Microbial Ecology of Biofiltration Units Used for the Desulfurization of Biogas
Abstract
:1. Introduction
2. The Biological Sulfur Cycle and the Sulfur-Oxidizing Bacteria
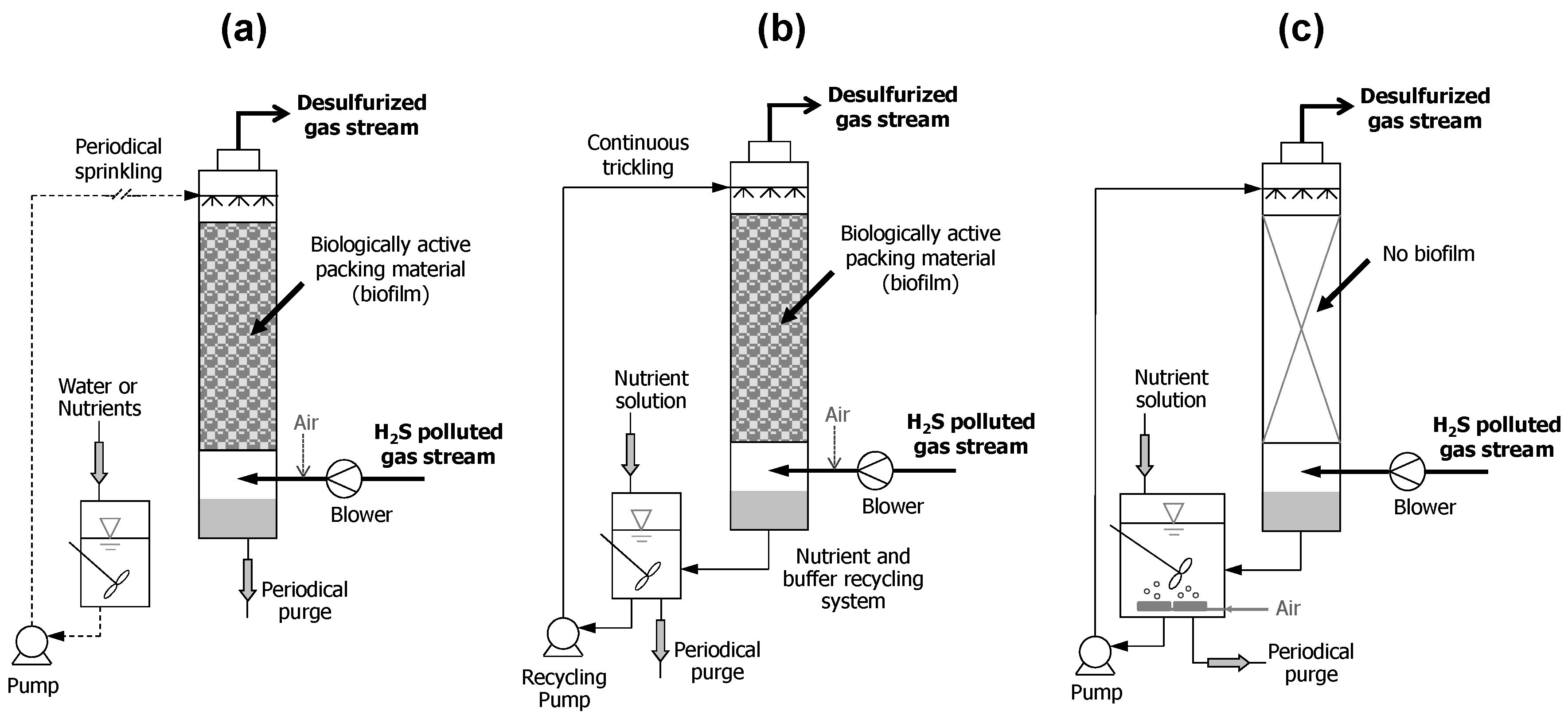
3. Biofiltration Technologies
4. Microbial Ecology Studies in Biofiltration Units for H2S Removal
4.1. Molecular Techniques for Characterizing Bacterial Communities in Biofilters
4.2. Aerobic Biofiltration
4.3. Anoxic Biofiltration
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Yentekakis, I.V.; Grammatiki, G. Biogas management: Advanced utilization for production of renewable energy and added-value chemicals. Front. Environ. Sci. 2017, 5, 7. [Google Scholar] [CrossRef]
- Plugge, C.M. Biogas. Microb. Biotechnol. 2017, 10, 1128–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biogas Renewable Energy. Available online: http://www.biogas-renewable-energy.info (accessed on 23 May 2019).
