Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome
Abstract
:1. Introduction
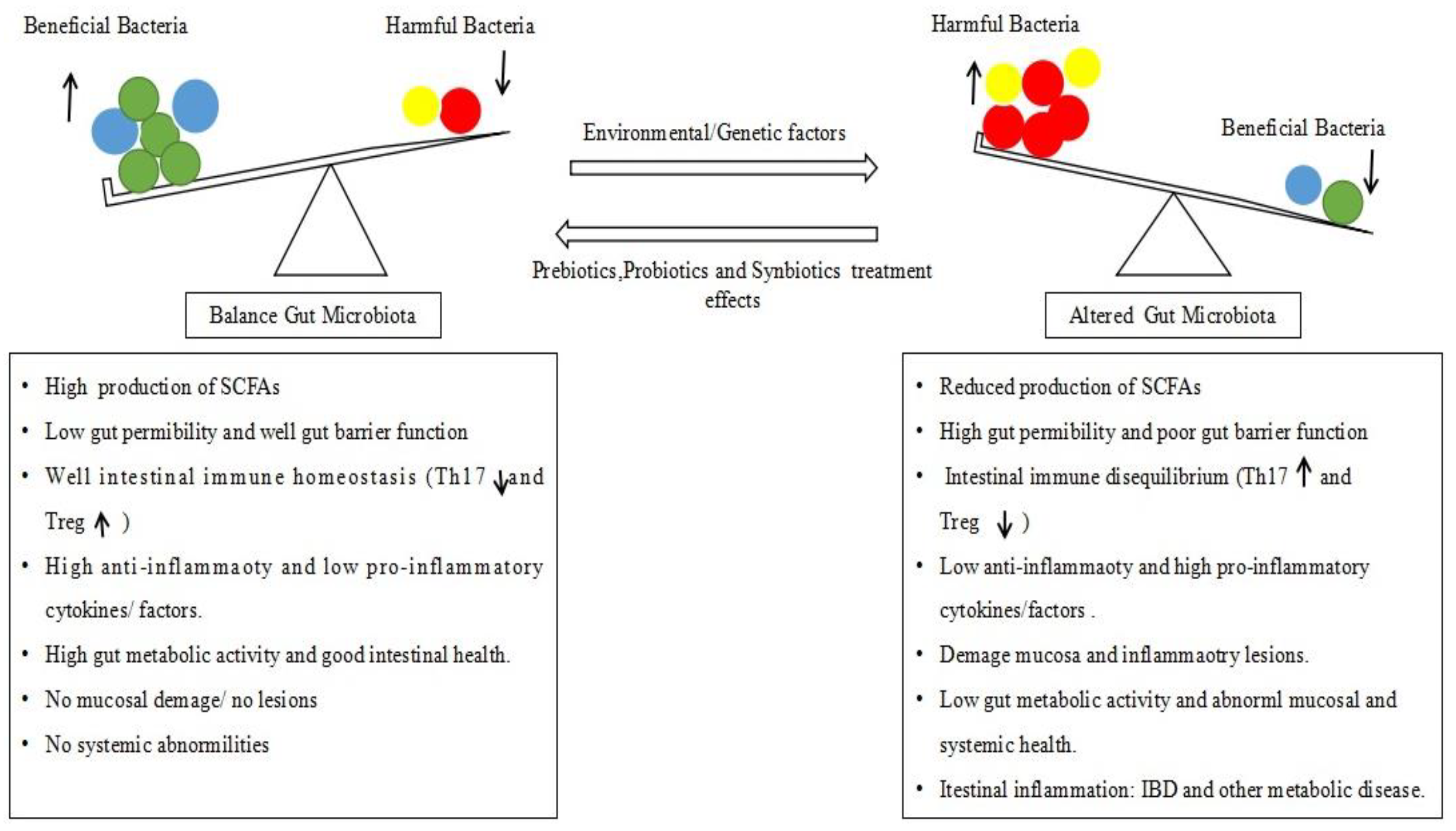
2. Alteration of Gut Microbiota (Dysbiosis) and IBD Pathology
3. Specific Individual Bacteria Species or Communities Involved in IBD
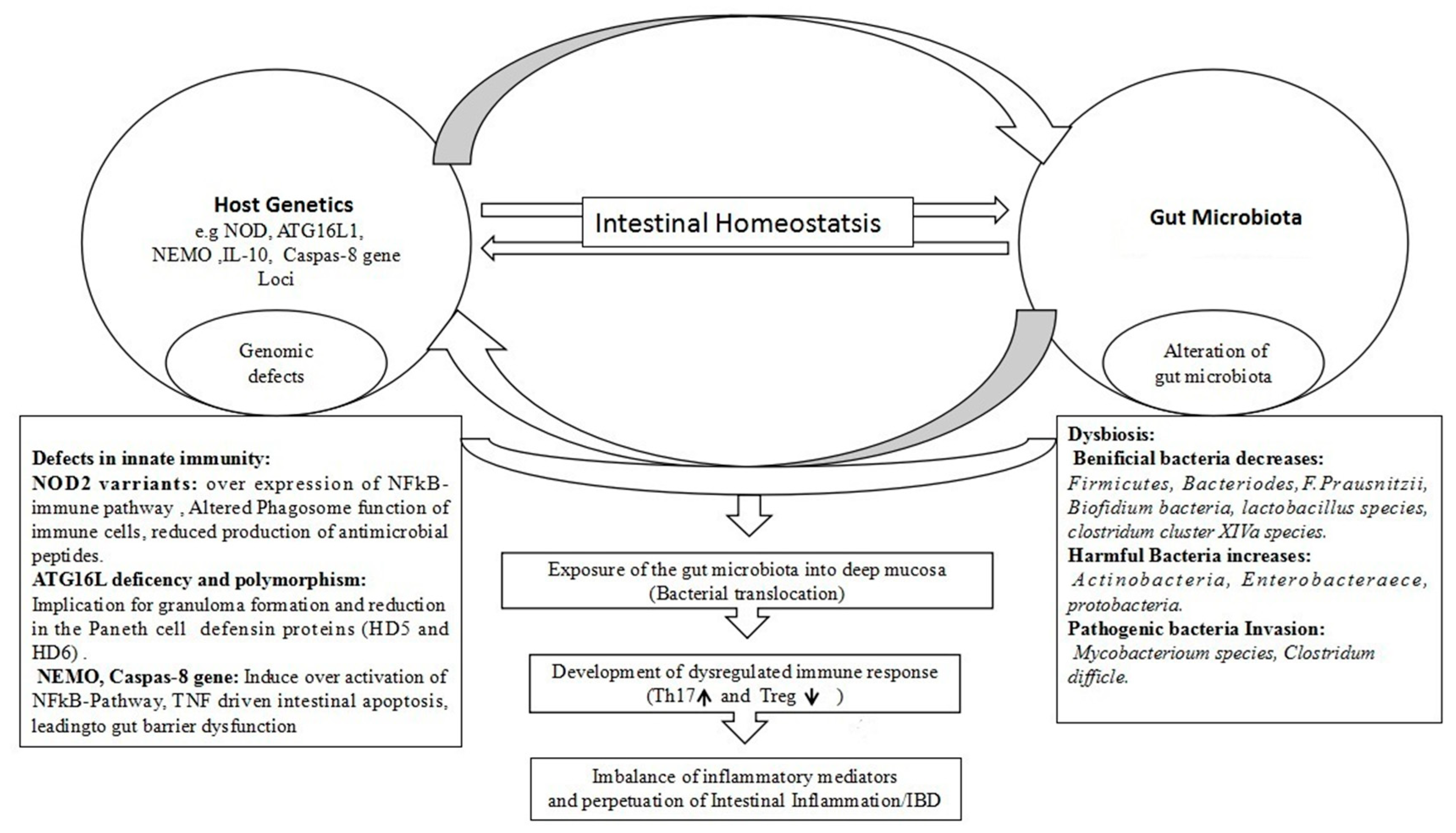
4. Genetics in Dysbiosis and IBD Pathology
5. Research Progress in IBD Treatment
5.1. Complementary and Alternative Medicine (CAM)
5.2. Traditional Chinese Herbal Medicine (TCMs) and IBD
5.3. Herbal Medicine as Prebiotics in IBD Treatment
5.4. Probiotics
5.5. Synbiotics
5.6. Fecal Microbiota Transplantation (FMT)
6. Conclusions
7. Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
Running Title
References
- Mulder, D.J.; Noble, A.J.; Justinich, C.J.; Duffin, J.M. A tale of two diseases: The history of inflammatory bowel disease. J. Crohn’s Colitis 2014, 8, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijmenga, C. Expressing the differences between Crohn’s disease and ulcerative colitis. PLoS Med. 2005, 2, e230. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720. [Google Scholar] [CrossRef] [PubMed]
- Chow, D.K.; Leong, R.W.; Tsoi, K.K.; Ng, S.S.; Leung, W.K.; Wu, J.C.; Wong, V.W.; Chan, F.K.; Sung, J.J. Long-term follow-up of ulcerative colitis in the Chinese population. Am. J. Gastroenterol. 2009, 104, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Ouyang, Q.; Hu, R.W. Progression of inflammatory bowel disease in China. J. Dig. Dis. 2010, 11, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.; Chan, F.K.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Thia, K.T.; Loftus, E.V., Jr.; Sandborn, W.J.; Yang, S.K. An update on the epidemiology of inflammatory bowel disease in Asia. Am. J. Gastroenterol. 2008, 103, 3167. [Google Scholar] [CrossRef] [PubMed]
- Victoria, C.R.; Sassak, L.Y.; Nunes, H.R.D.C. Incidence and prevalence rates of inflammatory bowel diseases, in midwestern of São Paulo State, Brazil. Arq. Gastroenterol. 2009, 46, 20–25. [Google Scholar] [CrossRef]
- Ouyang, Q.; Tandon, R.; Goh, K.L.; Pan, G.Z.; Fock, K.M.; Fiocchi, C.; Lam, S.K.; Xiao, S.D. Management consensus of inflammatory bowel disease for the Asia–Pacific. J. Gastroenterol. Hepatol. 2006, 21, 1772–1782. [Google Scholar] [CrossRef]
- Foster, A.; Jacobson, K. Changing incidence of inflammatory bowel disease: Environmental influences and lessons learnt from the South Asian population. Front. Pediatr. 2013, 1, 34. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- De Souza, H.S.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Rosen, C.E.; Palm, N.W. Navigating the Microbiota Seas: Triangulation Finds a Way Forward. Cell Host Microbe 2018, 23, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gkouskou, K.; Deligianni, C.; Tsatsanis, C.; Eliopoulos, A.G. The gut microbiota in mouse models of inflammatory bowel disease. Front. Cell. Infect. Microbiol. 2014, 4, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanda, K.; Moss, A.C. Update on the management of ulcerative colitis: Treatment and maintenance approaches focused on MMX® mesalamine. Clin. Pharmacol. Adv. Appl. 2012, 4, 41–50. [Google Scholar]
- MacDermott, R.P.; Green, J.A. Refractory ulcerative colitis treatment. Gastroenterol. Hepatol. 2007, 3, 64–69. [Google Scholar]
- Rhen, T.; Cidlowski, J.A. Antiinflammatory action of glucocorticoids—New mechanisms for old drugs. N. Engl. J. Med. 2005, 353, 1711–1723. [Google Scholar] [CrossRef]
- Habens, F.; Srinivasan, N.; Oakley, F.; Mann, D.A.; Ganesan, A.; Packham, G. Novel sulfasalazine analogues with enhanced NF-kB inhibitory and apoptosis promoting activity. Apoptosis 2005, 10, 481–491. [Google Scholar] [CrossRef]
- Nugent, S.G.; Kumar, D.; Rampton, D.S.; Evans, D.F. Intestinal luminal pH in inflammatory bowel disease: Possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 2001, 48, 571–577. [Google Scholar] [CrossRef]
- Van Dieren, J.M.; Kuipers, E.J.; Samsom, J.N.; Nieuwenhuis, E.E.; van der Woude, C.J. Revisiting the immunomodulators tacrolimus, methotrexate, and mycophenolate mofetil: Their mechanisms of action and role in the treatment of IBD. Inflamm. Bowel Dis. 2006, 12, 311–327. [Google Scholar] [CrossRef]
- Willot, S.; Noble, A.; Deslandres, C. Methotrexate in the treatment of inflammatory bowel disease: An 8-year retrospective study in a Canadian pediatric IBD center. Inflamm. Bowel Dis. 2011, 17, 2521–2526. [Google Scholar] [CrossRef]
- Ratner, M. IL-17–targeting biologics aim to become standard of care in psoriasis. Nat. Biotechnol. 2015, 33, 3–4. [Google Scholar] [CrossRef]
- Ogata, H.; Matsui, T.; Nakamura, M.; Iida, M.; Takazoe, M.; Suzuki, Y.; Hibi, T. A randomised dose finding study of oral tacrolimus (FK506) therapy in refractory ulcerative colitis. Gut 2006, 55, 1255–1262. [Google Scholar] [CrossRef] [Green Version]
- Triantafillidis, J.K.; Merikas, E.; Georgopoulos, F. Current and emerging drugs for the treatment of inflammatory bowel disease. Drug Des. Dev. Ther. 2011, 5, 185–210. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Tlaskalová-Hogenová, H.; Štěpánková, R.; Kozáková, H.; Hudcovic, T.; Vannucci, L.; Tučková, L.; Rossmann, P.; Hrnčíř, T.; Kverka, M.; Zákostelská, Z.; et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 2011, 8, 110–120. [Google Scholar] [CrossRef]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Frank, D.N.; Amand, A.L.S.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Trans. Med. 2009, 1, 6–14. [Google Scholar] [CrossRef]
- Koboziev, I.; Reinoso Webb, C.; Furr, K.L.; Grisham, M.B. Role of the enteric microbiota in intestinal homeostasis and inflammation. Free Radic. Biol. Med. 2014, 68, 122–133. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Wu, W.; Chen, F.; Liu, Z.; Cong, Y. Microbiota-specific Th17 cells: Yin and Yang in regulation of inflammatory bowel disease. Inflamm. Bowel Dis. 2016, 22, 1473–1482. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Torow, N.; Hornef, M.W. The neonatal window of opportunity: Setting the stage for life-long host-microbial interaction and immune homeostasis. J. Immunol. 2017, 198, 557–563. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; Liu, H.; Cross, J.R.; Pfeffer K.; Coffer, P.J.; Rudensky, A.Y. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Hooper, L.V.; Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010, 10, 159–169. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Lee, Y.K.; Mazmanian, S.K. Microbial learning lessons: SFB educate the immune system. Immunity 2014, 40, 457–459. [Google Scholar] [CrossRef]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.; Ng, D.C.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. T reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232. [Google Scholar] [CrossRef]
