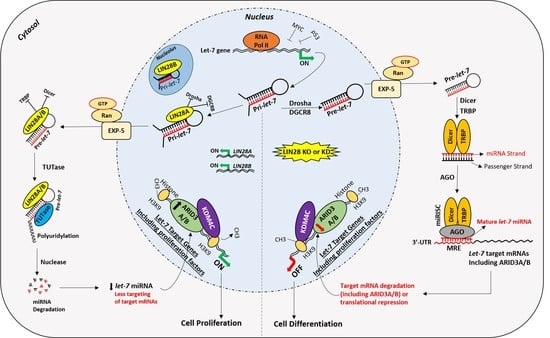
The Role of LIN28-let-7-ARID3B Pathway in Placental Development
Abstract
1. Introduction
2. Early Placental Development and Trophoblast Cells
2.1. Syncytial Pathway
2.2. Invasive Pathway
3. Functional Analysis of microRNAs in Trophoblast Cells
4. Let-7 miRNAs
5. Suppression of let-7 miRNAs by LIN28
6. Gene Regulation by LIN28-let-7 miRNA Axis in Trophoblast Cells
7. LIN28-let-7-ARID3B Pathway in Trophoblast Cells
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Carter, A.M. Evolution of Placental Function in Mammals: The Molecular Basis of Gas and Nutrient Transfer, Hormone Secretion, and Immune Responses. Physiol. Rev. 2012, 92, 1543–1576. [Google Scholar] [CrossRef] [PubMed]
- Gude, N.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, W.J.; Boyd, J.D. Development of the human placenta in the first three months of gestation. J. Anat. 1960, 94, 297–328. [Google Scholar]
- Crocker, I.P.; Cooper, S.; Ong, S.C.; Baker, P.N. Differences in Apoptotic Susceptibility of Cytotrophoblasts and Syncytiotrophoblasts in Normal Pregnancy to Those Complicated with Preeclampsia and Intrauterine Growth Restriction. Am. J. Pathol. 2003, 162, 637–643. [Google Scholar] [CrossRef]
- Longtine, M.S.; Chen, B.; Odibo, A.; Zhong, Y.; Nelson, D. Villous trophoblast apoptosis is elevated and restricted to cytotrophoblasts in pregnancies complicated by preeclampsia, IUGR, or preeclampsia with IUGR. Placenta 2012, 33, 352–359. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L.; Thornburg, K.L. Placental Origins of Chronic Disease. Physiol. Rev. 2016, 96, 1509–1565. [Google Scholar] [CrossRef]
- Barker, D.J.P. The developmental origins of well-being. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.P. The developmental origins of chronic adult disease. Acta Paediatr. 2004, 93, 26–33. [Google Scholar] [CrossRef]
- Jiang, S. A Regulator of Metabolic Reprogramming: MicroRNA Let-7. Transl. Oncol. 2019, 12, 1005–1013. [Google Scholar] [CrossRef]
- Boyerinas, B.; Park, S.-M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr.-Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef]
- Ali, A.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. LIN28-let-7 axis regulates genes in immortalized human trophoblast cells by targeting the ARID3B-complex. FASEB J. 2019, 33, 12348–12363. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.E.; Finnegan, E.F.; Zisoulis, D.G.; Lovci, M.T.; Melnik-Martinez, K.V.; Yeo, G.W.; Pasquinelli, A.E. Functional Genomic Analysis of the let-7 Regulatory Network in Caenorhabditis elegans. PLoS Genet. 2013, 9, e1003353. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Canfield, J.; Arlıer, S.; Mong, E.F.; Lockhart, J.; Van Wye, J.; Guzeloglu-Kayisli, O.; Schatz, F.; Magness, R.R.; Lockwood, C.J.; Tsibris, J.C.M.; et al. Decreased LIN28B in preeclampsia impairs human trophoblast differentiation and migration. FASEB J. 2018, 33, 2759–2769. [Google Scholar] [CrossRef]
- Bazer, F.W.; Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Wu, G. Comparative aspects of implantation. Reproduction 2009, 138, 195–209. [Google Scholar] [CrossRef]
- Herzog, M. A contribution to our knowledge of the earliest known stages of placentation and embryonic development in man. Am. J. Anat. 2005, 9, 361–400. [Google Scholar] [CrossRef]
- Pötgens, A.; Schmitz, U.; Bose, P.; Versmold, A.; Kaufmann, P.; Frank, H.-G. Mechanisms of Syncytial Fusion: A Review. Placenta 2002, 23, S107–S113. [Google Scholar] [CrossRef]
- Kim, S.-M.; Kim, J.-S. A Review of Mechanisms of Implantation. Dev. Reprod. 2017, 21, 351–359. [Google Scholar] [CrossRef]
- Benirschke, K.; Burton, G.J.; Baergen, R.N. The Pathology of the Human Placenta; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2012; pp. 97–571. [Google Scholar]
- Frank, H.-G. 10-Placental Development. In Fetal and Neonatal Physiology, 5th ed.; Polin, R.A., Abman, S.H., Rowitch, D.H., Benitz, W.E., Fox, W.W., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 101–113. [Google Scholar]
- Chen, D.-B.; Zheng, J. Regulation of placental angiogenesis. Microcirculation 2014, 21, 15–25. [Google Scholar] [CrossRef]
