Corynebacterium glutamicum CrtR and Its Orthologs in Actinobacteria: Conserved Function and Application as Genetically Encoded Biosensor for Detection of Geranylgeranyl Pyrophosphate
Abstract
:1. Introduction
2. Results
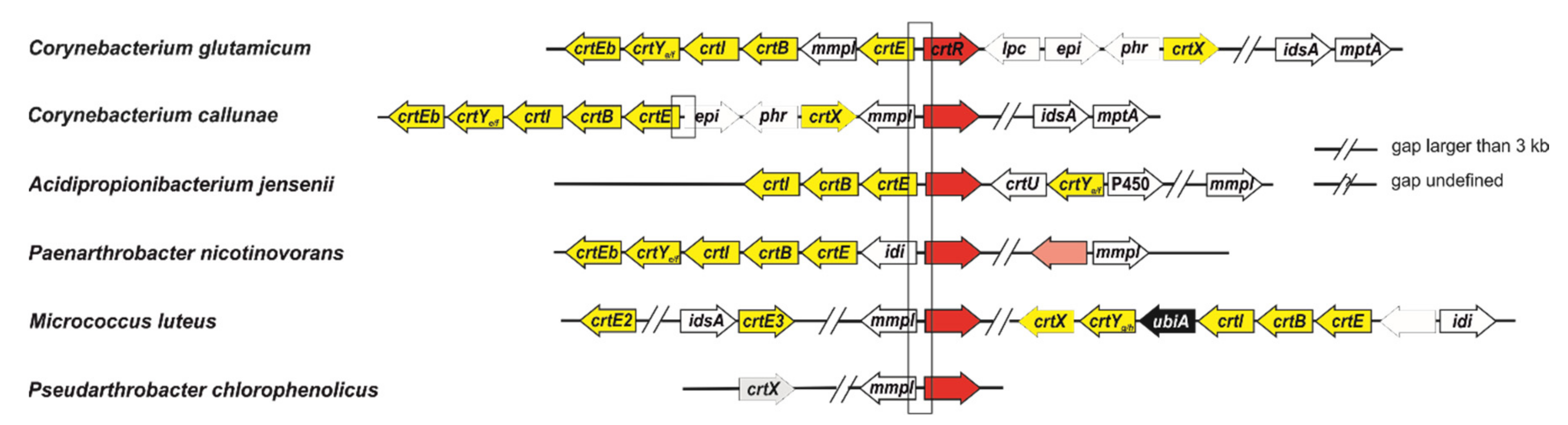
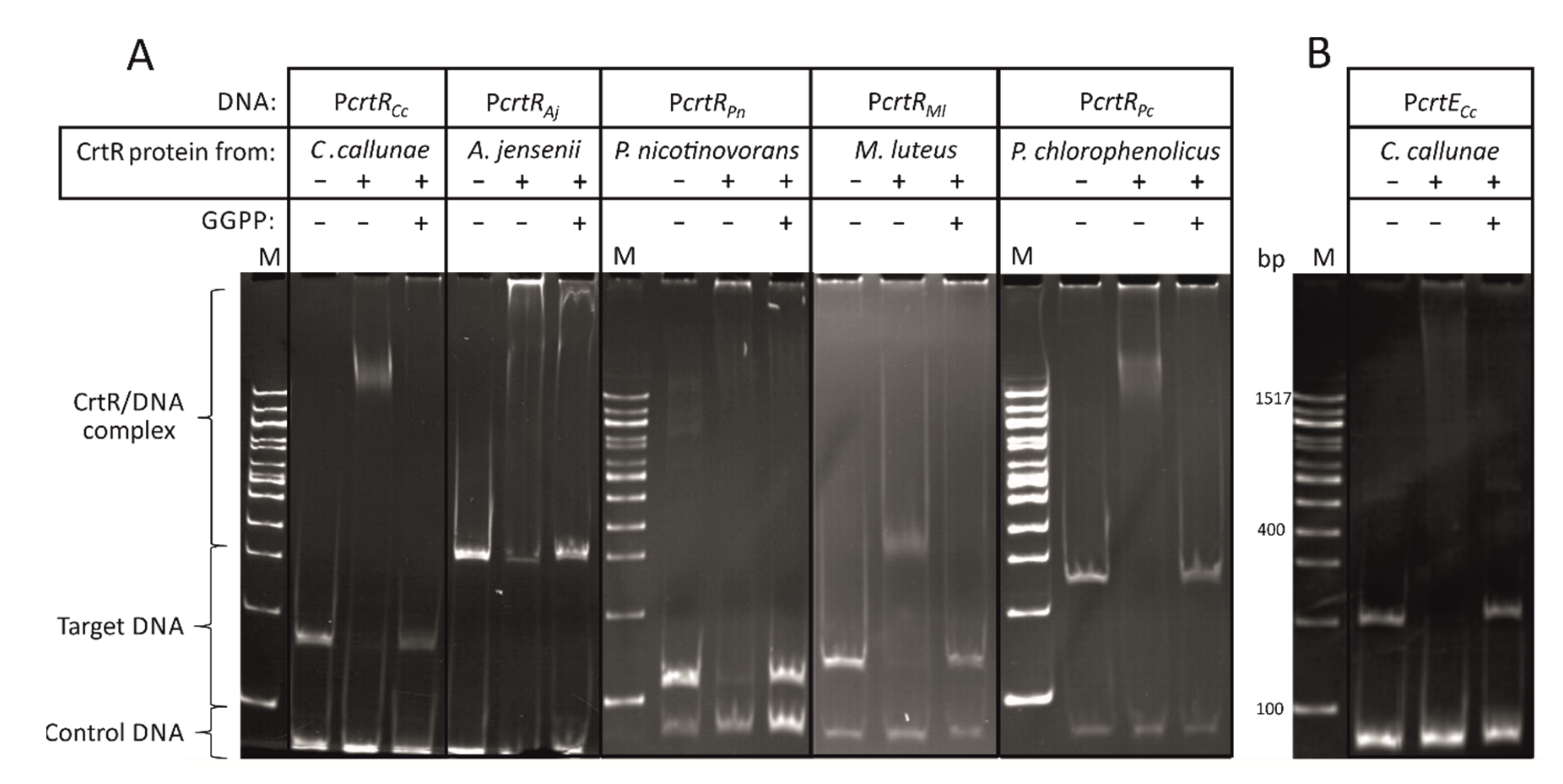
2.1. CrtR Orthologs from Actinobacteria Showed Binding to Their Own Promoters and Derepression by GGPP
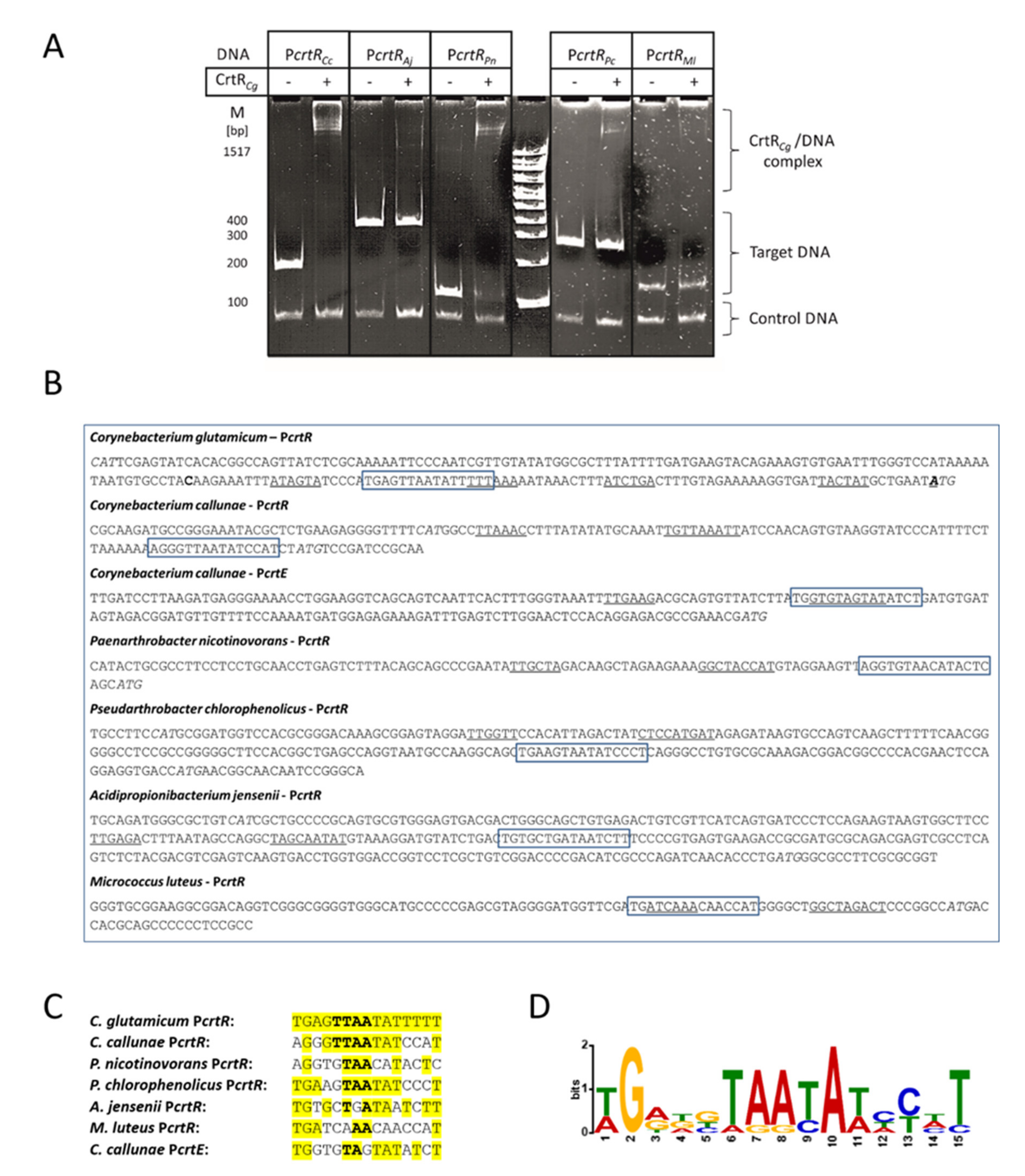
2.2. CrtR from C. glutamicum Binds to Heterologous crtR Promoter DNA Sequences
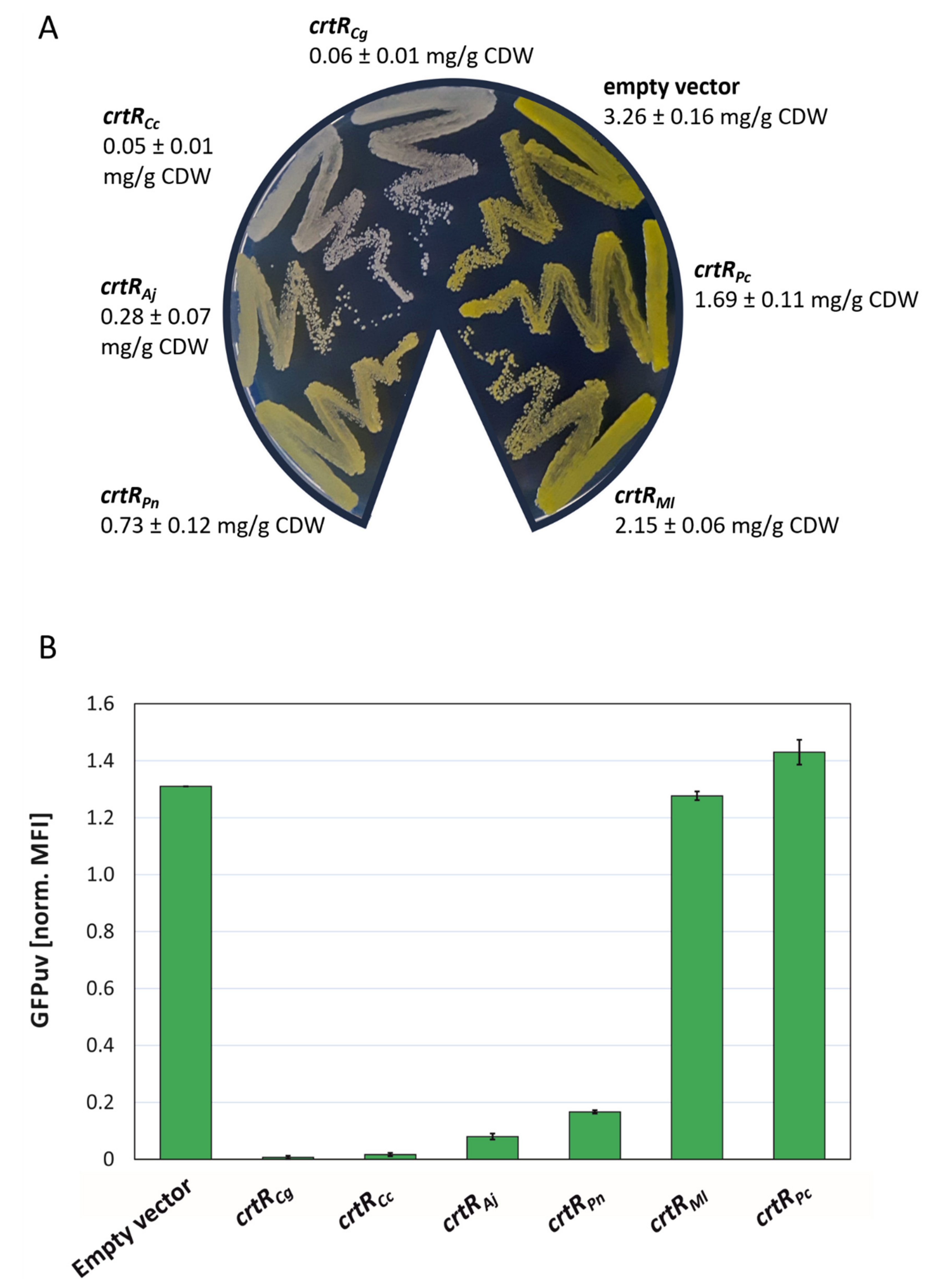
2.3. CrtR Orthologs from Actinobacteria Affected Carotenogenesis and Expression of a CrtE Transcriptional Fusion in C. glutamicum In Vivo
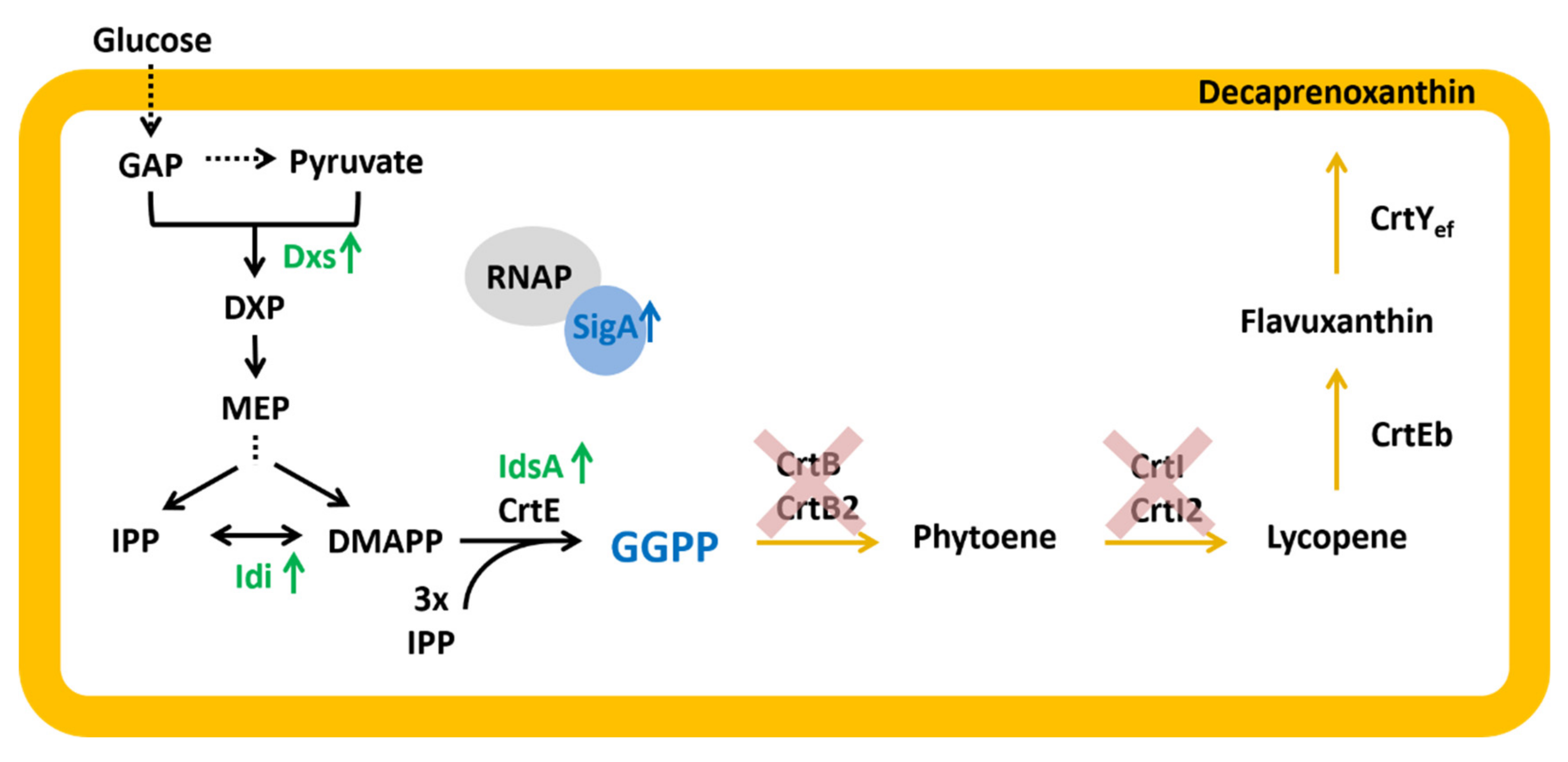
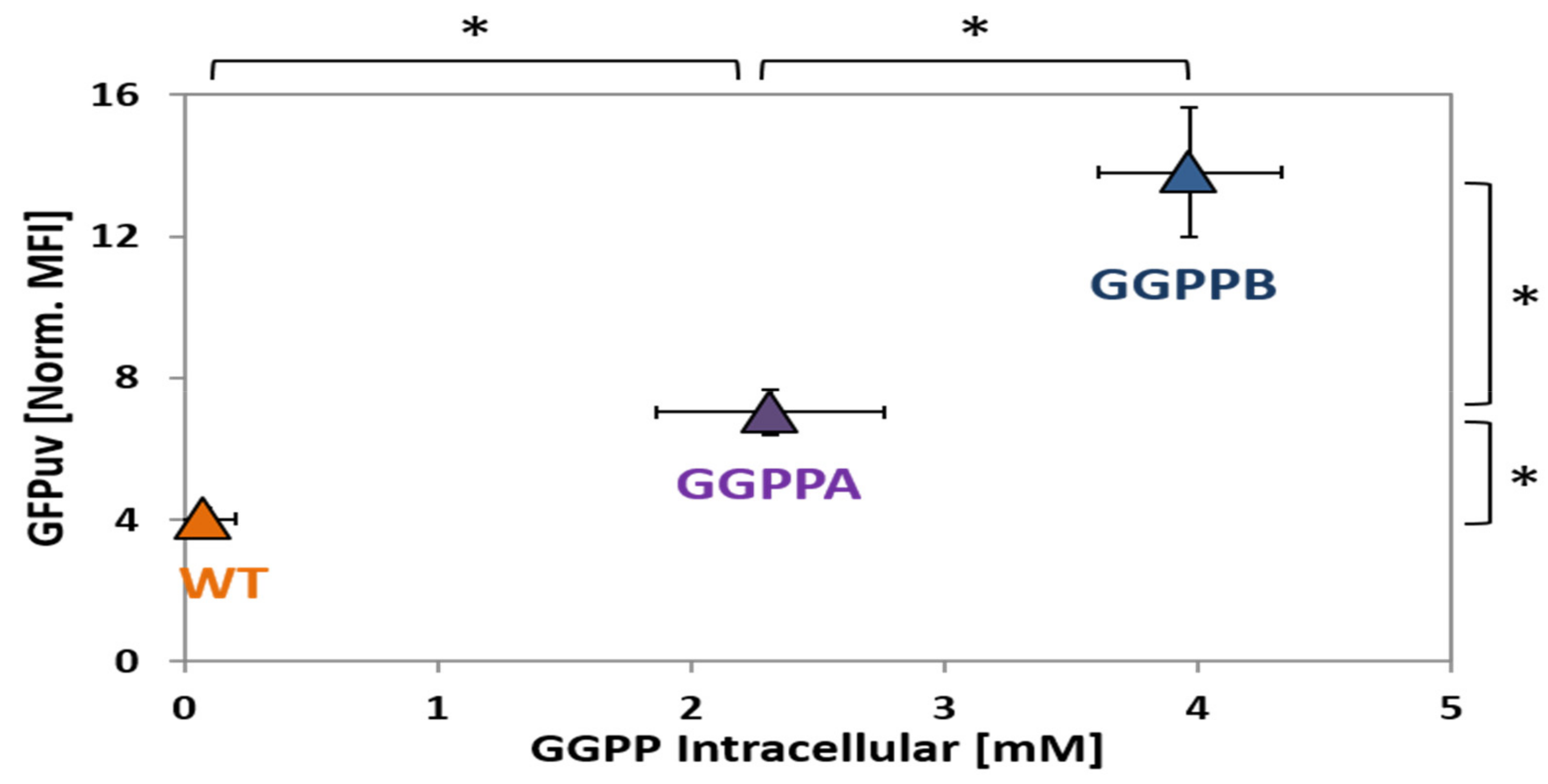
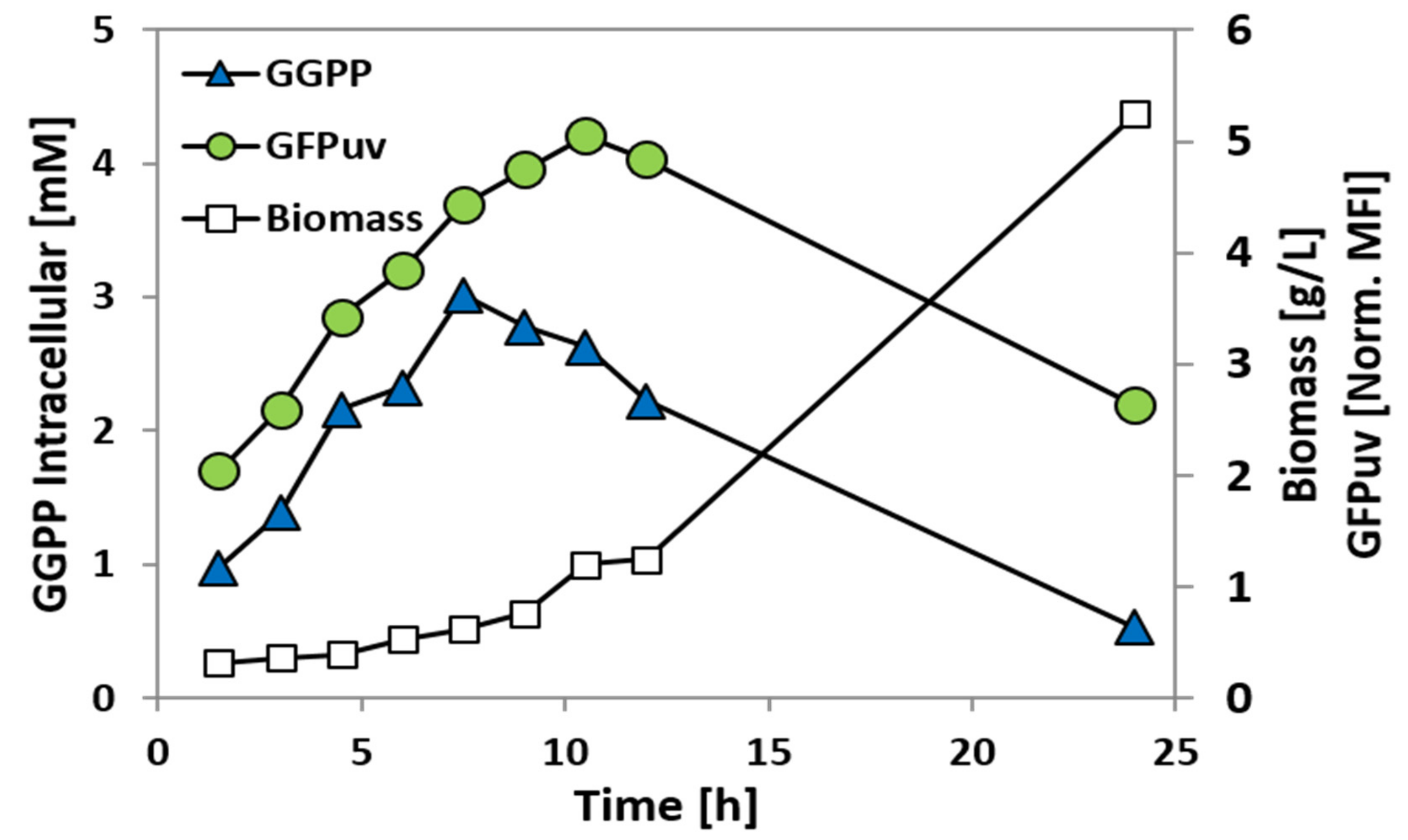
2.4. Construction and Analysis of a GGPP Biosensor
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media and Growth Conditions
4.2. Recombinant DNA Work and Gene Expression
4.3. Extraction of Carotenoids from Bacterial Cells and HPLC Analysis
4.4. Analysis of Fluorescence via Flow Cytometry
4.5. Overproduction and Purification of the Transcriptional Regulator CrtR
4.6. Electrophoretic Mobility Shift Assay (EMSA)
4.7. Extraction of GGPP and LC-MS Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mahr, R.; Frunzke, J. Transcription factor-based biosensors in biotechnology: Current state and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, J.K.; Church, G.M. Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. USA 2016, 113, 2388–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, Y.; Wang, M. Design, Optimization and Application of Small Molecule Biosensor in Metabolic Engineering. Front. Microbiol. 2017, 8, 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Jensen, M.K.; Keasling, J.D. Development of biosensors and their application in metabolic engineering. Curr. Opin. Chem. Biol. 2015, 28, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, B.; Peters, G.; Coussement, P.; Maertens, J.; De Mey, M. Tailor-made transcriptional biosensors for optimizing microbial cell factories. J. Ind. Microbiol. Biotechnol. 2017, 44, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Rohmer, M.; Seemann, M.; Horbach, H.; Bringer-Meyer, S.; Sahm, H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 1996, 118, 2564–2566. [Google Scholar] [CrossRef]
- Britton, L.-J. Pfander. In Carotenoids Handbook; Birkhauser Verlag: Basel, Switzerland, 2004. [Google Scholar] [CrossRef]
- Henke, N.A.; Wichmann, J.; Baier, T.; Frohwitter, J.; Lauersen, K.J.; Risse, J.M.; Peters-Wendisch, P.; Kruse, O.; Wendisch, V.F. Patchoulol Production with Metabolically Engineered Corynebacterium glutamicum. Genes (Basel) 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Schempp, F.M.; Drummond, L.; Buchhaupt, M.; Schrader, J. Microbial cell factories for the production of terpenoid flavor and fragrance compounds. J. Agric. Food Chem. 2017. [Google Scholar] [CrossRef]