- Mucida, D.; Park, Y.; Kim, G.; Turovskaya, O.; Scott, I.; Kronenberg, M.; Cheroutre, H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 2007, 317, 256–260. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Qiu, X.; Zhang, M.; Yang, X.; Hong, N.; Yu, C. Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating TNBS-induced colitis. J. Crohn’s Colitis 2013, 7, e558–e568. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Buttó, L.F.; Haller, D. Dysbiosis in intestinal inflammation: Cause or consequence. Int. J. Med Microbiol. 2016, 306, 302–309. [Google Scholar] [CrossRef]
- Buttó, L.F.; Schaubeck, M.; Haller, D. Mechanisms of microbehost interaction in Crohn’s disease: Dysbiosis vs pathobiont selection. Front. Immunol. 2015, 6, 1–20. [Google Scholar] [CrossRef]
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A.; et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef]
- Ohata, A.; Usami, M.; Miyoshi, M. Short-chain fatty acids alter tight junction permeability in intestinal monolayer cells via lipoxygenase activation. Nutrition 2005, 21, 838–847. [Google Scholar] [CrossRef] [Green Version]
- Vinolo, M.A.; Ferguson, G.J.; Kulkarni, S.; Damoulakis, G.; Anderson, K.; Bohlooly-Y, M.; Stephens, L.; Hawkins, P.T.; Curi, R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS ONE 2011, 6, e21205. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; ter Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016, 167, 1125–1136. [Google Scholar] [CrossRef]
- Luo, A.; Leach, S.T.; Barres, R.; Hesson, L.B.; Grimm, M.C.; Simar, D. The microbiota and epigenetic regulation of T helper 17/regulatory T cells: In search of a balanced immune system. Front. Immunol. 2017, 8, 417. [Google Scholar] [CrossRef]
- Swidsinski, A.; Loening–Baucke, V.; Verstraelen, H.; Osowska, S.; Doerffel, Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology 2008, 135, 568–579. [Google Scholar] [CrossRef]
- Abu-Shanab, A.; Quigley, E.M. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 691–701. [Google Scholar] [CrossRef]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–325. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [Green Version]
- Serino, M.; Blasco-Baque, V.; Nicolas, S.; Burcelin, R. Far from the eyes, close to the heart: Dysbiosis of gut microbiota and cardiovascular consequences. Curr. Cardiol. Rep. 2014, 16, 540. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef]
- Casen, C.; Vebø, H.C.; Sekelja, M.; Hegge, F.T.; Karlsson, M.K.; Ciemniejewska, E.; Dzankovic, S.; Frøyland, C.; Nestestog, R.; Engstrand, L.; et al. Deviations in human gut microbiota: A novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment. Pharmacol. Ther. 2015, 42, 71–83. [Google Scholar] [CrossRef]
- Wright, E.K.; Kamm, M.A.; Teo, S.M.; Inouye, M.; Wagner, J.; Kirkwood, C.D. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: A systematic review. Inflamm. Bowel Dis. 2015, 21, 1219–1228. [Google Scholar]
- Huttenhower, C.; Kostic, A.D.; Xavier, R.J. Inflammatory bowel disease as a model for translating the microbiome. Immunity 2014, 40, 843–854. [Google Scholar] [CrossRef]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, B.; Zhang, Y.; Wei, H.; Lei, Y.; Zhao, L. Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J. Clin. Microbiol. 2007, 45, 496–500. [Google Scholar] [CrossRef]
- Tamboli, C.P.; Neut, C.; Desreumaux, P.; Colombel, J.F. Dysbiosis in inflammatory bowel disease. Gut 2004, 53, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef]
- Nagalingam, N.A.; Lynch, S.V. Role of the microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2011, 18, 968–984. [Google Scholar] [CrossRef]
- Mondot, S.; Kang, S.; Furet, J.P.; Aguirre de Cárcer, D.; McSweeney, C.; Morrison, M.; Marteau, P.; Dore, J.; Leclerc, M. Highlighting new phylogenetic specificities of Crohn’s disease microbiota. Inflamm. Bowel Dis. 2010, 17, 185–192. [Google Scholar] [CrossRef]
- Hedin, C.R.; McCarthy, N.E.; Louis, P.; Farquharson, F.M.; McCartney, S.; Taylor, K.; Prescott, N.J.; Murrells, T.; Stagg, A.J.; Whelan, K.; et al. Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut 2014, 63, 1578–1586. [Google Scholar] [CrossRef]
- Nguyen, G.C. Bugs and drugs: Insights into the pathogenesis of inflammatory bowel disease. Am. J. Gastroenterol. 2011, 106, 2143–2145. [Google Scholar] [CrossRef]
- Bien, J.; Palagani, V.; Bozko, P. The intestinal microbiota dysbiosis and Clostridium difficile infection: Is there a relationship with inflammatory bowel disease? Ther. Adv. Gastroenterol. 2013, 6, 53–68. [Google Scholar] [CrossRef]
- Li, J.; Butcher, J.; Mack, D.; Stintzi, A. Functional impacts of the intestinal microbiome in the pathogenesis of inflammatory bowel disease. Inflamm. Bowel Dis. 2014, 21, 139–153. [Google Scholar] [CrossRef]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631–637. [Google Scholar]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Morgan, X.C. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Gatto, D.; Sainsbury, E.; Harriman, G.R.; Hengartner, H.; Zinkernagel, R.M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000, 288, 2222–2226. [Google Scholar] [CrossRef]
- Sartor, R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef]
- Nemoto, H.; Kataoka, K.; Ishikawa, H.; Ikata, K.; Arimochi, H.; Iwasaki, T.; Yasutomo, K. Reduced diversity and imbalance of fecal microbiota in patients with ulcerative colitis. Dig. Dis. Sci. 2012, 57, 2955–2964. [Google Scholar] [CrossRef]
- Sha, S.; Xu, B.; Wang, X.; Zhang, Y.; Wang, H.; Kong, X.; Wu, K. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn. Microbiol. Infect. Dis. 2013, 75, 245–251. [Google Scholar] [CrossRef]
- Schultsz, C.; van den Berg, F.M.; Fiebo, W.; Tytgat, G.N.; Dankert, J. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 1999, 117, 1089–1097. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655. [Google Scholar] [CrossRef]
- Yilmaz, B.; Juillerat, P.; Øyås, O.; Ramon, C.; Bravo, F.D.; Franc, Y.; Fournier, N.; Michetti, P.; Mueller, C.; Geuking, M.; et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat. Med. 2019, 25, 323. [Google Scholar] [CrossRef]
- Brown, E.M.; Wlodarska, M.; Willing, B.P.; Vonaesch, P.; Han, J.; Reynolds, L.A.; Arrieta, M.C.; Uhrig, M.; Scholz, R.; Partida, O.; et al. Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nat. Commun. 2015, 6, 7806. [Google Scholar] [CrossRef]
- Keely, S.; Walker, M.M.; Marks, E.; Talley, N.J. Immune dysregulation in the functional gastrointestinal disorders. Eur. J. Clin. Investig. 2015, 45, 1350–1359. [Google Scholar] [CrossRef] [Green Version]
- D’argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Blanco, G.D.V.; et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 2016, 111, 879. [Google Scholar] [CrossRef]
- Distrutti, E.; Monaldi, L.; Ricci, P.; Fiorucci, S. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J. Gastroenterol. 2016, 22, 2219–2241. [Google Scholar] [CrossRef]
- Larcombe, S.; Hutton, M.L.; Lyras, D. Involvement of bacteria other than Clostridium difficile in antibiotic-associated diarrhoea. Trends Microbiol. 2016, 24, 463–476. [Google Scholar] [CrossRef]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef]
- Bibiloni, R.; Mangold, M.; Madsen, K.L.; Fedorak, R.N.; Tannock, G.W. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J. Med. Microbiol. 2006, 55, 1141–1149. [Google Scholar] [CrossRef]