- Vicovac, L. Trophoblast differentiation during formation of anchoring villi in a model of the early human placenta in vitro. Placenta 1995, 16, 41–56. [Google Scholar] [CrossRef]
- Damsky, C.H.; Fitzgerald, M.L.; Fisher, S.J. Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J. Clin. Investig. 1992, 89, 210–222. [Google Scholar] [CrossRef]
- Prakobphol, A.; Genbacev, O.; Gormley, M.; Kapidzic, M.; Fisher, S.J. A role for the L-selectin adhesion system in mediating cytotrophoblast emigration from the placenta. Dev. Biol. 2006, 298, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Kertschanska, S.; Kadyrov, M.; Rath, W.; Kaufmann, P.; Huppertz, B. Invasive depth of extravillous trophoblast correlates with cellular phenotype:A comparison of intra- and extrauterine implantation sites. Histochem. Cell Biol. 2002, 117, 401–414. [Google Scholar] [CrossRef]
- Espinoza, J.; Romero, R.; Kim, Y.M.; Kusanovic, J.P.; Hassan, S.; Erez, O.; Gotsch, F.; Than, N.G.; Papp, Z.; Kim, C.J. Normal and abnormal transformation of the spiral arteries during pregnancy. J. Périnat. Med. 2006, 34, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Anin, S.; Vince, G.; Quenby, S. Trophoblast invasion. Hum. Fertil. 2004, 7, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Pijnenborg, R.; Vercruysse, L.; Hanssens, M. The Uterine Spiral Arteries in Human Pregnancy: Facts and Controversies. Placenta 2006, 27, 939–958. [Google Scholar] [CrossRef]
- Lyall, F.; Bulmer, J.N.; Duffie, E.; Cousins, F.; Thériault, A.; Robson, S.C. Human Trophoblast Invasion and Spiral Artery Transformation. Am. J. Pathol. 2001, 158, 1713–1721. [Google Scholar] [CrossRef]
- Red-Horse, K.; Zhou, Y.; Genbacev, O.; Prakobphol, A.; Foulk, R.; McMaster, M.; Fisher, S.J. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Investig. 2004, 114, 744–754. [Google Scholar] [CrossRef]
- Reynolds, L.; Redmer, D.A. Angiogenesis in the Placenta1. Biol. Reprod. 2001, 64, 1033–1040. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.-T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef]
- Christie, M.; Boland, A.; Huntzinger, E.; Weichenrieder, O.; Izaurralde, E. Structure of the PAN3 Pseudokinase Reveals the Basis for Interactions with the PAN2 Deadenylase and the GW182 Proteins. Mol. Cell 2013, 51, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genome Res. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs:Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Doridot, L.; Miralles, F.; Barbaux, S.; Vaiman, D. Trophoblasts, invasion, and microRNA. Front. Genet. 2013, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, H.; Yang, J.; Long, W.; Zhang, B.; Liu, J.; Yu, B. Down-regulated circPAPPA suppresses the proliferation and invasion of trophoblast cells via the miR-384/STAT3 pathway. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.M.; Liu, Y.H.; Zhang, Z.; Han, N.; Xue, S.H.; Wang, P. Effect of microRNA-106b on the invasion and proliferation of trophoblasts through targeting MMP-2. Zhonghua Fu Chan Ke Za Zhi 2017, 52, 327–332. [Google Scholar]
- Liu, F.; Wu, K.; Wu, W.; Chen, Y.; Wu, H.; Wang, H.; Zhang, W. miR-203 contributes to pre-eclampsia via inhibition of VEGFA expression. Mol. Med. Rep. 2018, 17, 5627–5634. [Google Scholar] [CrossRef]
- Jiang, L.; Long, A.; Tan, L.; Hong, M.; Wu, J.; Cai, L.; Li, Q. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 2017, 51, 70–75. [Google Scholar] [CrossRef]
- Wang, R.; Liu, W.; Liu, X.; Liu, X.; Tao, H.; Wu, D.; Zhao, Y.; Zou, L. MicroRNA-210 regulates human trophoblast cell line HTR-8/SVneo function by attenuating Notch1 expression: Implications for the role of microRNA-210 in pre-eclampsia. Mol. Reprod. Dev. 2019, 86, 896–907. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, X.; Sun, Q.; Dai, X.; Cai, Y. MicroRNA-16 is involved in the pathogenesis of pre-eclampsia via regulation of Notch2. J. Cell. Physiol. 2019, 235, 4530–4544. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Jia, Z.; Li, L. miR-320a upregulation contributes to the development of preeclampsia by inhibiting the growth and invasion of trophoblast cells by targeting interleukin 4. Mol. Med. Rep. 2019, 20, 3256–3264. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Meng, Q.; Shi, Y.-P.; Xu, H.-S. Regulatory role of microRNA-320a in the proliferation, migration, invasion, and apoptosis of trophoblasts and endothelial cells by targeting estrogen-related receptor γ. J. Cell. Physiol. 2018, 234, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, X.; He, Z.; Xiong, Y.; Fang, Q. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J. Cell. Mol. Med. 2019, 23, 4422–4433. [Google Scholar] [CrossRef]