- Arellano, L.; Dorado, A.D.; Fortuny, M.; Gabriel, D.; Gamisans, X.; González-Sánchez, A.; Hernández, S.; Lafuente, J.; Monroy, O.; Mora, M.; et al. Purificación y Usos del Biogás, 1st ed.; Universitat Autònoma de Barcelona: Barcelona, Spain, 2017; pp. 25–27. [Google Scholar]
- Muñoz, R.; Meier, L.; Diaz, I.; Jeison, D. A review on the state-of-the-art of physical/chemical and biological technologies for biogas upgrading. Rev. Environ. Sci. Biotechnol. 2015, 14, 727–759. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ye, G.; Sun, D.; Sun, G.; Zeng, X.; Xu, J.; Liang, S. Performances of two biotrickling filters in treating H₂S-containing waste gases and analysis of corresponding bacterial communities by pyrosequencing. Appl. Microbiol. Biotechnol. 2012, 95, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Syed, M.; Soreanu, G.; Falletta, P.; Béland, M. Removal of hydrogen sulfide from gas streams using biological processes—A review. Can. Biosyst. Eng. 2006, 48, 2. [Google Scholar]
- Niesner, J.; Jecha, D.; Stehlik, P. Biogas upgrading technologies: State of the art review in European region. Chem. Eng. Trans. 2013, 35, 517–522. [Google Scholar]
- Chung, W.J.; Griebel, J.J.; Kim, E.T.; Yoon, H.; Simmonds, A.G.; Ji, H.J.; Dirlam, P.T.; Glass, R.S.; Wie, J.J.; Nguyen, N.A. The use of elemental sulfur as an alternative feedstock for polymeric materials. Nat. Chem. 2013, 5, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Vikrant, K.; Kailasa, S.K.; Tsang, D.C.W.; Lee, S.S.; Kumar, P.; Giri, B.S.; Singh, R.S.; Kim, K.-H. Biofiltration of hydrogen sulfide: Trends and challenges. J. Clean. Prod. 2018, 187, 131–147. [Google Scholar] [CrossRef]
- Tortora, G.J.; Funke, B.R.; Case, C.L. Microbiology: An Introduction, 13th ed.; Pearson: New York, NY, USA, 2019. [Google Scholar]
- Maier, R.M. Biogeochemical cycling. In Environmental Microbiology, 3rd ed.; Pepper, I.L., Gerba, C.P., Gentry, T.J., Eds.; Elsevier: San Diego, CA, USA, 2015; pp. 339–373. [Google Scholar]
- Lens, P.N.; Kuenen, J.G. The biological sulfur cycle: Novel opportunities for environmental biotechnology. Water Sci. Technol. 2001, 44, 57–66. [Google Scholar] [CrossRef]
- Moestedt, J.; Påledal, S.N.; Schnürer, A. The effect of substrate and operational parameters on the abundance of sulphate-reducing bacteria in industrial anaerobic biogas digesters. Bioresour. Technol. 2013, 132, 327–332. [Google Scholar] [CrossRef] [Green Version]
- Vergara-Fernández, A.; Scott, F.; Moreno-Casas, P.; Revah, S. Removal of gaseous pollutants from air by fungi. In Fungal Bioremediation: Fundamentals and Applications; Tomasini, A., León-Santiesteban, H.H., Eds.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2019; pp. 264–284. [Google Scholar]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Stahl, D.A. Brock Biology of Microorganisms, 15th ed.; Pearson: New York, NY, USA, 2019. [Google Scholar]
- Willey, J.M.; Sherwood, L.M.; Woolverton, C.J. Prescott’s Microbiology, 10th ed.; Mc Graw Hill Education: New York, NY, USA, 2017. [Google Scholar]
- Dahl, C.; Friedrich, C.; Kletzin, A. Sulfur oxidation in prokaryotes. In Encyclopedia of Life Sciences (ELS); John Wiley & Sons, Ltd.: Chichester, UK, 2008; Available online: http://www.els.net/10.1002/9780470015902.a0021155 (accessed on 23 May 2019).
- Tang, K.; Baskaran, V.; Nemati, M. Bacteria of the sulphur cycle: An overview of microbiology, biokinetics and their role in petroleum and mining industries. Biochem. Eng. J. 2009, 44, 73–94. [Google Scholar] [CrossRef]
- Dopson, M.; Johnson, D.B. Biodiversity, metabolism and applications of acidophilic sulfur-metabolizing microorganisms. Environ. Microbiol. 2012, 14, 2620–2631. [Google Scholar] [CrossRef] [PubMed]
- Huber, B.; Herzog, B.; Drewes, J.E.; Koch, K.; Müller, E. Characterization of sulfur oxidizing bacteria related to biogenic sulfuric acid corrosion in sludge digesters. BMC Microbiol. 2016, 16, 153. [Google Scholar] [CrossRef] [PubMed]
- Czaja, A.-D.; Beukes, N.J.; Osterhout, J.T. Sulfur-oxidizing bacteria prior to the Great Oxidation Event from the 2.52 Ga Gamohaan Formation of South Africa. Geology 2016, 44, 983–986. [Google Scholar] [CrossRef] [Green Version]
- Forte, E.; Giuffrè, A. How Bacteria Breathe in Hydrogen Sulfide-Rich Environments. Biochem. Soc. 2016, 38, 8–11. Available online: http://www.biochemist.org/bio/03805/0008/038050008.pdf (accessed on 23 May 2019).