- Schrader, J.; Bohlmann, J. Biotechnology of Isoprenoids; Springer International Publishing: New York, NY, USA, 2015. [Google Scholar] [CrossRef]
- Misawa, N. Pathway engineering for functional isoprenoids. Curr. Opin. Biotechnol. 2011, 22, 627–633. [Google Scholar] [CrossRef]
- George, K.W.; Alonso-Gutierrez, J.; Keasling, J.D.; Lee, T.S. Isoprenoid drugs, biofuels, and chemicals--artemisinin, farnesene, and beyond. Adv. Biochem. Eng. Biotechnol. 2015, 148, 355–389. [Google Scholar] [CrossRef]
- Novoveská, L.; Ross, M.E.; Stanley, M.S.; Pradelles, R.; Wasiolek, V.; Sassi, J.-F. Microalgal Carotenoids: A Review of Production, Current Markets, Regulations, and Future Direction. Mar. Drugs 2019, 17, 640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Saiz, M.; de la Fuente, J.L.; Barredo, J.L. Xanthophyllomyces dendrorhous for the industrial production of astaxanthin. Appl. Microbiol. Biotechnol. 2010, 88, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Tsubokura, A.; Yoneda, H.; Mizuta, H. Paracoccus carotinifaciens sp. nov., a new aerobic gram-negative astaxanthin-producing bacterium. Int. J. Syst. Bacteriol. 1999, 49(Pt. 1), 277–282. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Swofford, C.A.; Sinskey, A.J. Modular engineering for microbial production of carotenoids. Metab. Eng. Commun. 2020, 10, e00118. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, B.F.; Pitera, D.J.; Newman, J.D.; Martin, V.J.; Keasling, J.D. Microbial sensors for small molecules: Development of a mevalonate biosensor. Metab. Eng. 2007, 9, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.Y.; Cirino, P.C. Design and application of a mevalonate-responsive regulatory protein. Angew. Chem. Int. Ed. Engl. 2011, 50, 1084–1086. [Google Scholar] [CrossRef]