- Shih, J.-C.; Lin, H.-H.; Hsiao, A.-C.; Su, Y.-T.; Tsai, S.; Chien, C.-L.; Kung, H.-N. Unveiling the role of microRNA-7 in linking TGF-β-Smad-mediated epithelial-mesenchymal transition with negative regulation of trophoblast invasion. FASEB J. 2019, 33, 6281–6295. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Wu, P.Y.; Gao, R.Q. MiR-218 inhibits HTR-8 cells migration and invasion by targeting SOX4. Zhongguo Ying Yong Sheng Li Xue Za Zhi 2017, 33, 169–173. [Google Scholar] [PubMed]
- Xue, F.; Yang, J.; Li, Q.; Zhou, H. Down-regulation of microRNA-34a-5p promotes trophoblast cell migration and invasion via targetting Smad4. Biosci. Rep. 2019, 39, 39. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Xu, J.; Zhang, X.; Zhang, H.; Xiang, Y.; Fang, C.; Wang, T.; Xia, S.; Zhang, Q.; et al. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signaling. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Sun, M.; Chen, H.; Liu, J.; Tong, C.; Meng, T. MicroRNA-34a inhibits human trophoblast cell invasion by targeting MYC. BMC Cell Biol. 2015, 16. [Google Scholar] [CrossRef]
- Ding, J.; Huang, F.; Wu, G.; Han, T.; Xu, F.; Weng, D.; Wu, C.; Zhang, X.; Yao, Y.; Zhu, X. MiR-519d-3p Suppresses Invasion and Migration of Trophoblast Cells via Targeting MMP-2. PLoS ONE 2015, 10, e0120321. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Li, J.; Zhong, Q.; Li, Y. MiR-101 inhibits migration and invasion of trophoblast HTR-8/SVneo cells by targeting CXCL6 in preeclampsia. Minerva Med. 2019. [Google Scholar] [CrossRef]
- Liu, J.-J.; Zhang, L.; Zhang, F.-F.; Luan, T.; Yin, Z.-M.; Rui, C.; Ding, H.-J. Influence of miR-34a on preeclampsia through the Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 923–931. [Google Scholar] [PubMed]
- Yang, X.; Meng, T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene 2018, 683, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Chen, Y.; Long, Y.; Yu, J.; Li, M. Tumor necrosis factor-alpha suppresses the invasion of HTR-8/SVneo trophoblast cells through microRNA-145-5p-mediated downregulation of Cyr61. Life Sci. 2018, 209, 132–139. [Google Scholar] [CrossRef]
- Zou, A.-X.; Chen, B.; Li, Q.-X.; Liang, Y.-C. MiR-134 inhibits infiltration of trophoblast cells in placenta of patients with preeclampsia by decreasing ITGB1 expression. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2199–2206. [Google Scholar]
- Ding, J.; Cheng, Y.; Zhang, Y.; Liao, S.; Yin, T.; Yang, J. The miR-27a-3p/USP25 axis participates in the pathogenesis of recurrent miscarriage by inhibiting trophoblast migration and invasion. J. Cell. Physiol. 2019, 234, 19951–19963. [Google Scholar] [CrossRef]
- Wang, N.; Feng, Y.; Xu, J.; Zou, J.; Chen, M.; He, Y.; Liu, H.; Xue, M.; Feng, Y.-L. miR-362-3p regulates cell proliferation, migration and invasion of trophoblastic cells under hypoxia through targeting Pax3. Biomed. Pharmacother. 2018, 99, 462–468. [Google Scholar] [CrossRef]
- Wu, L.; Song, W.-Y.; Xie, Y.; Hu, L.-L.; Hou, X.-M.; Wang, R.; Gao, Y.; Zhang, J.-N.; Zhang, L.; Li, W.-W.; et al. miR-181a-5p suppresses invasion and migration of HTR-8/SVneo cells by directly targeting IGF2BP2. Cell Death Dis. 2018, 9, 16. [Google Scholar] [CrossRef]
- Peng, H.-Y.; Li, M.-Q.; Li, H.-P. MiR-137 Restricts the Viability and Migration of HTR-8/SVneo Cells by Downregulating FNDC5 in Gestational Diabetes Mellitus. Curr. Mol. Med. 2019, 19, 494–505. [Google Scholar] [CrossRef]
- Qian, S.; Liu, R. miR-30b facilitates preeclampsia through targeting MXRA5 to inhibit the viability, invasion and apoptosis of placental trophoblast cells. Int. J. Clin. Exp. Pathol. 2019, 12, 4057–4065. [Google Scholar]