- Rath, H.C.; Herfarth, H.H.; Ikeda, J.S.; Grenther, W.B.; Hamm, T.E.; Balish, E.; Taurog, J.D.; Hammer, R.E.; Wilson, K.H.; Sartor, R.B. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J. Clin. Investig. 1996, 98, 945–953. [Google Scholar] [CrossRef]
- Sellon, R.K.; Tonkonogy, S.; Schultz, M.; Dieleman, L.A.; Grenther, W.; Balish, E.; Rennick, D.M.; Sartor, R.B. Resident enteric bacteria are necessary for the development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998, 66, 5224–5231. [Google Scholar]
- Strober, W.; Fuss, I.; Mannon, P. The fundamental basis of inflammatory bowel disease. J. Clin. Investig. 2007, 117, 514–521. [Google Scholar] [CrossRef] [Green Version]
- Taurog, J.D.; Richardson, J.A.; Croft, J.T.; Simmons, W.A.; Zhou, M.; Fernández-Sueiro, J.L.; Balish, E.; Hammer, R.E. The germ free state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 1994, 180, 2359–2364. [Google Scholar] [CrossRef]
- Veltkamp, C.; Tonkonogy, S.L.; De Jong, Y.P.; Albright, C.; Grenther, W.B.; Balish, E.; Terhorst, C.; Sartor, R.B. Continuous stimulation by normal luminal bacteria is essential for the development and perpetuation of colitis in Tgϵ26 mice. Gastroenterology 2001, 120, 900–913. [Google Scholar] [CrossRef]
- Pils, M.C.; Bleich, A.; Prinz, I.; Fasnacht, N.; Bollati-Fogolin, M.; Schippers, A.; Rozell, B.; Müller, W. Commensal gut flora reduces susceptibility to experimentally induced colitis via T-cell-derived interleukin-10. Inflamm. Bowel Dis. 2010, 17, 2038–2046. [Google Scholar] [CrossRef]
- Yang, I.; Eibach, D.; Kops, F.; Brenneke, B.; Woltemate, S.; Schulze, J.; Bleich, A.; Gruber, A.D.; Muthupalani, S.; Fox, J.G.; et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS ONE 2013, 8, e70783. [Google Scholar]
- Garrett, W.S.; Lord, G.M.; Punit, S.; Lugo-Villarino, G.; Mazmanian, S.K.; Ito, S.; Glickman, J.N.; Glimcher, L.H. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 2007, 131, 33–45. [Google Scholar] [CrossRef]
- Ott, S.J.; Waetzig, G.H.; Rehman, A.; Moltzau-Anderson, J.; Bharti, R.; Grasis, J.A.; Cassidy, L.; Tholey, A.; Fickenscher, H.; Seegert, D.; et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 2017, 152, 799–811. [Google Scholar] [CrossRef]
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W.; et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015, 149, 102–109. [Google Scholar] [CrossRef]
- Rioux, K.P.; Fedorak, R.N. Probiotics in the treatment of inflammatory bowel disease. J. Clin. Gastroenterol. 2006, 40, 260–263. [Google Scholar] [CrossRef]
- Bengmark, S. Bioecological control of inflammatory bowel disease. Clin. Nutr. 2007, 26, 169181. [Google Scholar] [CrossRef]
- Roberfroid, M. Prebiotics: The concept revisited. J. Nutr. 2007, 137, 830–837. [Google Scholar] [CrossRef]
- Bengmark, S. Pre-, pro-and synbiotics. Curr. Opin. Clin. Nutr. Metab. Care 2001, 4, 571–579. [Google Scholar] [CrossRef]
- Gionchetti, P.; Rizzello, F.; Helwig, U.; Venturi, A.; Lammers, K.M.; Brigidi, P.; Vitali, B.; Poggioli, G.; Miglioli, M.; Campieri, M. Prophylaxis of pouchitis onset with probiotic therapy: A double-blind, placebo-controlled trial. Gastroenterology 2003, 124, 1202–1209. [Google Scholar] [CrossRef]
- Perencevich, M.; Burakoff, R. Use of antibiotics in the treatment of inflammatory bowel disease. Inflamm. Bowel Dis. 2006, 12, 651–664. [Google Scholar] [CrossRef]
- Damaskos, D.; Kolios, G. Probiotics and prebiotics in inflammatory bowel disease: Microflora ‘on the scope’. Br. J. Clin. Pharmacol. 2008, 65, 453–467. [Google Scholar] [CrossRef]
- Matijašić, M.; Meštrović, T.; Perić, M.; Čipčić Paljetak, H.; Panek, M.; Vranešić Bender, D.; Ljubas Kelečić, D.; Krznarić, Ž.; Verbanac, D. Modulating composition and metabolic activity of the gut microbiota in IBD patients. Int. J. Mol. Sci. 2016, 17, 578. [Google Scholar] [CrossRef]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The gut microbiota in immune-mediated inflammatory diseases. Front. Microbiol. 2016, 7, 1081. [Google Scholar] [CrossRef]
- Nell, S.; Suerbaum, S.; Josenhans, C. The impact of the microbiota on the pathogenesis of IBD: Lessons from mouse infection models. Nat. Rev. Microbiol. 2010, 8, 564–567. [Google Scholar] [CrossRef]
- Rhee, K.J.; Wu, S.; Wu, X.; Huso, D.L.; Karim, B.; Franco, A.A.; Rabizadeh, S.; Golub, J.E.; Mathews, L.E.; Shin, J.; et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 2009, 77, 1708–1718. [Google Scholar] [CrossRef]
- Hold, G.L.; Smith, M.; Grange, C.; Watt, E.R.; El-Omar, E.M.; Mukhopadhya, I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. WJG 2014, 20, 1192–1210. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef]
- Simpson, K.W.; Dogan, B.; Rishniw, M.; Goldstein, R.E.; Klaessig, S.; McDonough, P.L.; German, A.J.; Yates, R.M.; Russell, D.G.; Johnson, S.E.; et al. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect. Immun. 2006, 74, 4778–4792. [Google Scholar] [CrossRef]
- Darfeuille-Michaud, A.; Neut, C.; Barnich, N.; Lederman, E.; Di Martino, P.; Desreumaux, P.; Gambiez, L.; Joly, B.; Cortot, A.; Colombel, J.F. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 1998, 115, 1405–1413. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Aldeguer, X.; Lopez-Siles, M.; González-Huix, F.; López-Oliu, C.; Dahbi, G.; Blanco, J.E.; Blanco, J.; Garcia-Gil, J.L.; Darfeuille-Michaud, A. Molecular diversity of Escherichia coli in the human gut: New ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 872–882. [Google Scholar] [CrossRef]
- Mann, E.A.; Saeed, S.A. Gastrointestinal infection as a trigger for inflammatory bowel disease. Curr. Opin. Gastroenterol. 2012, 28, 24–29. [Google Scholar] [CrossRef]
- Pierce, E.S. Ulcerative colitis and Crohn’s disease: Is Mycobacterium avium subspecies paratuberculosis the common villain? Gut Pathog. 2010, 2, 21. [Google Scholar] [CrossRef]
- Elguezabal, N.; Chamorro, S.; Molina, E.; Garrido, J.M.; Izeta, A.; Rodrigo, L.; Juste, R.A. Lactase persistence, NOD2 status and Mycobacterium avium subsp. paratuberculosis infection associations to Inflammatory Bowel Disease. Gut Pathog. 2012, 4, 6. [Google Scholar] [CrossRef]
- Kirkwood, C.D.; Wagner, J.; Boniface, K.; Vaughan, J.; Michalski, W.P.; Catto-Smith, A.G.; Cameron, D.J.; Bishop, R.F. Mycobacterium avium subspecies paratuberculosis in children with early onset Crohn’s disease. Inflamm. Bowel Dis. 2009, 15, 1643–1655. [Google Scholar] [CrossRef]
- Naser, S.A.; Ghobrial, G.; Romero, C.; Valentine, J.F. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn’s disease. Lancet 2004, 364, 1039–1044. [Google Scholar] [CrossRef]
- Sanderson, J.D.; Moss, M.T.; Tizard, M.L.; Hermon-Taylor, J. Mycobacterium paratuberculosis DNA in Crohn’s disease tissue. Gut 1992, 33, 890–896. [Google Scholar] [CrossRef]
- Friswell, M.; Campbell, B.; Rhodes, J. The role of bacteria in the pathogenesis of inflammatory bowel disease. Gut Liver 2010, 4, 295–306. [Google Scholar] [CrossRef]