- Mohapatra, B.R.; Gould, W.D.; Dinardo, O.; Koren, D.W. An overview of the biochemical and molecular aspects of microbial oxidation of inorganic sulfur compounds. CLEAN 2008, 36, 823–829. [Google Scholar] [CrossRef]
- Pokorna, D.; Zabranska, J. Sulfur-oxidizing bacteria in environmental technology. Biotechnol. Adv. 2015, 33, 1246–1259. [Google Scholar] [CrossRef]
- Lin, S.; Mackey, H.R.; Hao, T.; Guo, G.; van Loosdrecht, M.C.M.; Chen, G. Biological sulfur oxidation in wastewater treatment: A review of emerging opportunities. Water Res. 2018, 143, 399–415. [Google Scholar] [CrossRef]
- Labrenz, M.; Grote, J.; Mammitzsch, K.; Boschker, H.T.; Laue, M.; Jost, G.; Glaubitz, S.; Jürgens, K. Sulfurimonas gotlandica sp. nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2013, 63, 4141–4148. [Google Scholar] [CrossRef]
- Schulz, H.N.; Jorgensen, B.B. Big bacteria. Annu. Rev. Microbiol. 2001, 55, 105–137. [Google Scholar] [CrossRef]
- Williams, T.M.; Unz, R.F.; Doman, J.T. Ultrastructure of Thiothrix spp. and “Type 021N” bacteria. Appl. Environ. Microbiol. 1987, 53, 1560–1570. [Google Scholar] [PubMed]
- Sorokin, D.Y.; Kuenen, J.G. Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol. Rev. 2005, 29, 685–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prange, A.; Chauvistré, R.; Modrow, H.; Hormes, J.; Trüper, H.G.; Dahl, C. Quantitative speciation of sulfur in bacterial sulfur globules: X-ray absorption spectroscopy reveals at least three different species of sulfur. Microbiology 2002, 148, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, M.F.; Zhang, T.; Fang, H.H. Sulfur-driven autotrophic denitrification: Diversity, biochemistry, and engineering applications. Appl. Microbiol. Biotechnol. 2010, 88, 1027–1042. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, W.; Dam, B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 2009, 33, 999–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, F.J.; Dmytrenko, O.; Delong, E.F.; Cavanaugh, C.M. Metatranscriptomic analysis of sulfur oxidation genes in the endosymbiont of Solemya velum. Front. Microbiol. 2011, 2, 134. [Google Scholar] [CrossRef] [PubMed]
- Harada, M.; Yoshida, T.; Kuwahara, H.; Shimamura, S.; Takaki, Y.; Kato, C.; Miwa, T.; Miyake, H.; Maruyama, T. Expression of genes for sulfur oxidation in the intracellular chemoautotrophic symbiont of the deep-sea bivalve Calyptogena okutanii. Extremophiles 2009, 13, 895–903. [Google Scholar] [CrossRef]
- Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Yang, C.L.; Gao, X.Y.; Lin, C.M.; Li, Y.Q.; Li, Y.; et al. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 2019, 9, 3290. [Google Scholar] [CrossRef]
- Muyzer, G.; Sorokin, D.Y.; Mavromatis, K.; Lapidus, A.; Clum, A.; Ivanova, N.; Pati, A.; d’Haeseleer, P.; Woyke, T.; Kyrpides, N.C. Complete genome sequence of “Thioalkalivibrio sulfidophilus” HL-EbGr7. Stand. Genom. Sci. 2011, 4, 23–35. [Google Scholar] [CrossRef]
- Muyzer, G.; Sorokin, D.Y.; Mavromatis, K.; Lapidus, A.; Foster, B.; Sun, H.; Ivanova, N.; Pati, A.; D’haeseleer, P.; Woyke, T.; et al. Complete genome sequence of Thioalkalivibrio sp. K90mix. Stand. Genom. Sci. 2011, 5, 341–355. [Google Scholar] [CrossRef]
- Sievert, S.M.; Scott, K.M.; Klotz, M.G.; Chain, P.S.; Hauser, L.J.; Hemp, J.; Hügler, M.; Land, M.; Lapidus, A.; Larimer, F.W.; et al. Genome of the Epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 2008, 74, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Omri, I.; Bouallagui, H.; Aouidi, F.; Godon, J.J.; Hamdi, M. H2S gas biological removal efficiency and bacterial community diversity in biofilter treating wastewater odor. Bioresour. Technol. 2011, 102, 10202–10209. [Google Scholar] [CrossRef]
- Muyzer, G.; Kuenen, J.G.; Robertson, L.A. Colorless sulfur bacteria. In The Prokaryotes; Springer: New York, NY, USA, 2013; pp. 555–588. [Google Scholar]
- Takai, K.; Suzuki, M.; Nakagawa, S.; Miyazaki, M.; Suzuki, Y.; Inagaki, F.; Horikoshi, K. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteo-bacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2006, 56, 1725–1733. [Google Scholar] [PubMed]