- Niu, Z.-R.; Han, T.; Sun, X.-L.; Luan, L.-X.; Gou, W.; Zhu, X. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am. J. Obstet. Gynecol. 2018, 218, 249. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, X.; Weng, Z.; Zhang, S.; Ning, H.; Li, B. Expression and role of microRNA 18b and hypoxia inducible factor-1α in placental tissues of preeclampsia patients. Exp. Ther. Med. 2017, 14, 4554–4560. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, Y.; She, R.; Wang, Q.; Li, Y.; Zhang, H. Up-regulation of miR-299 suppressed the invasion and migration of HTR-8/SVneo trophoblast cells partly via targeting HDAC2 in pre-eclampsia. Biomed. Pharmacother. 2018, 97, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; She, K.; Li, H.; Yuan, X.; Han, X.; Wang, Y. MicroRNA-454 contributes to sustaining the proliferation and invasion of trophoblast cells through inhibiting Nodal/ALK7 signaling in pre-eclampsia. Chem. Interact. 2019, 298, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Zhang, M. Exploration of the regulation and control mechanisms of miR-145 in trophoblast cell proliferation and invasion. Exp. Ther. Med. 2018, 16, 5298–5304. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, H.; Ma, Y.; Zhu, X.; Zhang, S.; Li, J. MiR-221-3p is down-regulated in preeclampsia and affects trophoblast growth, invasion and migration partly via targeting thrombospondin 2. Biomed. Pharmacother. 2019, 109, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, J.; Cao, Z.; Gao, L.; Zheng, Y.; Wang, J.; Liu, X. miR-126a-3p induces proliferation, migration and invasion of trophoblast cells in pre-eclampsia-like rats by inhibiting A Disintegrin and Metalloprotease 9. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Li, J.; Fu, Z.; Jiang, H.; Chen, L.; Wu, X.; Ding, H.; Xia, Y.; Wang, X.; Tang, Q.; Wu, W. IGF2-derived miR-483-3p contributes to macrosomia through regulating trophoblast proliferation by targeting RB1CC1. Mol. Hum. Reprod. 2018, 24, 444–452. [Google Scholar] [CrossRef]
- Xiao, J.; Tao, T.; Yin, Y.; Zhao, L.; Yang, L.; Hu, L. miR-144 may regulate the proliferation, migration and invasion of trophoblastic cells through targeting PTEN in preeclampsia. Biomed. Pharmacother. 2017, 94, 341–353. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Y.; Lu, H.; Wang, H.; Shi, X.; Shao, X.; Li, Y.-X.; Zhao, Y.; Wang, Y.-L. miR-518b Enhances Human Trophoblast Cell Proliferation Through Targeting Rap1b and Activating Ras-MAPK Signal. Front. Endocrinol. 2018, 9, 100. [Google Scholar] [CrossRef]
- Brkić, J.; Dunk, C.; O’Brien, J.; Fu, G.; Nadeem, L.; Wang, Y.-L.; Rosman, D.; Salem, M.; Shynlova, O.; Yougbaré, I.; et al. MicroRNA-218-5p Promotes Endovascular Trophoblast Differentiation and Spiral Artery Remodeling. Mol. Ther. 2018, 26, 2189–2205. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, Y.; Luo, R.; Bian, X.; Wang, Y.; Shao, X.; Li, Y.-X.; Liu, M.; Wang, Y.-L. A positive feedback self-regulatory loop between miR-210 and HIF-1α mediated by CPEB2 is involved in trophoblast syncytiolization:Implication of trophoblast malfunction in preeclampsia. Biol. Reprod. 2019. [Google Scholar] [CrossRef]
- Kumar, P.; Luo, Y.; Tudela, C.; Alexander, J.M.; Mendelson, C.R. The c-Myc-Regulated MicroRNA-17∼92 (miR-17∼92) and miR-106a∼363 Clusters Target hCYP19A1 and hGCM1 To Inhibit Human Trophoblast Differentiation. Mol. Cell. Biol. 2013, 33, 1782–1796. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Jiang, S.; Chen, J.; Li, J.; Ao, L.; Zhang, Y. Upregulated long noncoding RNA Linc00261 in pre-eclampsia and its effect on trophoblast invasion and migration via regulating miR-558/TIMP4 signaling pathway. J. Cell. Biochem. 2019, 120, 13243–13253. [Google Scholar] [CrossRef]
- Wang, X.; Peng, S.; Cui, K.; Hou, F.; Ding, J.; Li, A.; Wang, M.; Geng, L. MicroRNA-576-5p enhances the invasion ability of trophoblast cells in preeclampsia by targeting TFAP2A. Mol. Genet. Genom. Med. 2019, 8, e1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, J.; Li, M.-Q.; Xu, J.; Zhang, J.-P.; Jin, L.-P. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis. 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-M.; Cao, P.; Xin, L.; Zhang, Y.; Liu, Z.; Yao, N.; Ma, Y.-Y. Effect of miR-133 on apoptosis of trophoblasts in human placenta tissues via Rho/ROCK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10600–10608. [Google Scholar] [PubMed]