- Ferwerda, G.; Girardin, S.E.; Kullberg, B.J.; Le Bourhis, L.; De Jong, D.J.; Langenberg, D.M.; Van Crevel, R.; Adema, G.J.; Ottenhoff, T.H.; Van der Meer, J.W. NOD2 and toll-like receptors are nonredundant recognition systems of Mycobacterium tuberculosis. PLoS Pathog. 2005, 1, e34. [Google Scholar] [CrossRef]
- Ferwerda, G.; Kullberg, B.J.; De Jong, D.J.; Girardin, S.E.; Langenberg, D.M.; Van Crevel, R.; Ottenhoff, T.H.; Van der Meer, J.W.; Netea, M.G. Mycobacterium paratuberculosis is recognized by Toll-like receptors and NOD2. J. Leukoc. Biol. 2007, 82, 1011–1018. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Master, S.S.; Singh, S.B.; Taylor, G.A.; Colombo, M.I.; Deretic, V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004, 119, 753–766. [Google Scholar] [CrossRef]
- Hansen, R.; Thomson, J.M.; El-Omar, E.M.; Hold, G.L. The role of infection in the aetiology of inflammatory bowel disease. J. Gastroenterol. 2010, 45, 266–276. [Google Scholar] [CrossRef]
- Juste, R.A.; Elguezabal, N.; Garrido, J.M.; Pavon, A.; Geijo, M.V.; Sevilla, I.; Cabriada, J.L.; Tejada, A.; García-Campos, F.; Casado, R.; et al. On the prevalence of M. avium subspecies paratuberculosis DNA in the blood of healthy individuals and patients with inflammatory bowel disease. PLoS ONE 2008, 3, e2537. [Google Scholar] [CrossRef]
- Selby, W.; Pavli, P.; Crotty, B.; Florin, T.; Radford-Smith, G.; Gibson, P.; Mitchell, B.; Connell, W.; Read, R.; Merrett, M.; et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn’s disease. Gastroenterology 2007, 132, 2313–2319. [Google Scholar] [CrossRef]
- Navaneethan, U.; Venkatesh, P.G.; Shen, B. Clostridium difficile infection and inflammatory bowel disease: Understanding the evolving relationship. World J. Gastroenterol. WJG 2010, 16, 4892–4904. [Google Scholar] [CrossRef]
- Kim, H.; Rhee, S.H.; Pothoulakis, C.; LaMont, J.T. Inflammation and apoptosis in Clostridium difficile enteritis is mediated by PGE2 up-regulation of Fas ligand. Gastroenterology 2007, 133, 875–886. [Google Scholar] [CrossRef]
- Issa, M.; Vijayapal, A.; Graham, M.B.; Beaulieu, D.B.; Otterson, M.F.; Lundeen, S.; Skaros, S.; Weber, L.R.; Komorowski, R.A.; Knox, J.F.; et al. Impact of Clostridium difficile on inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 2007, 5, 345–351. [Google Scholar] [CrossRef]
- Fujimoto, T.; Imaeda, H.; Takahashi, K.; Kasumi, E.; Bamba, S.; Fujiyama, Y.; Andoh, A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of C rohn’s disease. J. Gastroenterol. Hepatol. 2013, 28, 613–619. [Google Scholar] [CrossRef]
- Fava, F.; Danese, S. Intestinal microbiota in inflammatory bowel disease: Friend of foe? World J. Gastroenterol. WJG 2011, 17, 557. [Google Scholar] [CrossRef]
- Seksik, P.; Rigottier-Gois, L.; Gramet, G.; Sutren, M.; Pochart, P.; Marteau, P.; Jian, R.; Dore, J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 2003, 52, 237–242. [Google Scholar] [CrossRef]
- Favier, C.; Neut, C.; Mizon, C.; Cortot, A.; Colombel, J.F.; Mizon, J. Fecal β-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig. Dis. Sci. 1997, 42, 817–822. [Google Scholar] [CrossRef]
- Bartels, L.E.; Jepsen, P.; Christensen, L.A.; Gerdes, L.U.; Vilstrup, H.; Dahlerup, J.F. Diagnosis of Helicobacter pylori infection is associated with lower prevalence and subsequent incidence of Crohn’s disease. J. Crohn’s Colitis 2015, 10, 443–448. [Google Scholar] [CrossRef]
- McMullen, L.; Leach, S.T.; Lemberg, D.A.; Day, A.S. Current roles of specific bacteria in the pathogenesis of inflammatory bowel disease. Microbiology 2015, 1, 82–91. [Google Scholar] [CrossRef]
- Zhang, L.; Lee, H.; Grimm, M.C.; Riordan, S.M.; Day, A.S.; Lemberg, D.A. Campylobacter concisus and inflammatory bowel disease. World J. Gastroenterol. WJG 2014, 20, 1259–1267. [Google Scholar] [CrossRef]
- Martin, H.M.; Campbell, B.J.; Hart, C.A.; Mpofu, C.; Nayar, M.; Singh, R.; Englyst, H.; Williams, H.F.; Rhodes, J.M. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer 1. Gastroenterology 2004, 127, 80–93. [Google Scholar] [CrossRef]
- De Hertogh, G.; Aerssens, J.; Geboes, K.P.; Geboes, K. Evidence for the involvement of infectious agents in the pathogenesis of Crohn’s disease. World J. Gastroenterol. WJG 2008, 14, 845. [Google Scholar] [CrossRef]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A microbial signature for Crohn’s disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef]
- Sokol, H.; Lay, C.; Seksik, P.; Tannock, G.W. Analysis of bacterial bowel communities of IBD patients: What has it revealed? Inflamm. Bowel Dis. 2008, 14, 858–867. [Google Scholar] [CrossRef]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef]
- Vrakas, S.; Mountzouris, K.C.; Michalopoulos, G.; Karamanolis, G.; Papatheodoridis, G.; Tzathas, C.; Gazouli, M. Intestinal bacteria composition and translocation of bacteria in inflammatory bowel disease. PLoS ONE 2017, 12, e0170034. [Google Scholar] [CrossRef]
- Walters, S.S.; Quiros, A.; Rolston, M.; Grishina, I.; Li, J.; Fenton, A.; DeSantis, T.Z.; Thai, A.; Andersen, G.L.; Papathakis, P.; et al. Analysis of gut microbiome and diet modification in patients with Crohn’s disease. SOJ Microbiol. Infect. Dis. 2014, 2, 1. [Google Scholar] [CrossRef]
- Jacobs, J.P.; Goudarzi, M.; Singh, N.; Tong, M.; McHardy, I.H.; Ruegger, P.; Asadourian, M.; Moon, B.H.; Ayson, A.; Borneman, J.; et al. Adisease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 750–766. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, 79. [Google Scholar] [CrossRef]
- Ott, S.J.; Musfeldt, M.; Wenderoth, D.F.; Hampe, J.; Brant, O.; Fölsch, U.R.; Timmis, K.N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.; Verma, A.K.; Ahuja, V.; Paul, J. Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. J. Clin. Microbiol. 2010, 48, 4279–4282. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vázquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef]
- Scaldaferri, F.; Gerardi, V.; Lopetuso, L.R.; Del Zompo, F.; Mangiola, F.; Boškoski, I.; Bruno, G.; Petito, V.; Laterza, L.; Cammarota, G.; et al. Gut microbial flora, prebiotics, and probiotics in IBD: Their current usage and utility. BioMed Res. Int. 2013, 2013, 9. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Wang, W.; Chen, L.; Zhou, R.; Wang, X.; Song, L.; Huang, S.; Wang, G.; Xia, B. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 2014, 52, 398–406. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Tong, M.; Li, X.; Parfrey, L.W.; Roth, B.; Ippoliti, A.; Wei, B.; Borneman, J.; McGovern, D.P.; Frank, D.N.; Li, E. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS ONE 2013, 8, e80702. [Google Scholar] [CrossRef]
- Okayasu, I.; Hatakeyama, S.; Yamada, M.; Ohkusa, T.; Inagaki, Y.; Nakaya, R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 1990, 98, 694–702. [Google Scholar] [CrossRef]
- Håkansson, Å.; Tormo-Badia, N.; Baridi, A.; Xu, J.; Molin, G.; Hagslätt, M.L.; Karlsson, C.; Jeppsson, B.E.N.G.T.; Cilio, C.M.; Ahrné, S. Immunological alteration and changes of gut microbiota after dextran sulfate sodium (DSS) administration in mice. Clin. Exp. Med. 2015, 15, 107–120. [Google Scholar] [CrossRef]