- Kellermann, C.; Griebler, C. Thiobacillus thiophilus sp. nov., a chemolithoautotrophic, thiosulfate-oxidizing bacterium isolated from contaminated aquifer sediments. Int. J. Syst. Evol. Microbiol. 2009, 59, 583–588. [Google Scholar] [CrossRef]
- Shapleigh, J.P. Denitrifying prokaryotes. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2013; pp. 405–425. [Google Scholar]
- Sorokin, D.Y.; Kuenen, J.G.; Jetten, M.S. Denitrification at extremely high pH values by the alkaliphilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium Thioalkalivibrio denitrificans strain ALJD. Arch. Microbiol. 2001, 175, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.A.; Kuenen, J.G. The colorless sulfur bacteria. In The Prokaryotes. A Handbook on the Biology of Bacteria; Springer: New York, NY, USA, 2006; pp. 985–1011. [Google Scholar]
- Cardoso, R.B.; Sierra-Alvarez, R.; Rowlette, P.; Flores, E.R.; Gómez, J.; Field, J.A. Sulfide oxidation under chemolithoautotrophic denitrifying conditions. Biotechnol. Bioeng. 2006, 95, 1148–1157. [Google Scholar] [CrossRef]
- Mahmood, Q.; Zheng, P.; Cai, J.; Wu, D.; Hu, B.; Islam, E.; Azim, M.R. Comparison of anoxic sulfide biooxidation using nitrate/nitrite as electron acceptor. Environ. Prog. Sustain. Energy 2007, 26, 169–177. [Google Scholar] [CrossRef]
- An, S.; Tang, K.; Nemati, M. Simultaneous biodesulphurization and denitrification using an oil reservoir microbial culture: Effects of sulphide loading rate and sulphide to nitrate loading ratio. Water Res. 2010, 44, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Aita, B.C.; Mayer, F.D.; Muratt, D.T.; Brondani, M.; Pujol, S.B.; Denardi, L.B.; Hoffmann, R.; da Silveria, D.D. Biofiltration of H2S-rich biogas using Acidithiobacillus thiooxidans. Clean Technol. Environ. Policy 2016, 18, 689–703. [Google Scholar] [CrossRef]
- Aizpuru, A.; Khammar, N.; Malhautier, L.; Fanlo, J. Biofiltration for the treatment of complex mixtures of VOC–influence of the packing material. Eng. Life Sci. 2003, 23, 211–226. [Google Scholar] [CrossRef]
- Khammar, N.; Malhautier, L.; Degrange, V.; Lensi, R.; Fanlo, J. Evaluation of dispersion methods for enumeration of microorganisms from peat and activated carbon biofilters treating volatile organic compounds. Chemosphere 2004, 54, 243–254. [Google Scholar] [CrossRef]
- Mudliar, S.; Giri, B.; Padoley, K.; Satpute, D.; Dixit, R.; Bhatt, P.; Pandey, R.; Juwarkar, A.; Vaidya, A. Bioreactors for treatment of VOCs and odours—A review. J. Environ. Manag. 2010, 91, 1039–1054. [Google Scholar] [CrossRef] [PubMed]
- Jaber, M.B.; Anet, B.; Amrane, A.; Couriol, C.; Lendormi, T.; Le Cloirec, P.; Cogny, G.; Fillières, R. Impact of nutrients supply and pH changes on the elimination of hydrogen sulfide, dimethyl disulfide and ethanethiol by biofiltration. Chem. Eng. J. 2014, 258, 420–426. [Google Scholar] [CrossRef]
- Rabbani, K.A.; Charles, W.; Kayaalp, A.; Cord-Ruwisch, R.; Ho, G. Pilot-scale biofilter for the simultaneous removal of hydrogen sulphide and ammonia at a wastewater treatment plant. Biochem. Eng. J. 2016, 107, 1–10. [Google Scholar] [CrossRef]
- Ben Jaber, M.; Couvert, A.; Amrane, A.; Le Cloirec, P.; Dumont, E. Removal of hydrogen sulfide in air using cellular concrete waste: Biotic and abiotic filtrations. Chem. Eng. J. 2017, 319, 268–278. [Google Scholar] [CrossRef]
- Barbusinski, K.; Kalemba, K.; Kasperczyk, D.; Urbaniec, K.; Kozik, V. Biological methods for odor treatment—A review. J. Clean. Prod. 2017, 152, 223–241. [Google Scholar] [CrossRef]
- Jin, J.; Veiga, M.C.; Kennes, C. Autotrophic deodorization o hydrogen sulfide in a biotrickling filter. J. Chem. Technol. Biotechnol. 2005, 80, 998–1004. [Google Scholar] [CrossRef]
- Allegue, L.B.; Hinge, J. Report: Biogas and Bio-Syngas Upgrading; Danish Technological Institute: Taastrup, Denmark, 2012; pp. 1–97. [Google Scholar]
- Iliuta, I.; Iliuta, M.C.; Larachi, F. Hydrodynamics modeling of bioclogging in waste gas treating trickle-bed bioreactors. Ind. Eng. Chem. Res. 2005, 44, 5044–5052. [Google Scholar] [CrossRef]
- Rybarczyk, P.; Szulczynski, B.; Gebicki, J.; Hupka, J. Treatment of malodorous air in biotrickling filters: A review. Biochem. Eng. J. 2018, 141, 146–162. [Google Scholar] [CrossRef]