- Zhang, L.; Yuan, J.-M.; Zhao, R.-H.; Wang, L.-M.; Tu, Z.-B. Correlation of MiR-152 expression with VEGF expression in placental tissue of preeclampsia rat and its influence on apoptosis of trophoblast cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3553–3560. [Google Scholar] [PubMed]
- Li, X.; Lu, J.; Dong, L.; Lv, F.; Liu, W.; Liu, G.; Zhu, W.; Diao, X. Effects of MiR-155 on trophoblast apoptosis in placental tissues of preeclampsia rats through HIF-1α signaling pathway. Panminerva Med. 2019. [Google Scholar] [CrossRef]
- Du, E.; Cao, Y.; Feng, C.; Lu, J.; Yang, H.; Zhang, Y. The Possible Involvement of miR-371a-5p Regulating XIAP in the Pathogenesis of Recurrent Pregnancy Loss. Reprod. Sci. 2019, 26, 1468–1475. [Google Scholar] [CrossRef]
- Dong, X.; Yang, L.; Wang, H. miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA). Gynecol. Endocrinol. 2016, 33, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhao, X.; Yuan, X.; Li, P. Elevated microRNA-34a contributes to trophoblast cell apoptosis in preeclampsia by targeting BCL-2. J. Hum. Hypertens. 2017, 31, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, S.; Li, Y.; Han, B.; Ma, Y. Inhibition of miR-18a increases expression of estrogen receptor 1 and promotes apoptosis in human HTR8 trophoblasts. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2017, 33, 1102–1107. [Google Scholar] [PubMed]
- West, R.; Russ, J.E.; Bouma, G.J.; Winger, Q.A. BRCA1 regulates HMGA2 levels in the Swan71 trophoblast cell line. Mol. Reprod. Dev. 2019, 86, 1663–1670. [Google Scholar] [CrossRef]
- Li, L.; Hou, A.; Gao, X.; Zhang, J.; Zhang, L.; Wang, J.; Li, H.; Song, Y. Lentivirus-mediated miR-23a overexpression induces trophoblast cell apoptosis through inhibiting X-linked inhibitor of apoptosis. Biomed. Pharmacother. 2017, 94, 412–417. [Google Scholar] [CrossRef]
- Mao, Z.; Yao, M.; Li, Y.; Fu, Z.; Li, S.; Zhang, L.; Zhou, Z.; Tang, Q.; Han, X.; Xia, Y. miR-96-5p and miR-101-3p as potential intervention targets to rescue TiO2 NP-induced autophagy and migration impairment of human trophoblastic cells. Biomater. Sci. 2018, 6, 3273–3283. [Google Scholar] [CrossRef]
- Zhang, X.; Ge, Y.-W.; Wang, Z.-X.; Xu, Q.-L.; Guo, R.; Xu, H.-Y. MiR-200c regulates apoptosis of placental trophoblasts in preeclampsia rats through Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7209–7216. [Google Scholar]
- Gu, Y.; Meng, J.; Zuo, C.; Wang, S.; Li, H.; Zhao, S.; Huang, T.; Wang, X.-T.; Yan, J. Downregulation of MicroRNA-125a in Placenta Accreta Spectrum Disorders Contributes Antiapoptosis of Implantation Site Intermediate Trophoblasts by Targeting MCL1. Reprod. Sci. 2019, 26, 1582–1589. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Liu, S.; Xia, Q.; Zhao, P.; Cheng, T.; Hong, K.; Xiang, Z. Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev. Biol. 2007, 7, 88. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.J.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.; Degnan, B.M.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2015, 7, 100–113. [Google Scholar] [CrossRef]
- Zhang, H.; Artiles, K.L.; Fire, A.Z. Functional relevance of “seed” and “non-seed” sequences in microRNA-mediated promotion of C. elegans developmental progression. RNA 2015, 21, 1980–1992. [Google Scholar] [CrossRef]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.-P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Zhu, H.; De Soysa, T.Y.; Shinoda, G.; Seligson, M.T.; Tsanov, K.M.; Nguyen, L.; Asara, J.M.; Cantley, L.C.; Daley, G.Q. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013, 155, 778–792. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Daley, G.Q. Lin28:Primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013, 12, 395–406. [Google Scholar] [CrossRef]
- Caygill, E.E.; Johnston, L.A. Temporal Regulation of Metamorphic Processes in Drosophila by the let-7 and miR-125 Heterochronic MicroRNAs. Curr. Biol. 2008, 18, 943–950. [Google Scholar] [CrossRef]
- Wagner, S.; Ngezahayo, A.; Murua Escobar, H.; Nolte, I. Role of miRNA let-7 and its major targets in prostate cancer. Biomed Res. Int. 2014, 2014, 14. [Google Scholar] [CrossRef]