- Ettreiki, C.; Gadonna-Widehem, P.; Mangin, I.; Coëffier, M.; Delayre-Orthez, C.; Anton, P.M. Juvenile ferric iron prevents microbiota dysbiosis and colitis in adult rodents. World J. Gastroenterol. WJG 2012, 18, 2619–2629. [Google Scholar] [CrossRef]
- Rooks, M.G.; Veiga, P.; Wardwell-Scott, L.H.; Tickle, T.; Segata, N.; Michaud, M.; Gallini, C.A.; Beal, C.; van Hylckama-Vlieg, J.E.; Ballal, S.A.; et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014, 8, 1403–1417. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G., III; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional characterization of inflammatory bowel disease–associated gut dysbiosis in gnotobiotic mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Dennis, K.L.; Wang, Y.; Blatner, N.R.; Wang, S.; Saadalla, A.; Trudeau, E.; Roers, A.; Weaver, C.T.; Lee, J.J.; Gilbert, J.A.; et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10–producing T cells. Cancer Res. 2013, 73, 5905–5913. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef]
- Duerr, R.H.; Taylor, K.D.; Brant, S.R.; Rioux, J.D.; Silverberg, M.S.; Daly, M.J.; Steinhart, A.H.; Abraham, C.; Regueiro, M.; Griffiths, A.; et al. A Genome-Wide Association Study Identifies IL23R as an Inflammatory Bowel Disease Gene. Science 2006, 314, 1461–1463. [Google Scholar] [CrossRef]
- A Fisher, S.; Tremelling, M.; A Anderson, C.; Gwilliam, R.; Bumpstead, S.; Prescott, N.J.; Nimmo, E.R.; Massey, D.; Berzuini, C.; Johnson, C.; et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat. Genet. 2008, 40, 710–712. [Google Scholar] [CrossRef]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef]
- Tanabe, T.; Chamaillard, M.; Ogura, Y.; Zhu, L.; Qiu, S.; Masumoto, J.; Ghosh, P.; Moran, A.; Predergast, M.M.; Tromp, G.; et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004, 23, 1587–1597. [Google Scholar] [CrossRef]
- Ogura, Y.; Inohara, N.; Benito, A.; Chen, F.F.; Yamaoka, S.; Núñez, G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 2001, 276, 4812–4818. [Google Scholar] [CrossRef]
- Watanabe, T.; Kitani, A.; Murray, P.J.; Strober, W. NOD2 is a negative regulator of Toll-like receptor 2–mediated T helper type 1 responses. Nat. Immunol. 2004, 5, 800–808. [Google Scholar] [CrossRef]
- Wehkamp, J.; Salzman, N.H.; Porter, E.; Nuding, S.; Weichenthal, M.; Petras, R.E.; Shen, B.; Schaeffeler, E.; Schwab, M.; Linzmeier, R.; et al. Reduced Paneth cell α-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 18129–18134. [Google Scholar] [CrossRef]
- Petnicki-Ocwieja, T.; Hrncir, T.; Liu, Y.-J.; Biswas, A.; Hudcovic, T.; Tlaskalova-Hogenova, H.; Kobayashi, K.S. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA 2009, 106, 15813–15818. [Google Scholar] [CrossRef] [Green Version]
- Frank, D.N.; Robertson, C.E.; Hamm, C.M.; Kpadeh, Z.; Zhang, T.; Chen, H.; Zhu, W.; Sartor, R.B.; Boedeker, E.C.; Harpaz, N.; et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2010, 17, 179–184. [Google Scholar] [CrossRef]
- Li, E.; Hamm, C.M.; Gulati, A.S.; Sartor, R.B.; Chen, H.; Wu, X.; Zhang, T.; Rohlf, F.J.; Zhu, W.; Gu, C.; et al. Inflammatory Bowel Diseases Phenotype, C. difficile and NOD2 Genotype Are Associated with Shifts in Human Ileum Associated Microbial Composition. PLoS ONE 2012, 7, e26284. [Google Scholar] [CrossRef]
- Couturier-Maillard, A.; Secher, T.; Rehman, A.; Normand, S.; De Arcangelis, A.; Häesler, R.; Huot, L.; Grandjean, T.; Bressenot, A.; Delanoye-Crespin, A.; et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Investig. 2013, 123, 700–711. [Google Scholar] [CrossRef] [Green Version]
- Rehman, A.; Sina, C.; Gavrilova, O.; Häsler, R.; Ott, S.; Baines, J.F.; Schreiber, S.; Rosenstiel, P. Nod2 is essential for temporal development of intestinal microbial communities. Gut 2011, 60, 1354–1362. [Google Scholar] [CrossRef]
- Hampe, J.; Franke, A.; Rosenstiel, P.; Till, A.; Teuber, M.; Huse, K.; Albrecht, M.; Mayr, G.; De La Vega, F.M.; Briggs, J.; et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007, 39, 207–211. [Google Scholar] [CrossRef]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Miyoshi, H.; Loh, J.; Lennerz, J.K.; Kishi, C.; Kc, W.; Carrero, J.A.; Hunt, S.; et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Gunther, C.; Martini, E.; Wittkopf, N.; Amann, K.; Weigmann, B.; Neumann, H.; Waldner, M.J.; Hedrick, S.M.; Tenzer, S.; Neurath, M.F.; et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 2011, 477, 335–339. [Google Scholar] [CrossRef]
- Ward, J.M.; Anver, M.R.; Haines, D.C.; Melhorn, J.M.; Gorelick, P.; Yan, L.; Fox, J.G. Imflammatory large bowel disease in immunodeficient mice naturally infected with. Helicobacter hepaticus. Lab. Anim. Sci. 1996, 46, 15–20. [Google Scholar]
- Cerf-Bensussan, N.; Gaboriau-Routhiau, V. The immune system and the gut microbiota: Friends or foes? Nat. Rev. Immunol. 2010, 10, 735–744. [Google Scholar] [CrossRef]
- Kullberg, M.C.; Jankovic, D.; Gorelick, P.L.; Caspar, P.; Letterio, J.J.; Cheever, A.W.; Sher, A. Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus–induced colitis. J. Exp. Med. 2002, 196, 505–515. [Google Scholar] [CrossRef]
- Cadwell, K.; Patel, K.K.; Maloney, N.S.; Liu, T.-C.; Ng, A.C.; Storer, C.E.; Head, R.D.; Xavier, R.; Stappenbeck, T.S.; Virgin, H.W. Virus-Plus-Susceptibility Gene Interaction Determines Crohn’s Disease Gene Atg16L1 Phenotypes in Intestine. Cell 2010, 141, 1135–1145. [Google Scholar] [CrossRef]
- Nenci, A.; Becker, C.; Wullaert, A.; Gareus, R.; Van Loo, G.; Danese, S.; Huth, M.; Nikolaev, A.; Neufert, C.; Madison, B.; et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 2007, 446, 557–561. [Google Scholar] [CrossRef]
- Rossi, M.; Corradini, C.; Amaretti, A.; Nicolini, M.; Pompei, A.; Zanoni, S.; Matteuzzi, D. Fermentation of Fructooligosaccharides and Inulin by Bifidobacteria: A Comparative Study of Pure and Fecal Cultures. Appl. Environ. Microbiol. 2005, 71, 6150–6158. [Google Scholar] [CrossRef]
- Bensoussan, M.; Jovenin, N.; Garcia, B.; Vandromme, L.; Jolly, D.; Bouché, O.; Thiéfin, G.; Cadiot, G. Complementary and alternative medicine use by patients with inflammatory bowel disease: Results from a postal survey. Gastroentérol. Clin. Biol. 2006, 30, 14–23. [Google Scholar] [CrossRef]
- Langhorst, J.; Anthonisen, I.B.; Steder-Neukamm, U.; Lüdtke, R.; Spahn, G.; Michalsen, A.; Dobos, G.J. Amount of Systemic Steroid Medication Is a Strong Predictor for the Use of Complementary and Alternative Medicine in Patients With Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2005, 11, 287–295. [Google Scholar] [CrossRef]
- Hilsden, R.J.; Verhoef, M.J.; Best, A.; Pocobelli, G. Complementary and alternative medicine use by Canadian patients with inflammatory bowel disease: Results from a national survey. Am. J. Gastroenterol. 2003, 98, 1563–1568. [Google Scholar] [CrossRef]
- Rahimi, R.; Mozaffari, S.; Abdollahi, M. On the use of herbal medicines in management of inflammatory bowel diseases: A systematic review of animal and human studies. Dig. Dis. Sci. 2009, 54, 471–480. [Google Scholar] [CrossRef]