- Cox, H.H.; Deshusses, M.A. Co-treatment of H2S and toluene in a biotrickling filter. Chem. Eng. J. 2002, 87, 101–110. [Google Scholar] [CrossRef]
- Lee, E.Y.; Lee, N.Y.; Cho, K.; Ryu, H.W. Removal of hydrogen sulfide by sulfate-resistant Acidithiobacillus thiooxidans AZ11. J. Biosci. Bioeng. 2006, 101, 309–314. [Google Scholar] [CrossRef]
- Gabriel, D.; Deshusses, M.A.; Gamisans, X. Desulfurization of biogas in biotrickling filters. In Air Pollution Prevention and Control; John Wiley & Sons, Ltd.: Chichester, UK, 2013; pp. 513–523. [Google Scholar]
- Almenglo, F.; Bezerra, T.; Lafuente, J.; Gabriel, D.; Ramirez, M.; Cantero, D. Effect of gas-liquid flow pattern and microbial diversity analysis of a pilot-scale biotrickling filter for anoxic biogas desulfurization. Chemosphere 2016, 157, 215–223. [Google Scholar] [CrossRef]
- Lebrero, R.; Toledo-Cervantes, A.; Muñoz, R.; del Nery, V.; Foresti, E. Biogas upgrading from vinasse digesters: A comparison between an anoxic biotrickling filter and an algal-bacterial photobioreactor. J. Chem. Technol. Biotechnol. 2016, 91, 2488–2495. [Google Scholar] [CrossRef]
- Ben Jaber, M.; Couvert, A.; Amrane, A.; Le Cloirec, P.; Dumont, E. Hydrogen sulfide removal from a biogas mimic by biofiltration under anoxic conditions. J. Environ. Chem. Eng. 2017, 5, 5617–5623. [Google Scholar] [CrossRef]
- Khanongnuch, R.; Di Capua, F.; Lakaniemi, A.-M.; Rene, E.R.; Lens, P.N.L. H2S removal and microbial community composition in an anoxic biotrickling filter under autotrophic and mixotrophic conditions. J. Hazard. Mater. 2019, 367, 397–406. [Google Scholar] [CrossRef]
- Lebrero, R.; Bouchy, L.; Stuetz, R.; Muñoz, R. Odor assessment and management in wastewater treatment plants: A review. Crit. Rev. Environ. Sci. Technol. 2011, 41, 915–950. [Google Scholar] [CrossRef]
- Gabriel, D.; Gamisans, X.; Muñoz, R. Technologies limiting gas and odor emissions. In Innovative Wastewater Treatment and Resource Recovery Technologies: Impacts on Energy, Economy and Environment; IWA Publishing: London, UK, 2017; pp. 233–254. [Google Scholar]
- Okabe, S.; Ito, T.; Sugita, K.; Satoh, H. Succession of internal sulfur cycles and sulfur-oxidizing bacterial communities in microaerophilic wastewater biofilms. Appl. Environ. Microbiol. 2005, 71, 2520–2529. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
- Pace, N.R. A molecular view of microbial diversity and the biosphere. Science 1997, 276, 734–740. [Google Scholar] [CrossRef]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar]
- Pace, N.R.; Stahl, D.A.; Olsen, G.J. Analyzing natural microbial populations by rRNA sequences. ASM News 1985, 51, 4–12. [Google Scholar]
- Smalla, K.; Oros-Sichler, M.; Milling, A.; Heuer, H.; Baumgarte, S.; Becker, R.; Neuber, G.; Kropf, S.; Ulrich, A.; Tebbe, C.C. Bacterial diversity of soils assessed by DGGE, T-RFLP and SSCP fingerprints of PCR-amplified 16S rRNA gene fragments: Do the different methods provide similar results? J. Microbiol. Methods 2007, 69, 470–479. [Google Scholar] [CrossRef]
- Reysenbach, A.L.; Wickham, G.S.; Pace, N.R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 1994, 60, 2113–2119. [Google Scholar]
- Wen, C.; Wu, L.; Qin, Y.; Van Nostrand, J.D.; Ning, D.; Sun, B.; Xue, K.; Liu, F.; Deng, Y.; Liang, Y.; et al. Evaluation of the reproducibility of amplicon sequencing with Illumina MiSeq platform. PLoS ONE 2017, 12, e0176716. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kubota, K. Visualization of microorganisms in bioprocesses. In Optimization and Applicability of Bioprocesses; Purohit, H.J., Kalia, V.C., Vaidya, A.N., Khardenavis, A.A., Eds.; Springer: Singapore, 2017; pp. 13–26. [Google Scholar]
- Sercu, B.; Núñez, D.; Van Langenhove, H.; Aroca, G.; Verstraete, W. Operational and microbiological aspects of a bioaugmented two-stage biotrickling filter removing hydrogen sulfide and dimethyl sulfide. Biotechnol. Bioeng. 2005, 90, 259–269. [Google Scholar] [CrossRef]
- Ramirez, M.; Gómez, J.M.; Cantero, D.; Páca, J.; Halecký, M.; Kozliak, E.I.; Sobotka, M. Hydrogen sulfide removal from air by Acidithiobacillus thiooxidans in a trickle bed reactor. Folia Microbiol. 2009, 54, 409–414. [Google Scholar] [CrossRef]
