Hydrog. Energy 2002, 27, 1209–1215. [Google Scholar]
- Tamagnini, P.; Axelsson, R.; Lindberg, P.; Oxelfelt, F.; Wünschiers, R.; Lindblad, P. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ghirardi, M.L.; Posewitz, M.C.; Maness, P.C.; Dubini, A.; Yu, J.; Seibert, M. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant. Biol. 2007, 58, 71–91. [Google Scholar] [CrossRef] [PubMed]
- McNeely, K.; Xu, Y.; Bennette, N.; Bryant, D.A.; Dismukes, G.C. Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 2010, 76, 5032–5038. [Google Scholar] [CrossRef]
- Ducat, D.C.; Sachdeva, G.; Silver, P.A. Rewiring hydrogenase-dependent redox circuits in cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 3941–3946. [Google Scholar] [CrossRef] [Green Version]
- Kumaraswamy, G.K.; Guerra, T.; Qian, X.; Zhang, S.; Bryant, D.A.; Dismukes, G.C. Reprogramming the glycolytic pathway for increased hydrogen production in cyanobacteria: Metabolic engineering of NAD+-dependent GAPDH. Energy Environ. Sci. 2013, 6, 3722–3731. [Google Scholar] [CrossRef]
- Maswanna, T.; Phunpruch, S.; Lindblad, P.; Maneeruttanarungroj, C. Enhanced hydrogen production by optimization of immobilized cells of the green alga Tetraspora sp. CU2551 grown under anaerobic condition. Biomass Bioenergy 2018, 111, 88–95. [Google Scholar] [CrossRef]
- Saxena, R.K.; Anand, P.; Saran, S.; Isar, J. Microbial production of 1, 3-propanediol: Recent developments and emerging opportunities. Biotechnol. Adv. 2009, 27, 895–913. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, Y.; Maki, Y.; Tatsuke, T.; Hanai, T. Cyanobacterial production of 1, 3-propanediol directly from carbon dioxide using a synthetic metabolic pathway. Metab. Eng. 2016, 34, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, Y.; Goto, R.; Umetani, Y.; Hanai, T. Construction of a novel d-lactate producing pathway from dihydroxyacetone phosphate of the Calvin cycle in cyanobacterium, Synechococcus elongatus PCC 7942. J. Biosci. Bioeng. 2017, 124, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sheng, J.; Curtiss, R., III. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruffing, A.M. Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Front. Bioeng. Biotechnol. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Yao, L.; Gao, Q.; Wang, W.; Qi, F.; Lu, X. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 2014, 13, 169–176. [Google Scholar] [CrossRef]
- Yoshino, T.; Liang, Y.; Arai, D.; Maeda, Y.; Honda, T.; Muto, M.; Kakunaka, N.; Tanaka, T. Alkane production by the marine cyanobacterium Synechococcus sp. NKBG15041c possessing the α-olefin biosynthesis pathway. Appl. Microbiol. Biotechnol. 2015, 99, 1521–1529. [Google Scholar] [CrossRef]
- Van der Woude, A.D.; Angermayr, S.A.; Veetil, V.P.; Osnato, A.; Hellingwerf, K.J. Carbon sink removal: Increased photosynthetic production of lactic acid by Synechocystis sp. PCC6803 in a glycogen storage mutant. J. Biotechnol. 2014, 184, 100–102. [Google Scholar] [CrossRef]