- Lukovac, S.; Belzer, C.; Pellis, L.; Keijser, B.J.; De Vos, W.M.; Montijn, R.C.; Roeselers, G. Differential Modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of Host Peripheral Lipid Metabolism and Histone Acetylation in Mouse Gut Organoids. mBio 2014, 5, e01438-14. [Google Scholar] [CrossRef]
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef]
- World Health Organization. General Guidelines for Methodologies on Research and Evaluation of Traditional Medicine; No. WHO/EDM/TRM/2000.1; World Health Organization: Geneva, Swizterland, 2000. [Google Scholar]
- Zhang, J.; Bai, G. Wumei pill decoction in the treatment of 72 cases of chronic nonspecific ulcerative colitis. Mod. J. Integr. Tradit. West. Med. 2007, 16, 5156–5157. [Google Scholar]
- Foley, M.H.; Cockburn, D.W.; Koropatkin, N.M. The Sus operon: A model system for starch uptake by the human gut Bacteroidetes. Cell. Mol. Life Sci. 2016, 73, 2603–2617. [Google Scholar] [CrossRef]
- Liang, J.H.; Zheng, K.W.; Sun, L.Q. Explore the regulative action of astragalus polysaccharide for intestinal dysbacteriosis in ulcerative colitis rat models. Stud. Trace Elem. Health. 2013. [Google Scholar]
- Zhou, Z.; Ma, T.L.; Feng, L. Effects of polysaccharides from Portulaca oleracea L. on intestinal flora and blood endotoxin in mice with ulcerative colitis. Chin. J. Microecol. 2014, 26, 646–648. [Google Scholar]
- Mahowald, M.A.; Rey, F.E.; Seedorf, H.; Turnbaugh, P.J.; Fulton, R.S.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.K.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Li, W.-Y.; Wan, Z.; Zhao, B.; He, Z.-W.; Wu, Z.-G.; Huang, G.-L.; Wang, J.; Li, B.-B.; Lu, Y.-J.; et al. Huangqin-Tang Ameliorates TNBS-Induced Colitis by Regulating Effector and Regulatory CD4+ T Cells. BioMed Res. Int. 2015, 2015, 1–13. [Google Scholar] [CrossRef]
- LI Cheng-De, Z.; Wen-Bin, S.; Yan, F.; YU, L.; Mao, J.S. Effect of Astragalus Polysaccharides on Th17/Treg Cytokines and Pulmonary Inflammation in Asthmatic Rats. Chin. Pharmacol. Bull. 2013, 29, 1275–1278. [Google Scholar]
- Govers, M.J.A.P.; Gannon, N.J.; Dunshea, F.; Gibson, P.R.; Muir, J.G. Wheat bran affects the site of fermentation of resistant starch and luminal indexes related to colon cancer risk: A study in pigs. Gut 1999, 45, 840–847. [Google Scholar] [CrossRef]
- Rombeau, J.L.; Kripke, S.A. Metabolic and Intestinal Effects of Short-Chain Fatty Acids. J. Parenter. Enter. Nutr. 1990, 14, 181S–185S. [Google Scholar] [CrossRef]
- Scheppach, W. Effects of short chain fatty acids on gut morphology and function. Gut 1994, 35, S35–S38. [Google Scholar] [CrossRef]
- Säemann, M.D.; Österreicher, C.H.; Diakos, C.; Stöckl, J.; Zlabinger, G.J.; Böhmig, G.A.; Burtscher, H.; Parolini, O.; Hörl, W.H. Anti-inflammatory effects of sodium butyrate on human monocytes: Potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000, 14, 2380–2382. [Google Scholar] [CrossRef]
- Segain, J.P.; De La Blétiere, D.R.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottiere, H.M.; Galmiche, J.P. Butyrate inhibits inflammatory responses through NFκB inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef]
- Kanauchi, O.; Matsumoto, Y.; Matsumura, M.; Fukuoka, M.; Bamba, T. The Beneficial Effects of Microflora, Especially Obligate Anaerobes, and Their Products on the Colonic Environment in Inflammatory Bowel Disease. Curr. Pharm. Des. 2005, 11, 1047–1053. [Google Scholar] [CrossRef]
- Lührs, H.; Gerke, T.; Muller, J.G.; Melcher, R.; Schauber, J.; Boxberger, F.; Scheppach, W.; Menzel, T. Butyrate Inhibits NF-κB Activation in Lamina Propria Macrophages of Patients with Ulcerative Colitis. Scand. J. Gastroenterol. 2002, 37, 458–466. [Google Scholar] [CrossRef]
- Scheppach, W. German-Austrian Scfa Study Group Treatment of distal ulcerative colitis with short-chain fatty acid enemas a placebo-controlled trial. Dig. Dis. Sci. 1996, 41, 2254–2259. [Google Scholar] [CrossRef]
- Kanauchi, O.; Agata, K. Protein, and Dietary Fiber-rich New Foodstuff from Brewer’s Spent Grain Increased Excretion of Feces and Jejunum Mucosal Protein Content in Rats. Biosci. Biotechnol. Biochem. 1997, 61, 29–33. [Google Scholar] [CrossRef]
- Kanauchi, O.; Mitsuyama, K.; Homma, T.; Takahama, K.; Fujiyama, Y.; Andoh, A.; Araki, Y.; Suga, T.; Hibi, T.; Naganuma, M.; et al. Treatment of ulcerative colitis patients by long-term administration of germinated barley foodstuff: Multi-center open trial. Int. J. Mol. Med. 2003, 12, 701–704. [Google Scholar] [CrossRef]
- Kanauchi, O.; Iwanaga, T.; Andoh, A.; Araki, Y.; Nakamura, T.; Mitsuyama, K.; Suzuki, A.; Hibi, T.; Bamba, T. Dietary fiber fraction of germinated barley foodstuff attenuated mucosal damage and diarrhea, and accelerated the repair of the colonic mucosa in an experimental colitis. J. Gastroenterol. Hepatol. 2001, 16, 160–168. [Google Scholar] [CrossRef]
- Araki, Y.; Andoh, A.; Koyama, S.; Fujiyama, Y.; Kanauchi, O.; Bamba, T. Effects of Germinated Barley Foodstuff on Microflora and Short Chain Fatty Acid Production in Dextran Sulfate Sodium-induced Colitis in Rats. Biosci. Biotechnol. Biochem. 2000, 64, 1794–1800. [Google Scholar] [CrossRef]
- Shiba, T.; Aiba, Y.; Ishikawa, H.; Ushiyama, A.; Takagi, A.; Mine, T.; Koga, Y. The Suppressive Effect of Bifidobacteria on Bacteroides vulgatus a Putative Pathogenic Microbe in Inflammatory Bowel Disease. Microbiol. Immunol. 2003, 47, 371–378. [Google Scholar] [CrossRef]
- Gupta, P.; Andrew, H.; Kirschner, B.S.; Guandalini, S. Is Lactobacillus GG Helpful in Children With Crohn’s Disease? Results of a Preliminary, Open-Label Study. J. Pediatr. Gastroenterol. Nutr. 2000, 31, 453–457. [Google Scholar] [CrossRef]
- Kanauchi, O.; Nakamura, T.; Agata, K.; Mitsuyama, K.; Iwanaga, T. Effects of germinated barley foodstuff on dextran sulfate sodium-induced colitis in rats. J. Gastroenterol. 1998, 33, 179–188. [Google Scholar] [CrossRef]
- Setoyama, H.; Imaoka, A.; Ishikawa, H.; Umesaki, Y. Prevention of gut inflammation by Bifidobacterium in dextran sulfate-treated gnotobiotic mice associated with Bacteroides strains isolated from ulcerative colitis patients. Microbes Infect. 2003, 5, 115–122. [Google Scholar] [CrossRef]
- Cherbut, C.; Michel, C.; Lecannu, G. The Prebiotic Characteristics of Fructooligosaccharides Are Necessary for Reduction of TNBS-Induced Colitis in Rats. J. Nutr. 2003, 133, 21–27. [Google Scholar] [CrossRef]
- Matsumoto, S.; Watanabe, N.; Imaoka, A.; Okabe, Y. Preventive Effects of Bifidobacterium- and Lactobacillus-Fermented Milk on the Development of Inflammatory Bowel Disease in Senescence-Accelerated Mouse P1/Yit Strain Mice. Digestion 2001, 64, 92–99. [Google Scholar] [CrossRef]
- Videla, S.; Vilaseca, J.; Antolín, M.; García-Lafuente, A.; Guarner, F.; Crespo, E.; Casalots, J.; Salas, A.; Malagelada, J.R. Dietary inulin improves distal colitis induced by dextran sodium sulfate in the rat. Am. J. Gastroenterol. 2001, 96, 1486–1493. [Google Scholar] [CrossRef]