- Takamizawa, J.; Chamoto, K.; Tsuji, T.; Funamoto, H.; Kosaka, A.; Matsuzaki, J.; Sato, T.; Konishi, H.; Fujio, K.; Yamamoto, K.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.-M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of Let-7-Regulated Oncofetal Genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef]
- Copley, M.R.; Babovic, S.; Benz, C.; Knapp, D.J.; Beer, P.A.; Kent, D.; Wohrer, S.; Treloar, D.Q.; Day, C.; Rowe, K.; et al. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013, 15, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Emmrich, S.; Rasche, M.; Schöning, J.; Reimer, C.; Keihani, S.; Maroz, A.; Xie, Y.; Li, Z.; Schambach, A.; Reinhardt, D.; et al. miR-99a/100∼125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 2014, 28, 858–874. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Ha, M.; Lim, J.; Yoon, M.-J.; Park, J.-E.; Kwon, S.C.; Chang, H.; Kim, V.N. Mono-Uridylation of Pre-MicroRNA as a Key Step in the Biogenesis of Group II let-7 MicroRNAs. Cell 2012, 151, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Büssing, I.; Slack, F.J.; Großhans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genome Res. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Nam, Y.; Chen, C.; Gregory, R.I.; Chou, J.J.; Sliz, P. Molecular Basis for Interaction of let-7 MicroRNAs with Lin28. Cell 2011, 147, 1080–1091. [Google Scholar] [CrossRef]
- Zhang, P.; Elabd, S.; Hammer, S.; Solozobova, V.; Yan, H.; Bartel, F.; Inoue, S.; Henrich, T.; Wittbrodt, J.; Loosli, F.; et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene 2015, 34, 5729–5738. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Publisher Correction: Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 321. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, Y.; Ito, H.; Watanabe, A.; Ge, S.X.; Kodama, T.; Aburatani, H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006, 384, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Baltimore, D. RNA-binding protein Lin28 in cancer and immunity. Cancer Lett. 2016, 375, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, G.; Shyh-Chang, N.; De Soysa, T.Y.; Zhu, H.; Seligson, M.T.; Shah, S.P.; Abo-Sido, N.; Yabuuchi, A.; Hagan, J.P.; Gregory, R.I.; et al. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 2013, 31, 1563–1573. [Google Scholar] [CrossRef]
- Mayr, F.; Heinemann, U. Mechanisms of Lin28-Mediated miRNA and mRNA Regulation—A Structural and Functional Perspective. Int. J. Mol. Sci. 2013, 14, 16532–16553. [Google Scholar] [CrossRef]
- Zhang, J.; Ratanasirintrawoot, S.; Chandrasekaran, S.; Wu, Z.; Ficarro, S.B.; Yu, C.; Ross, C.A.; Cacchiarelli, D.; Xia, Q.; Seligson, M.; et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 2016, 19, 66–80. [Google Scholar] [CrossRef]
- Viswanathan, S.; Daley, G.Q.; Gregory, R. Selective Blockade of MicroRNA Processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef]
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V.N. Lin28 Mediates the Terminal Uridylation of let-7 Precursor MicroRNA. Mol. Cell 2008, 32, 276–284. [Google Scholar] [CrossRef]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; Lapierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R. Lin28A and Lin28B Inhibit let-7 MicroRNA Biogenesis by Distinct Mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.E.; Chang, H.-M.; Piskounova, E.; Gregory, R. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 2012, 18, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Hagan, J.P.; Piskounova, E.; Gregory, R. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009, 16, 1021–1025. [Google Scholar] [CrossRef]
- Faehnle, C.R.; Walleshauser, J.; Joshua-Tor, L. Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway. Nature 2014, 514, 252–256. [Google Scholar] [CrossRef]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Ustianenko, D.; Chiu, H.-S.; Treiber, T.; Weyn-Vanhentenryck, S.M.; Treiber, N.; Meister, G.; Sumazin, P.; Zhang, C. LIN28 Selectively Modulates a Subclass of Let-7 MicroRNAs. Mol. Cell 2018, 71, 271–283.e5. [Google Scholar] [CrossRef]
- West, R.; McWhorter, E.S.; Ali, A.; Goetzman, L.N.; Russ, J.; Gonzalez-Berrios, C.L.; Anthony, R.V.; Bouma, G.J.; A Winger, Q. HMGA2 is regulated by LIN28 and BRCA1 in human placental cells. Biol. Reprod. 2018, 100, 227–238. [Google Scholar] [CrossRef]