- Chou, H.H.; Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Wendisch, V.F. Production of amino acids—Genetic and metabolic engineering approaches. Bioresour. Technol. 2017, 245, 1575–1587. [Google Scholar] [CrossRef]
- Kinoshita, S.; Udaka, S.; Shimono, M. Studies on the amino acid fermentation. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 1957, 3, 193–205. [Google Scholar] [CrossRef]
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34. [Google Scholar] [CrossRef]
- Henke, N.A.; Heider, S.A.E.; Hannibal, S.; Wendisch, V.F.; Peters-Wendisch, P. Isoprenoid Pyrophosphate-Dependent Transcriptional Regulation of Carotenogenesis in Corynebacterium glutamicum. Front. Microbiol. 2017, 8, 633. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 2012, 12, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krubasik, P.; Takaichi, S.; Maoka, T.; Kobayashi, M.; Masamoto, K.; Sandmann, G. Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch. Microbiol. 2001, 176, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Peters-Wendisch, P.; Beekwilder, J.; Wendisch, V.F. IdsA is the major geranylgeranyl pyrophosphate synthase involved in carotenogenesis in Corynebacterium glutamicum. FEBS J. 2014, 281, 4906–4920. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F.; Beekwilder, J.; Brautaset, T. Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl. Microbiol. Biotechnol. 2014, 98, 4355–4368. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Wolf, N.; Hofemeier, A.; Peters-Wendisch, P.; Wendisch, V.F. Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2014, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Peters-Wendisch, P.; Netzer, R.; Stafnes, M.; Brautaset, T.; Wendisch, V.F. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014, 98, 1223–1235. [Google Scholar] [CrossRef]
- Henke, N.A.; Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F. Production of the Marine Carotenoid Astaxanthin by Metabolically Engineered Corynebacterium glutamicum. Mar. Drugs 2016, 14, 124. [Google Scholar] [CrossRef]
- Henke, N.A.; Wendisch, V.F. Improved Astaxanthin Production with Corynebacterium glutamicum by Application of a Membrane Fusion Protein. Mar. Drugs 2019, 17, 621. [Google Scholar] [CrossRef] [Green Version]
- Sprenger, G.A.; Schorken, U.; Wiegert, T.; Grolle, S.; de Graaf, A.A.; Taylor, S.V.; Begley, T.P.; Bringer-Meyer, S.; Sahm, H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 1997, 94, 12857–12862. [Google Scholar] [CrossRef] [Green Version]
- Frohwitter, J.; Heider, S.A.; Peters-Wendisch, P.; Beekwilder, J.; Wendisch, V.F. Production of the sesquiterpene (+)-valencene by metabolically engineered Corynebacterium glutamicum. J. Biotechnol. 2014, 191, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.P.; Grove, A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Boil. 2006, 8, 51–62. [Google Scholar]