- Ramírez, M.; Fernández, M.; Granada, C.; Le Borgne, S.; Gómez, J.M.; Cantero, D. Biofiltration of reduced sulphur compounds and community analysis of sulphur-oxidizing bacteria. Bioresour. Technol. 2011, 102, 4047–4053. [Google Scholar] [CrossRef] [Green Version]
- Maestre, J.P.; Rovira, R.; Gamisans, X.; Kinney, K.A.; Kirisits, M.J.; Lafuente, J.; Gabriel, D. Characterization of the bacterial community in a biotrickling filter treating high loads of H2S by molecular biology tools. Water Sci. Technol. 2009, 59, 1331–1337. [Google Scholar] [CrossRef]
- Fortuny, M.; Baeza, J.A.; Gamisans, X.; Casas, C.; Lafuente, J.; Deshusses, M.A.; Gabriel, D. Biological sweetening of energy gases mimics in biotrickling filters. Chemosphere 2008, 71, 10–17. [Google Scholar] [CrossRef]
- Henriet, O.; Meunier, C.; Henry, P.; Mahillon, J. Filamentous bulking caused by Thiothrix species is efficiently controlled in full-scale wastewater treatment plants by implementing a sludge densification strategy. Sci. Rep. 2017, 7, 1430. [Google Scholar] [CrossRef]
- Han, Y.; Perner, M. The globally widespread genus Sulfurimonas: Versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 2015, 6, 989. [Google Scholar] [CrossRef]
- Beller, H.R.; Chain, P.S.; Letain, T.E.; Chakicherla, A.; Larimer, F.W.; Richardson, P.M.; Coleman, M.A.; Wood, A.P.; Kelly, D.P. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 2006, 188, 1473–1488. [Google Scholar] [CrossRef]
- Macalady, J.L.; Dattagupta, S.; Schaperdoth, I.; Jones, D.S.; Druschel, G.K.; Eastman, D. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2008, 2, 590–601. [Google Scholar] [CrossRef]
- Maestre, J.P.; Rovira, R.; Alvarez-Hornos, F.J.; Fortuny, M.; Lafuente, J.; Gamisans, X.; Gabriel, D. Bacterial community analysis of a gas-phase biotrickling filter for biogas mimics desulfurization through the rRNA approach. Chemosphere 2010, 80, 872–880. [Google Scholar] [CrossRef]
- Moreira, D.; Amils, R. Phylogeny of Thiobacillus cuprinus and other mixotrophic Thiobacilli: Proposal for Thiomonas gen. nov. Int. J. Syst. Evol. Microbiol. 1997, 47, 522–528. [Google Scholar] [CrossRef]
- Chen, X.G.; Geng, A.L.; Yan, R.; Gould, W.D.; Ng, Y.L.; Liang, D.T. Isolation and characterization of sulphur-oxidizing Thiomonas sp. and its potential application in biological deodorization. Lett. Appl. Microbiol. 2004, 39, 495–503. [Google Scholar] [CrossRef]
- Chung, Y.-C.; Huang, C.-P.; Pan, J.R.; Tseng, C.-P. Comparison of autotrophic and mixotrophic biofilters for H2S removal. J. Environ. Eng. ASCE 1998, 124, 362–367. [Google Scholar] [CrossRef]
- Campbell, B.J.; Engel, A.S.; Porter, M.L.; Takai, K. The versatile ε-proteobacteria: Key players in sulphidic habitats. Nat. Rev. Microbiol. 2006, 4, 458–468. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov (accessed on 23 May 2019).
- Okabe, S.; Odagiri, M.; Ito, T.; Satoh, H. Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems. Appl. Environ. Microbiol. 2007, 73, 971–980. [Google Scholar] [CrossRef]
- Goncalves, J.J.; Govind, R. Enhanced biofiltration using cell attachment promotors. Environ. Sci. Technol. 2009, 43, 1049–1054. [Google Scholar] [CrossRef]
- Dec, W.; Cwalina, B.; Michalska, J.; Merkuda, D. Growth of Acidithiobacillus thiooxidans biofilm on glass, concrete and stoneware. Solid State Phenom. 2015, 227, 286–289. [Google Scholar] [CrossRef]
- Chouari, R.; Dardouri, W.; Sallami, F.; Ben Rais, M.; Le Paslier, D.; Sghir, A. Microbial analysis and efficiency of biofiltration packing systems for hydrogen sulfide removal from wastewater off gas. Environ. Eng. Sci. 2015, 32, 121–128. [Google Scholar] [CrossRef]
- Dul’tseva, N.M.; Turova, T.P.; Spiridonova, E.M.; Kolganova, T.V.; Osipov, G.A.; Gorlenko, V.M. Thiobacillus sajanensis sp. nov., a new obligately autotrophic sulfur-oxidizing bacterium isolated from Khoito-Gol hydrogen-sulfide springs, Buryatia. Mikrobiologiia 2006, 75, 670–681. [Google Scholar]
- Lebrero, R.; Rodríguez, E.; Estrada, J.M.; García-Encina, P.A.; Muñoz, R. Odor abatement in biotrickling filters: Effect of the EBRT on methyl mercaptan and hydrophobic VOCs removal. Bioresour. Technol. 2012, 109, 38–45. [Google Scholar] [CrossRef]