- Schneeman, B.O. Fiber, Inulin and Oligofructose: Similarities and Differences. J. Nutr. 1999, 129, 1424S–1427S. [Google Scholar] [CrossRef] [Green Version]
- Kleessen, B.; Hartmann, L.; Blaut, M. Oligofructose and long-chain inulin: Influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 2001, 86, 291–300. [Google Scholar] [CrossRef]
- Hoentjen, F.; Welling, G.W.; Harmsen, H.J.M.; Zhang, X.; Snart, J.; Tannock, G.W.; Lien, K.; A Churchill, T.; Lupicki, M.; A Dieleman, L. Reduction of Colitis by Prebiotics in HLA-B27 Transgenic Rats Is Associated with Microflora Changes and Immunomodulation. Inflamm. Bowel Dis. 2005, 11, 977–985. [Google Scholar] [CrossRef]
- Dignass, A.; Lindsay, J.O.; Sturm, A.; Windsor, A.; Colombel, J.-F.; Allez, M.; D’Haens, G.; D’Hoore, A.; Mantzaris, G.; Novacek, G.; et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis Part 2: Current management. J. Crohn’s Coliti 2012, 6, 991–1030. [Google Scholar] [CrossRef] [Green Version]
- Nerstedt, A.; Nilsson, E.C.; Ohlson, K.; Håkansson, J.; Svensson, L.T.; Löwenadler, B.; Svensson, U.K.; Mahlapuu, M. Administration of Lactobacillus evokes coordinated changes in the intestinal expression profile of genes regulating energy homeostasis and immune phenotype in mice. Br. J. Nutr. 2007, 97, 1117–1127. [Google Scholar] [CrossRef]
- Barbosa, T.M.; Serra, C.R.; La Ragione, R.M.; Woodward, M.J.; Henriques, A.O. Screening for Bacillus Isolates in the Broiler Gastrointestinal Tract. Appl. Environ. Microbiol. 2005, 71, 968–978. [Google Scholar] [CrossRef]
- Resta-Lenert, S.; E Barrett, K. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 2003, 52, 988–997. [Google Scholar] [CrossRef] [Green Version]
- Peluso, I.; Fina, D.; Caruso, R.; Stolfi, C.; Caprioli, F.; Fantini, M.C.; Caspani, G.; Grossi, E.; Di Iorio, L.; Paone, F.M.; et al. Lactobacillus paracasei subsp. paracasei B21060 Suppresses Human T-Cell Proliferation. Infect. Immun. 2007, 75, 1730–1737. [Google Scholar] [CrossRef]
- Cary, V.A.; Boullata, J. What is the evidence for the use of probiotics in the treatment of inflammatory bowel disease? J. Clin. Nurs. 2010, 19, 904–916. [Google Scholar] [CrossRef]
- Zocco, M.A.; Verme, L.Z.D.; Cremonini, F.; Piscaglia, A.C.; Nista, E.C.; Candelli, M.; Novi, M.; Rigante, D.; Cazzato, I.A.; Ojetti, V.; et al. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 2006, 23, 1567–1574. [Google Scholar] [CrossRef]
- Oliva, S.; Di Nardo, G.; Ferrari, F.; Mallardo, S.; Rossi, P.; Patrizi, G.; Cucchiara, S.; Stronati, L. Randomised clinical trial: The effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment. Pharmacol. Ther. 2012, 35, 327–334. [Google Scholar] [CrossRef]
- Henker, J.; Laass, M.W.; Schreiner, A.; Schulze, J.; Müller, S. Probiotic Escherichia coli Nissle 1917 (EcN) for Successful Remission Maintenance of Ulcerative Colitis in Children and Adolescents: An Open-Label Pilot Study. Z. Gastroenterol. 2008, 46, 874–875. [Google Scholar] [CrossRef]
- Kruis, W.; Schütz, E.; Fric, P.; Fixa, B.; Judmaier, G.; Stolte, M. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 1997, 11, 853–858. [Google Scholar] [CrossRef]
- Kato, K.; Mizuno, S.; Umesaki, Y.; Ishii, Y.; Sugitani, M.; Imaoka, A.; Otsuka, M.; Hasunuma, O.; Kurihara, R.; Iwasaki, A.; et al. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment. Pharmacol. Ther. 2004, 20, 1133–1141. [Google Scholar] [CrossRef]
- Guslandi, M. Saccharomyces Boulardii Plus Rifaximin in Mesalamine-intolerant Ulcerative Colitis. J. Clin. Gastroenterol. 2010, 44, 1. [Google Scholar] [CrossRef]
- Venturi; Gionchetti, P.; Rizzello, F.; Johansson, R.; Zucconi, E.; Brigidi, P.; Matteuzzi, D.; Campieri, M. Impact on the composition of the faecal flora by a new probiotic preparation: Preliminary data on maintenance treatment of patients with ulcerative colitis. Aliment. Pharmacol. Ther. 1999, 13, 1103–1108. [Google Scholar] [CrossRef]
- Sood, A.; Midha, V.; Makharia, G.K.; Ahuja, V.; Singal, D.; Goswami, P.; Tandon, R.K. The Probiotic Preparation, VSL#3 Induces Remission in Patients With Mild-to-Moderately Active Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 2009, 7, 1202–1209. [Google Scholar]
- Tursi, A.; Brandimarte, G.; Papa, A.; Giglio, A.; Elisei, W.; Giorgetti, G.M.; Forti, G.; Morini, S.; Hassan, C.; Pistoia, M.A.; et al. Treatment of Relapsing Mild-to-Moderate Ulcerative Colitis With the Probiotic VSL#3 as Adjunctive to a Standard Pharmaceutical Treatment: A Double-Blind, Randomized, Placebo-Controlled Study. Am. J. Gastroenterol. 2010, 105, 2218–2227. [Google Scholar]
- Mardini, H.E.; Grigorian, A.Y. Probiotic mix VSL# 3 is effective adjunctive therapy for mild to moderately active ulcerative colitis: A meta-analysis. Inflamm. Bowel Dis. 2014, 20, 1562–1567. [Google Scholar]
- Jijon, H.; Backer, J.; Diaz, H.; Yeung, H.; Thiel, D.; McKaigney, C.; De Simone, C.; Madsen, K. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology 2004, 126, 1358–1373. [Google Scholar] [CrossRef]
- Lee, J.; Rachmilewitz, D.; Raz, E. Homeostatic Effects of TLR9 Signaling in Experimental Colitis. Ann. N. Y. Acad. Sci. 2006, 1072, 351–355. [Google Scholar] [CrossRef]
- McCarthy, J.; Sheil, B.; O’Mahony, L.; Bennett, M.; Ryan, P.; FitzGibbon, J.; Kiely, B.; Collins, J.; Shanahan, F. Is the Mucosal Route of Administration Essential for Probiotic Function? Subcutaneous Administration is Associated with Attenuation of Murine Colitis and Arthritis. Endoscopy 2004, 36, 694–700. [Google Scholar] [CrossRef]
- Roller, M.; Rechkemmer, G.; Watzl, B. Prebiotic Inulin Enriched with Oligofructose in Combination with the Probiotics Lactobacillus rhamnosus and Bifidobacterium lactis Modulates Intestinal Immune Functions in Rats. J. Nutr. 2004, 134, 153–156. [Google Scholar] [CrossRef]
- Ishikawa, H.; Akedo, I.; Umesaki, Y.; Tanaka, R.; Imaoka, A.; Otani, T. Randomized Controlled Trial of the Effect of Bifidobacteria-Fermented Milk on Ulcerative Colitis. J. Am. Coll. Nutr. 2003, 22, 56–63. [Google Scholar] [CrossRef]
- Laake, K.O.; Line, P.-D.; Aabakken, L.; Løtveit, T.; Bakka, A.; Eide, J.; Røsetti, A.; Grzyb, K.; Bjørneklett, A.; Vatn, M.H.; et al. Assessment of Mucosal Inflammation and Circulation in Response to Probiotics in Patients Operated with Ileal Pouch Anal Anastomosis for Ulcerative Colitis. Scand. J. Gastroenterol. 2003, 38, 409–414. [Google Scholar] [CrossRef]
- Furrie, E.; Macfarlane, S.; Kennedy, A.; Cummings, J.H.; Walsh, S.V.; A O’Neil, D.; Macfarlane, G.T. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: A randomised controlled pilot trial. Gut 2005, 54, 242–249. [Google Scholar] [CrossRef]
- Ishikawa, H.; Matsumoto, S.; Ohashi, Y.; Imaoka, A.; Setoyama, H.; Umesaki, Y.; Tanaka, R.; Otani, T. Beneficial Effects of Probiotic Bifidobacterium and Galacto-Oligosaccharide in Patients with Ulcerative Colitis: A Randomized Controlled Study. Digestion 2011, 84, 128–133. [Google Scholar] [CrossRef]