- Sumi, S.; Suzuki, Y.; Matsuki, T.; Yamamoto, T.; Tsuruta, Y.; Mise, K.; Kawamura, T.; Ito, Y.; Shimada, Y.; Watanabe, E.; et al. Light-inducible carotenoid production controlled by a MarR-type regulator in Corynebacterium glutamicum. Sci. Rep. 2019, 9, 13136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viljoen, A.; Dubois, V.; Girard-Misguich, F.; Blaise, M.; Herrmann, J.L.; Kremer, L. The diverse family of MmpL transporters in mycobacteria: From regulation to antimicrobial developments. Mol. Microbiol. 2017, 104, 889–904. [Google Scholar] [CrossRef] [Green Version]
- Knoppova, M.; Phensaijai, M.; Vesely, M.; Zemanova, M.; Nesvera, J.; Patek, M. Plasmid vectors for testing in vivo promoter activities in Corynebacterium glutamicum and Rhodococcus erythropolis. Curr. Microbiol. 2007, 55, 234–239. [Google Scholar] [CrossRef]
- Banerjee, A.; Wu, Y.; Banerjee, R.; Li, Y.; Yan, H.; Sharkey, T.D. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J. Biol. Chem. 2013, 288, 16926–16936. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Xu, H.; Yu, H. Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2357–2365. [Google Scholar] [CrossRef]
- Lv, X.; Gu, J.; Wang, F.; Xie, W.; Liu, M.; Ye, L.; Yu, H. Combinatorial pathway optimization in Escherichia coli by directed co-evolution of rate-limiting enzymes and modular pathway engineering. Biotechnol. Bioeng. 2016, 113, 2661–2669. [Google Scholar] [CrossRef]