- Rathnasingh, C.; Raj, S.M.; Jo, J.E.; Park, S. Development and evaluation of efficient recombinant Escherichia coli strains for the production of 3-hydroxypropionic acid from glycerol. Biotechnol. Bioeng. 2009, 104, 729–739. [Google Scholar] [PubMed]
- Werpy, T.; Petersen, G.; Aden, A.; Bozell, J.; Holladay, J.; White, J.; Manheim, A.; Eliot, D.; Lasure, L.; Jones, S. Top Value Added Chemicals from Biomass. Volume 1—Results of Screening for Potential Candidates from Sugars and Synthesis Gas (No. DOE/GO-102004-1992); Department of Energy: Washington, DC, USA, 2004. [Google Scholar]
- Lan, E.I.; Chuang, D.S.; Shen, C.R.; Lee, A.M.; Ro, S.Y.; Liao, J.C. Metabolic engineering of cyanobacteria for photosynthetic 3-hydroxypropionic acid production from CO2 using Synechococcus elongatus PCC 7942. Metab. Eng. 2015, 31, 163–170. [Google Scholar] [CrossRef]
- Oliver, J.W.; Atsumi, S. A carbon sink pathway increases carbon productivity in cyanobacteria. Metab. Eng. 2015, 29, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Badary, A.; Takamatsu, S.; Nakajima, M.; Ferri, S.; Lindblad, P.; Sode, K. Glycogen production in marine cyanobacterial strain Synechococcus sp. NKBG 15041c. Mar. Biotechnol. 2018, 20, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Klähn, S.; Hagemann, M. Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 2011, 13, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Liang, F.; Duan, Y.; Tan, X.; Lu, X. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab. Eng. 2013, 19, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.H.; Frigaard, N.U. Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 2014, 21, 60–70. [Google Scholar] [CrossRef]




| Host Strain | Engineering Strategies | Growth Conditions | Chemicals | Production (mg/L) | Refs |
|---|---|---|---|---|---|
| S. elongatus UTEX 2973 | ΔM744_RS12430:P lac - cscB -Cmr | 38 °C, 3% CO2, 250 μE m−2 s−1 light, 150 mM NaCl | Sucrose | 35.5/h | Song, et al. [15] |
| S. elongatus PCC 7942 | cscB- ΔInvA – ΔGlgC-CMr | 35°C, 2% CO2, 65 μE m−2 s−1 light, 150 mM NaCl | Sucrose | 36.1/h | Ducat et al. [16] |
| S. elongatus PCC 7942 | CscB overexpression | 32 °C, 2% CO2, ~80 μE m−2s−1 PAR | Sucrose | 28.3/d | Weiss et al. [17] |
| Synechocystis sp. PCC6803 | slr9394: Kan Prbc pdc and slr1192 slr0168: Omega Prbc pdc and slr1192 | 32 °C, 5% CO2, 100 μE m−2 s−1 light, | Ethanol | 212/d | Gao et al. [18] |
| S. elongatus PCC 7942 | NSI: Bb1s-dxs-idi-ispA NSII: k- PcpcB1-cpcB1·SF·SQS NSIII:c-PcpcB1-cpcB1·SF·SQS | 30 °C, 5% CO2, 100 μE m−2 s−1 light, 10 mM MOPS | Squalene | 7.08/OD730 | Choi et al. [19] |
| Synechococcus sp. PCC 7002 | 30 °C, 2% CO2, 600 μmol photons m−2 s−1 | Glycogen | 3500 | Aikawa et al. [20] | |
| Synechocystis sp. PCC6803 | 5′-NS Ptrc10-lims (Ms)–ter-kmR- 3′-NS | 30 °C, 2% CO2, 50 μmol photons m−2 s−1 | Limonene | 6.7 | Lin et al. [21] |
| S. elongatus PCC 7942 | NSI:Ptrc10- ls | 30 °C, 5% CO2, 100 μE m−2 s−1 light, 10 mM N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid | Limonene | 5 | Wang et al. [22] |