- Montebello, A.M.; Bezerra, T.; Rovira, R.; Rago, L.; Lafuente, J.; Gamisans, X.; Campoy, S.; Baeza, M.; Gabriel, D. Operational aspects, pH transition and microbial shifts of a H2S desulfurizing biotrickling filter with random packing material. Chemosphere 2013, 93, 2675–2682. [Google Scholar] [CrossRef]
- Norlund, K.L.; Southam, G.; Tyliszczak, T.; Hu, Y.; Karunakaran, C.; Obst, M.; Hitchcock, A.P.; Warren, L.A. Microbial architecture of environmental sulfur processes: A novel syntrophic sulfur-metabolizing consortia. Environ. Sci. Technol. 2009, 43, 8781–8796. [Google Scholar] [CrossRef]
- Li, X.; Kappler, U.; Jiang, G.; Bond, P.L. The ecology of acidophilic microorganisms in the corroding concrete sewer environment. Front. Microbiol. 2017, 8, 683. [Google Scholar] [CrossRef]
- Tu, X.; Li, J.; Feng, R.; Sun, G.; Guo, J. Comparison of removal behavior of two biotrickling filters under transient condition and effect of pH on the bacterial communities. PLoS ONE 2016, 11, e0155593. [Google Scholar] [CrossRef]
- Fernández, M.; Ramírez, M.; Pérez, R.M.; Gómez, J.M.; Cantero, D. Hydrogen sulphide removal from biogas by an anoxic biotrickling filter packed with Pall rings. Chem. Eng. J. 2013, 225, 456–463. [Google Scholar] [CrossRef]
- Flood, B.E.; Jones, D.S.; Bailey, J.V. Sedimenticola thiotaurini sp. nov., a sulfur-oxidizing bacterium isolated from salt marsh sediments, and emended descriptions of the genus Sedimenticola and Sedimenticola selenatireducens. Int. J. Syst. Evol. Microbiol. 2015, 65, 2522–2530. [Google Scholar] [CrossRef]
- Cytryn, E.; van Rijn, J.; Schramm, A.; Gieseke, A.; de Beer, D.; Minz, D. Identification of bacteria potentially responsible for oxic and anoxic sulfide oxidation in biofilters of a recirculating mariculture system. Appl. Environ. Microbiol. 2005, 71, 6134–6141. [Google Scholar] [CrossRef]
- Bengtsson, J.; Eriksson, K.M.; Hartmann, M.; Wang, Z.; Shenoy, B.D.; Grelet, G.A.; Abarenkov, K.; Petri, A.; Rosenblad, M.A.; Nilsson, R.H. Metaxa: A software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie Leeuwenhoek 2011, 100, 471. [Google Scholar] [CrossRef]
- Valle, A.; Fernández, M.; Ramírez, M.; Rovira, R.; Gabriel, D.; Cantero, D. A comparative study of eubacterial communities by PCR-DGGE fingerprints in anoxic and aerobic biotrickling filters used for biogas desulfurization. Bioprocess Biosyst. Eng. 2018, 41, 1165–1175. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Bezsoudnova, E.Y.; Pol, A.; Muyzer, G. Denitrification in a binary culture and thiocyanate metabolism in Thiohalophilus thiocyanoxidans gen. nov. sp. nov.—A moderately halophilic chemolithoautotrophic sulfur-oxidizing Gammaproteobacterium from hypersaline lakes. Arch. Microbiol. 2007, 187, 441–450. [Google Scholar] [CrossRef]
- Brito, J.; Valle, A.; Almenglo, F.; Ramírez, M.; Cantero, D. Progressive change from nitrate to nitrite as the electron acceptor for the oxidation of H2S under feedback control in an anoxic biotrickling filter. Biochem. Eng. J. 2018, 139, 154–161. [Google Scholar] [CrossRef]


| Biogas From | H2S (ppm) |
|---|---|
| Wastewater AD plants | 0–4000 |
| Household waste | 72–648 |
| Agrifood industry | 288 |
| Agricultural waste | 2160–7200 |
| Landfill sites | 0–100 |
| Natural gas $ | 1.1–5.9 |
| SOB | Optimum pH Range | Anaerobic/Aerobic | Sulfur Oxidation Genes or Enzymes |
|---|---|---|---|
| Thiobacillus denitrificans | 6.8–7.4 | AN/AE | sqr, fcc, sox without soxCD, dsr, apr |
| Acidithiobacillus spp. | 2–2.5 | AN/AE * | tet, tqo, sqr, sdo, tst, hdr, sox without soxCD * |
| Thioalkalivibrio | 9–10 | AN/AE | fcc, sox without soxCD, hdr, dsr & |
| Sulfurimonas denitrificans | 7 | AN/AE | sox, sqr # |
| Conventional Biofilters | BTFs | Bioscrubbers | |
|---|---|---|---|
| Advantages |
|
|
|
| Drawbacks |
|
|
|
| Operational Behavior | ||||||
| Biofilter | BTFa (pH 4) | BTFn (pH 7) | ||||
| EBRT (s) | 60 | 30 | 15 | 60 | 30 | 15 |
| Maximum RE at the inlet (%) | 99 | 95 | 70 | 87.5 | 90 | 65 |
| Average pH and Bacterial Populations | ||||||