- Seabrook, J.L.; Cantlon, J.D.; Cooney, A.J.; McWhorter, E.E.; Fromme, B.A.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Role of LIN28A in mouse and human trophoblast cell differentiation. Biol. Reprod. 2013, 89, 95. [Google Scholar] [CrossRef]
- Chan, H.-W.; Lappas, M.; Yee, S.Y.; Vaswani, K.; Mitchell, M.; Rice, G.E. The expression of the let-7 miRNAs and Lin28 signalling pathway in human term gestational tissues. Placenta 2013, 34, 443–448. [Google Scholar] [CrossRef]
- Barbaux, S.; Gascoin-Lachambre, G.; Buffat, C.; Monnier, P.; Mondon, F.; Tonanny, M.-B.; Pinard, A.; Auer, J.; Bessières, B.; Barlier, A.; et al. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics 2012, 7, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Monk, D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 2015, 213, S152–S162. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, X.; Wang, R.; Lu, X.; Dang, Y.-L.; Wang, H.; Lin, H.-Y.; Zhu, C.; Ge, H.; Cross, J.C.; et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018, 28, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.A.; Brizot, M.L.; Biancolin, S.E.; Schultz, R.; De Carvalho, M.H.B.; Francisco, R.P.V.; Zugaib, M. Placental weight and birth weight to placental weight ratio in monochorionic and dichorionic growth-restricted and non-growth-restricted twins. Clinics 2017, 72, 265–271. [Google Scholar] [CrossRef]
- Hiden, U.; Wadsack, C.; Prutsch, N.; Gauster, M.; Weiss, U.; Frank, H.-G.; Schmitz, U.; Fast-Hirsch, C.; Hengstschläger, M.; Pötgens, A.; et al. The first trimester human trophoblast cell line ACH-3P: A novel tool to study autocrine/paracrine regulatory loops of human trophoblast subpopulations – TNF-α stimulates MMP15 expression. BMC Dev. Biol. 2007, 7, 137. [Google Scholar] [CrossRef] [PubMed]
- Chavez, S.L.; Abrahams, V.M.; Alvero, A.B.; Aldo, P.B.; Ma, Y.; Guller, S.; Romero, R.; Mor, G. The Isolation and Characterization of a Novel Telomerase Immortalized First Trimester Trophoblast Cell Line, Swan 71. Placenta 2009, 30, 939–948. [Google Scholar] [CrossRef]
- McWhorter, E.S.; West, R.; Russ, J.E.; Ali, A.; Winger, Q.A.; Bouma, G.J. LIN28B regulates androgen receptor in human trophoblast cells through Let-7c. Mol. Reprod. Dev. 2019, 86, 1086–1093. [Google Scholar] [CrossRef]
- Madison, B.B.; Jeganathan, A.N.; Mizuno, R.; Winslow, M.M.; Castells, A.; Cuatrecasas, M.; Rustgi, A.K. Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genome Res. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Stenglein, M.D.; Spencer, T.E.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus. Int. J. Mol. Sci. 2020, 21, 2549. [Google Scholar] [CrossRef] [PubMed]
- Wilsker, D.; Patsialou, A.; Dallas, P.; Moran, E. ARID proteins:A diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 2002, 13, 95–106. [Google Scholar]
- Hong, Z.; Wei, S.; Bi, X.; Zhao, J.; Huang, Z.; Li, Z.; Zhou, J.; Cai, J.; Chen, L.; Lin, C.; et al. Recent advances in the ARID family:Focusing on roles in human cancer. OncoTargets Ther. 2014, 7, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Takahashi, A.; Hara, E.; Horikoshi, N.; Pandita, T.K.; Nakajima, T. E2FBP1 antagonizes the p16INK4A-Rb tumor suppressor machinery for growth suppression and cellular senescence by regulating promyelocytic leukemia protein stability. Int. J. Oral Sci. 2011, 3, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Mogi, K.; Tsunematsu, Y.; Nakajima, T. E2FBP1/hDril1 modulates cell growth through downregulation of promyelocytic leukemia bodies. Cell Death Differ. 2004, 11, 747–759. [Google Scholar] [CrossRef]
- Kobayashi, K.; Jakt, L.M.; Nishikawa, S.-I. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene 2012, 32, 2640–2648. [Google Scholar] [CrossRef][Green Version]
- Bobbs, A.; Gellerman, K.; Hallas, W.M.; Joseph, S.; Yang, C.; Kurkewich, J.; Dahl, K.C. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PLoS ONE 2015, 10, e0131961. [Google Scholar] [CrossRef]
- Nakahara, S.; Fukushima, S.; Yamashita, J.; Kubo, Y.; Tokuzumi, A.; Miyashita, A.; Harada, M.; Nakamura, K.; Jinnin, M.; Ihn, H. AT-rich Interaction Domain-containing Protein 3B is a New Tumour Marker for Melanoma. Acta Derm. Venereol. 2017, 97, 112–114. [Google Scholar] [CrossRef]