| Synechococcus sp. PCC 7002 | NSI:ΔglgC:LS | 37 °C, 1% CO2, 250 μmol photons m−2 s−1 | Limonene | 4 | Davies et al. [23] |
| Synechococcus sp. PCC 7002 | ΔSYNPCC7002_A2842:PtetO2-DHDPS-aacC1ΔSYNPCC7002_A2542:Pclac94-ybjE-aphII | 37 °C, 1% CO2 and 200 µmol photons m−2 s−1 | Lysine | 400 | Korosh et al. [24] |
| Synechocystis sp. PCC6803 | pEEK2-Ptrccore- kivd- ADHΔddh | 37 °C, 50 mM NaHCO3 and 50 µmol photons m−2 s−1 | Isobutanol | 600 | Miao et al. [25] |
| S. elongatus PCC 7942 | AL257+NSIII:lacIq; Ptrc: alsD-alsS-adh; gentR + NSI:lacIq; Ptrc: galP-zwf-gnd; specR +cp12: lacIq; Ptrc: prk-rbcLXS; kanR | 30 °C, glucose (10 or 15 g/L), 50 mM NaHCO3, 30 μmol photons·m−2 s−1 | 2,3-butanediol | 12,600 | Kanno et al. [26] |
| Strain | Genotype/Growth | Stress Conditions | Target | Growth System | Results | Refs. |
|---|---|---|---|---|---|---|
| Spirulina subsalsa | Industrial wastewater (25%) | Protein | batch reactors | 166.20 mg L−1d−1 | Jiang et al. [74] | |
| Industrial wastewater (25%) | Lipid | 64.23 mg L−1d−1 | ||||
| Industrial wastewater (50%) | Carbohydrates | 48.98 mg L−1d−1 | ||||
| Synechocystis sp. PCC 6803 | 30 °C, 1% (v/v) CO2, 50–70 μmol photons m−2 s−1 | Artificial Sea water + Nitrogen + Phosphorus | Glycogen | Closed | Iijima et al. [75] | |
| Artificial Sea water + NPHEPHES media | ||||||
| BGG-11 media | ||||||
| Synechocystis sp. PCC 6803 | 28 °C, 150 μmol photons m−2 s−1 | pH-7.5 | Growth | Continuous culture | 12.1 mg L−1d−1 | Touloupakis et al. [76] |
| pH-8.5 | 11.7 mg L−1d−1 | |||||
| pH-9.5 | 11.8 mg L−1d−1 | |||||
| pH-10.0 | 11.5 mg L−1d−1 | |||||
| pH-10.5 | 10.6 mg L−1d−1 | |||||
| pH-11.0 | 8.2 mg L−1d−1 | |||||
| Synechocystis Syn-HZ24 | pH-11+ NaCl (300mM) | Ethanol | Closed | 0.9 g/L | Zhu et al. [77] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.Z.; Bilal, M.; Mehmood, S.; Sharma, A.; Iqbal, H.M.N. State-of-the-Art Genetic Modalities to Engineer Cyanobacteria for Sustainable Biosynthesis of Biofuel and Fine-Chemicals to Meet Bio–Economy Challenges. Life 2019, 9, 54. https://doi.org/10.3390/life9030054
Khan AZ, Bilal M, Mehmood S, Sharma A, Iqbal HMN. State-of-the-Art Genetic Modalities to Engineer Cyanobacteria for Sustainable Biosynthesis of Biofuel and Fine-Chemicals to Meet Bio–Economy Challenges. Life. 2019; 9(3):54. https://doi.org/10.3390/life9030054
Chicago/Turabian StyleKhan, Aqib Zafar, Muhammad Bilal, Shahid Mehmood, Ashutosh Sharma, and Hafiz M. N. Iqbal. 2019. "State-of-the-Art Genetic Modalities to Engineer Cyanobacteria for Sustainable Biosynthesis of Biofuel and Fine-Chemicals to Meet Bio–Economy Challenges" Life 9, no. 3: 54. https://doi.org/10.3390/life9030054
APA StyleKhan, A. Z., Bilal, M., Mehmood, S., Sharma, A., & Iqbal, H. M. N. (2019). State-of-the-Art Genetic Modalities to Engineer Cyanobacteria for Sustainable Biosynthesis of Biofuel and Fine-Chemicals to Meet Bio–Economy Challenges. Life, 9(3), 54. https://doi.org/10.3390/life9030054