| Layer | Upper | Middle | Bottom | Upper | Middle | Bottom |
| pH | 4.04 | 2.79 | 1.83 | 7.19 | 4.97 | 2.03 |
| Abundance of β-proteobacteria (%) | 20.0 | 32.9 | 23.6 | 25.2 | 29.9 | 19.1 |
| Abundance of β-proteobacteria (%) | 31.6 | 29.4 | 46.7 | 13.1 | 18.1 | 32.7 |
| Application | Scale | Process Type | Inlet Gas | H2S Load (ppmv) | Packing Material | Inoculum | Molecular Technique | Main Outcome from Microbial Ecology Studies | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Odor abatement | Lab |
| Air supplemented with pure H2S and DMS | 1220–4037 | Polyethylene carrier rings | A.t | DGGE |
| [80] |
| Odor abatement | Lab |
| Air supplemented with H2S and other organic RSC | 23–1320 | Polyurethane foam | A.t | DGGE |
| [82] |
| Biogas desulfurization | Lab |
| Biogas mimic (mixture of H2S, N2 and air) | 2000 | High density polypropylene grids | Culture from full-scale biogas desulfurization column (pH 1.6) adapted to pH 6 |
|
| [83] [89] |
| Biogas desulfurization | Lab | Aerobic BTF with pH control at 6.5–7 in the recirculating liquid | Biogas mimic (mixture of H2S, N2 and air) | 2000 | High density polypropylene grids | Aerobic sludge from a municipal WWTP at pH | |||
| Odor abatement | Pilot |
| Waste gases from municipal WWTP | 2.037 |
| Activated sludge from municipal WWTP |
|
| [6] [98] |
| Odor abatement | Large |
| Used air from stabilizer or primary decanter of WWTP | >500 |
| Not described | |||
| Odor abatement | Pilot |
| Odorous gas from a WWTP | ≅163–815 | Peat | “Self-inoculated” | SSCP |
| [40] |
| Biogas desulfurization | Lab |
| Reference synthetic gas | 2000 | Steel pall rings | Aerobic sludge from a local municipal WWTP | 16S rRNA gene amplicons pyrosequencing |
| [101] |
| Odor abatement | Bench |
| Synthetic polluted gases generated by mixing H2S vapors with fresh air | 121–4200 | Volcanic rock | Microbial consortium from biofilter treating landfill leachate waste gases + activated sludge from WWTP | MiSeq sequencing of 16S rRNA gene amplicons |
| [104] |
| Biogas desulfurization | Lab |
| Biogas from UASB reactor | 1400–14,000 | Polypropylene Pall rings | Biomass from open-pore polyurethane foam of a previous BTF | DGGE | Specialized bacterial community | [105] |
| Biogas desulfurization | Pilot |
| Biogas split from anaerobic digester from WWTP | 4490 | Open-pore polyurethane foam | Wastewater from degritter-degreasing unit of WWTP | 16S rRNA gene amplicons pyrosequencing |
| [66] |
| Biogas desulfurization | Lab |
| Biogas from UASB reactor | Not reported |
| Biomass from open-pore polyurethane foam of a previous BTF | DGGE | No influence of the packing material and operation time on bacterial diversity | [109] |
| Biogas desulfurization | Lab |
| Synthetic biogas (N2 and H2S) | 710–3564 | Polypropylene Pall rings | Not described | DGGE | Bacterial diversity reduced during the progressive adaptation from NO3− to NO2− | [111] |
| Odor abatement | Lab |
| Mixture of N2 gas and H2S generated using solutions of Na2S and H2SO4 | 100–500 | Polyurethane foam | Biofilm from a Thiobacillus-dominated lab-scale moving bed biofilm reactor | DGGE | Heterotrophic/mixotrophic denitrifying bacteria outcompete autotrophic denitrifying SOBs leading to an increased accumulation of biomass and decrease in the RE under mixotrophic conditions | [68] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Borgne, S.; Baquerizo, G. Microbial Ecology of Biofiltration Units Used for the Desulfurization of Biogas. ChemEngineering 2019, 3, 72. https://doi.org/10.3390/chemengineering3030072
Le Borgne S, Baquerizo G. Microbial Ecology of Biofiltration Units Used for the Desulfurization of Biogas. ChemEngineering. 2019; 3(3):72. https://doi.org/10.3390/chemengineering3030072
Chicago/Turabian StyleLe Borgne, Sylvie, and Guillermo Baquerizo. 2019. "Microbial Ecology of Biofiltration Units Used for the Desulfurization of Biogas" ChemEngineering 3, no. 3: 72. https://doi.org/10.3390/chemengineering3030072
APA StyleLe Borgne, S., & Baquerizo, G. (2019). Microbial Ecology of Biofiltration Units Used for the Desulfurization of Biogas. ChemEngineering, 3(3), 72. https://doi.org/10.3390/chemengineering3030072