- Taniguchi, H.; Henke, N.A.; Heider, S.A.E.; Wendisch, V.F. Overexpression of the primary sigma factor gene sigA improved carotenoid production by Corynebacterium glutamicum: Application to production of β-carotene and the non-native linear C50 carotenoid bisanhydrobacterioruberin. Metab. Eng. Commun. 2017, 4, 1–11. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Puhler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotech. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Abe, S.; Takayarna, K.; Kinoshita, S. Taxonomical studies on glutamic acid producing bacteria. J. Gener. Appl. Microbial. 1967, 13, 279–301. [Google Scholar] [CrossRef]
- Johnson, J.L.; Cummins, C.S. Cell wall composition and deoxyribonucleic acid similarities among the anaerobic coryneforms, classical propionibacteria, and strains of Arachnia propionica. J. Bacteriol. 1972, 109, 1047–1066. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Komagata, K. Taxonomic Studies on Coryneform Bacteria. J. Gen. Appl. Microbial. 1970, 16, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Schleifer, K.H.; Kloos, W.E.; Moore, A. Toxonomic Status of Micrococcus luteus (Schroeter 1872) Cohn 1872: Correlation Between Peptidoglycan Type and Genetic Compatibility. Int. J. Syst. Evolut. Microbiol. 1972, 22, 224–227. [Google Scholar]
- Kodama, Y.; Yamamoto, H.; Amano, N.; Amachi, T. Reclassification of two strains of Arthrobacter oxydans and proposal of Arthrobacter nicotinovorans sp. nov. Int. J. Syst. Bacteriol. 1992, 42, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Westerberg, K.; Elvang, A.M.; Stackebrandt, E.; Jansson, J.K. Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 2000, 50(Pt. 6), 2083–2092. [Google Scholar] [CrossRef]