- Kanamori, Y.; Hashizume, K.; Sugiyama, M.; Morotomi, M.; Yuki, N. CASE REPORT: Combination Therapy with Bifidobacterium breve, Lactobacillus casei, and Galactooligosaccharides Dramatically Improved the Intestinal Function in a Girl with Short Bowel Syndrome: A Novel Synbiotics Therapy for Intestinal Failure. Dig. Dis. Sci. 2001, 46, 2010–2016. [Google Scholar] [CrossRef]
- Berg, D.; Clemente, J.C.; Colombel, J.-F. Can inflammatory bowel disease be permanently treated with short-term interventions on the microbiome? Expert Rev. Gastroenterol. Hepatol. 2015, 9, 781–795. [Google Scholar] [CrossRef]
- Van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; De Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal Infusion of Donor Feces for RecurrentClostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef]
- Bennet, J.; Brinkman, M. Treatment of Ulcerative Colitis by Implantation of Normal Colonic Flora. Lancet 1989, 333, 164. [Google Scholar] [CrossRef]
- Cui, B.; Feng, Q.; Wang, H.; Wang, M.; Peng, Z.; Li, P.; Huang, G.; Liu, Z.; Wu, P.; Fan, Z.; et al. Fecal microbiota transplantation through mid-gut for refractory C rohn’s disease: Safety, feasibility, and efficacy trial results. J. Gastroenterol. Hepatol. 2015, 30, 51–58. [Google Scholar] [CrossRef]
- Anderson, J.L.; Edney, R.J.; Whelan, K. Systematic review: Faecal microbiota transplantation in the management of inflammatory bowel disease. Aliment. Pharmacol. Ther. 2012, 36, 503–516. [Google Scholar] [CrossRef]
- Kunde, S.; Pham, A.; Bonczyk, S.; Crumb, T.; Duba, M.; Conrad, H., Jr.; Cloney, D.; Kugathasan, S. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 597–601. [Google Scholar] [CrossRef]
- Rossen, N.G.; Fuentes, S.; Van Der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.; Duflou, A.; Lowenberg, M.; Brink, G.R.V.D.; Mathus-Vliegen, E.M.; De Vos, W.M.; et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 2015, 149, 110–118. [Google Scholar] [CrossRef]
- Angelberger, S.; Reinisch, W.; Makristathis, A.; Lichtenberger, C.; Dejaco, C.; Papay, P.; Novacek, G.; Trauner, M.; Loy, A.; Berry, D. Temporal Bacterial Community Dynamics Vary Among Ulcerative Colitis Patients After Fecal Microbiota Transplantation. Am. J. Gastroenterol. 2013, 108, 1620–1630. [Google Scholar] [CrossRef]
- Kump, P.K.; Gröchenig, H.-P.; Lackner, S.; Trajanoski, S.; Reicht, G.; Hoffmann, K.M.; Deutschmann, A.; Wenzl, H.H.; Petritsch, W.; Gorkiewicz, G.; et al. Alteration of Intestinal Dysbiosis by Fecal Microbiota Transplantation Does not Induce Remission in Patients with Chronic Active Ulcerative Colitis. Inflamm. Bowel Dis. 2013, 19, 2155–2165. [Google Scholar] [CrossRef] [Green Version]
- Rossen, N.G.; Macdonald, J.K.; De Vries, E.M.; D’Haens, G.R.; De Vos, W.M.; Zoetendal, E.G.; Ponsioen, C.Y. Fecal microbiota transplantation as novel therapy in gastroenterology: A systematic review. World J. Gastroenterol. 2015, 21, 5359–5371. [Google Scholar] [CrossRef]

| Model | Bacteria | Comments | Ref. |
|---|---|---|---|
| Human | Firmicutes (F. prausnitzii) | Decrease | [141] |
| Human | Bifidobacterium (Bifidobacterium adolescentis, D. invisus R. gnavus | Decrease Increase | [82] |
| Human | Clostridium clusters (IV and IXV) | Decrease | [142] |
| Human | Enterobacteriaceae (E. coli) | Increase | [143] |
| Human | Lactobacillus | Decrease | [144] |
| Human | Bacteroidetes, Enterobacteriaceae Firmicutes | Increase Decrease | [71] |
| Human | Helicobacter species (H. pylori) | Increase | [145] |
| Human | Mycobacterial species (MAP) | Increase | [146] |
| Human | Proteobacteria (E. coli and non-jejuni Campylobacter) | Increase | [147,148] |
| Human | Firmicutes and Bacteroidetes, Lachnospiracea Proteobacteria, Actinobacteria, Bacillus | Decrease Increase | [28] |
| Human | Staphylococcaceae, Strepotococcaceae, P. maltophilia, Klebsiella, Salmonella | Increase | [149] |
| Human | Faecalibacterium prausnitzii Escherichia coli, Fusobacterium | Decrese Increase | [150] |
| Human | Firmicutes, Faecalibacterium prausnitzii, Bifidobacteria | Decrease | [151] |
| Human | Bacteroidetes, Bacteroides, Flavobacterium, and Oscillospira Proteobacteria, Verrucomicrobia, Fusobacteria, Escherichia, Faecalibacterium, Streptococcus | Decrease | [152] |
| Human | Clostridium leptum, Faecalibacterium prausnitzi. Bacteroides spp | Decrease Increase | [153] |
| Human | Clostridia spp., Bacteroides Bacteroides fragilis, proteobacteria | Decrease Increase | [154] |
| Human | enterobactreiace | Increase | [155] |
| Human | Firmicutes: Roseburia, Phascolarctobacterium Enterobacteriaceae: Escherichia/Shigella, Ruminococcus gnavus | Decrease | [156] |
| Human | Firmicutes, Bacteroides species, Eubacterium species, Lactobacillus species Proteobacteria Enterobacteriaceae, | Decrease Increase | [157] |
| Human | Faecalibacterium and Roseburia Enterobacteriaceae and Ruminococcus gnavus | Decrease Increase | [52] |
| Human | Bacteroides, Lactobacillus, Ruminococcus, and Bifidobacterium Peptostreptococcus, Campylobacter, Methanobrevibacter | Decrease Increase | [158] |
| Human | Prevotella copri, Faecalibacterium prauznitzii Enterobacteriaceae | Decrease Increase | [159] |
| Human | Pseudomonas | Increase | [160] |
| Human | Roseburia hominis, Faecalibacterium prausnitzii | Decrease | [161] |
| Human | Proteobacteria, Bacteroidetes, Clostridia | Unchanged | [143] |
| Human | Bifidobacterium, E. coli Firmicutes, C. Coccoides, C. leptum, | Increase | [162] |
| Human | Bacteroidetes Firmicutes, Enterobacteriaceae | Decrease Increase | [163] |
| Human | Firmicutes, F. prausnitzii Bacteroidetes, Proteobacteria | Decrease Increase | [71,164] |
| Human | Firmicutes, Lachnospiraceae, Bacteroidetes, Actinobacteria, Bifidobacteriaceae, Proteobacteria | Decrease Increase | [83] |
| Model | Bacteria | Comments | Ref. |
|---|---|---|---|
| DSS-colitis | Bacteroides distasonis, Clostridium ramosum, Akkermansia muciniphila, Enterobacteriaceae | Increase | [165,166] |
| TNBS colitis | Enterobacteriaceae, Bacteroides | Increase | [167] |
| T-bet−/−1, Rag2−/− mice* | Mucispirillum, Desulfovibrio, and Helicobacteraceae | Increase | [168] |
| Gonobiotic mice | change in species diversity | Species diversity decrease | [169] |
| Colitis in IL-10−/− mice* | Enterobacteriaceae and adherent-invasive E. coli | Increased | [106,170] |
| Colitis inApc468/IL-10−/− mice* | Bacteroides and Porphyromonas genera | Increased | [171] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. https://doi.org/10.3390/pathogens8030126
Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, Che T, Zhang C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens. 2019; 8(3):126. https://doi.org/10.3390/pathogens8030126
Chicago/Turabian StyleKhan, Israr, Naeem Ullah, Lajia Zha, Yanrui Bai, Ashiq Khan, Tang Zhao, Tuanjie Che, and Chunjiang Zhang. 2019. "Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome" Pathogens 8, no. 3: 126. https://doi.org/10.3390/pathogens8030126
APA StyleKhan, I., Ullah, N., Zha, L., Bai, Y., Khan, A., Zhao, T., Che, T., & Zhang, C. (2019). Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens, 8(3), 126. https://doi.org/10.3390/pathogens8030126