- Kim, D.; Probst, L.; Das, C.; Tucker, P.W. REKLES Is an ARID3-restricted Multifunctional Domain. J. Biol. Chem. 2007, 282, 15768–15777. [Google Scholar] [CrossRef]
- Webb, C.F.; Bryant, J.; Popowski, M.; Allred, L.; Kim, D.; Harriss, J.; Schmidt, C.; Miner, C.A.; Rose, K.; Cheng, H.-L.; et al. The ARID Family Transcription Factor Bright Is Required for both Hematopoietic Stem Cell and B Lineage Development. Mol. Cell. Biol. 2011, 31, 1041–1053. [Google Scholar] [CrossRef]
- Liao, T.-T.; Hsu, W.-H.; Ho, C.-H.; Hwang, W.-L.; Lan, H.-Y.; Lo, T.; Chang, C.-C.; Tai, S.; Yang, M.-H. Let-7 Modulates Chromatin Configuration and Target Gene Repression through Regulation of the ARID3B Complex. Cell Rep. 2016, 14, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Lee, B.-K.; Beck, S.; Anjum, A.; Cook, K.R.; Popowski, M.; Tucker, H.O.; Kim, J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genome Res. 2014, 28, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Edwards, M.; Dang, C.; Harris, J.; Brown, M.; Kim, J.; Tucker, H.O. ARID3A is required for mammalian placenta development. Dev. Biol. 2016, 422, 83–91. [Google Scholar] [CrossRef]





| miRNA | Target Genes | Reference | Effect of Higher miRNA Expression |
|---|---|---|---|
| let-7 | ARID3A, ARID3B, HMGA1, cMYC | [11] | Reduces proliferation and invasiveness of trophoblast cells |
| miR-384 | STAT3 | [38] | |
| miR-106b | MMP-2 | [39] | |
| miR-203 | VEGFA | [40] | |
| miR-520g | MMP-2 | [41] | |
| miR-210 | Notch1 | [42] | |
| miR-16 | Notch2 | [43] | |
| miR-320a | IL-4 | [44] | |
| miR-320a | ERRγ | [45] | |
| miR-210-3p | FGF1 | [46] | |
| miR-7 | EMT-related TFs | [47] | Reduces migration and invasion of trophoblast cells |
| miR-218 | SOX4 | [48] | |
| miR-34a-5p | Smad4 | [49] | |
| miR-193b-3p | TGF-β2 | [50] | |
| miR-34a | MYC | [51] | |
| miR-519d | MMP-2 | [52] | |
| miR-101 | CXCL6 | [53] | |
| miR-34a | Notch | [54] | |
| miR-431 | ZEB1 | [55] | |
| miR-145-5p | Cyr61 | [56] | |
| miR-134 | ITGB1 | [57] | |
| miR-27a-3p | USP25 | [58] | |
| miR-362-3p | Pax3 | [59] | |
| miR-181a-5p | IGF2BP2 | [60] | |
| miR-137 | FNDC5 | [61] | |
| miR-30b | MXRA5 | [62] | |
| miR-30a-3p | IGF1 | [63] | |
| miR-18b | HIF-1α | [64] | |
| miR-299 | HDAc2 | [65] | |
| miR-454 | ALK7 | [66] | Increases proliferation of trophoblast cells |
| miR-145 | MUC1 | ([67] | |
| miR-221-3p | THBS2 | [68] | |
| miR-126a-3p | ADAM9 | [69] | |
| miR-483-3p | RB1CC1 | [70] | |
| miR-144 | PTEN | [71] | |
| miR-518b | Rap1b | [72] | |
| miR-218-5p | TGFB2 | [73] | Promotes endovascular extravillous trophoblast cells (enEVTs) and spiral artery remodeling |
| miR-210 | CPEB2 | [74] | Inhibits trophoblast syncytialization |
| miR-106a | hCYP19A1, hGCM1 | [75] | |
| miR-558 | TIMP4 | [76] | Enhances invasion of trophoblast cells |
| miR-576-5p | TFAP2A | [77] | |
| miR-184 | WIG1 | [78] | Promotes apoptosis of trophoblast cells |
| miR-133 | Rho/ROCK | [79] | |
| miR-152 | Bax, Bcl-2 | [80] | |
| miR-155 | HIF-1α | [81] | |
| miR-371a-5p | XIAP | [82] | |
| miR-520 | PARP1 | [83] | |
| miR-34a | BCL-2 | [84] | |
| miR-18a | ER1 | [85] | |
| miR-182 | BRCA1 | [86] | |
| miR-23a | XIAP | [87] | |
| miR-101-3p | mTOR | [88] | |
| miR-96-5p | mTOR, Bcl-2 | [88] | |
| miR-200c | Wnt/β-catenin | [89] | |
| miR-125a | MCL1 | [90] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, A.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. The Role of LIN28-let-7-ARID3B Pathway in Placental Development. Int. J. Mol. Sci. 2020, 21, 3637. https://doi.org/10.3390/ijms21103637
Ali A, Bouma GJ, Anthony RV, Winger QA. The Role of LIN28-let-7-ARID3B Pathway in Placental Development. International Journal of Molecular Sciences. 2020; 21(10):3637. https://doi.org/10.3390/ijms21103637
Chicago/Turabian StyleAli, Asghar, Gerrit J. Bouma, Russell V. Anthony, and Quinton A. Winger. 2020. "The Role of LIN28-let-7-ARID3B Pathway in Placental Development" International Journal of Molecular Sciences 21, no. 10: 3637. https://doi.org/10.3390/ijms21103637
APA StyleAli, A., Bouma, G. J., Anthony, R. V., & Winger, Q. A. (2020). The Role of LIN28-let-7-ARID3B Pathway in Placental Development. International Journal of Molecular Sciences, 21(10), 3637. https://doi.org/10.3390/ijms21103637