- Stansen, C.; Uy, D.; Delaunay, S.; Eggeling, L.; Goergen, J.L.; Wendisch, V.F. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 2005, 71, 5920–5928. [Google Scholar] [CrossRef] [Green Version]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
- Netzer, R.; Stafsnes, M.H.; Andreassen, T.; Goksoyr, A.; Bruheim, P.; Brautaset, T. Biosynthetic pathway for gamma-cyclic sarcinaxanthin in Micrococcus luteus: Heterologous expression and evidence for diverse and multiple catalytic functions of C(50) carotenoid cyclases. J. Bacteriol. 2010, 192, 5688–5699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanberg, C.; Lutnaes, B.F.; Langsrud, T.; Nes, I.F.; Holo, H. Propionibacterium jensenii produces the polyene pigment granadaene and has hemolytic properties similar to those of Streptococcus agalactiae. Appl. Environ. Microbial. 2007, 73, 5501–5506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monnet, C.; Loux, V.; Gibrat, J.-F.; Spinnler, E.; Barbe, V.; Vacherie, B.; Gavory, F.; Gourbeyre, E.; Siguier, P.; Chandler, M.; et al. The arthrobacter arilaitensis Re117 genome sequence reveals its genetic adaptation to the surface of cheese. PLoS ONE 2010, 5, e15489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutthiwong, N.; Dufossé, L. Production of carotenoids by Arthrobacter arilaitensis strains isolated from smear-ripened cheeses. FEMS Microbiol. Lett. 2014, 360, 174–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Huang, Y.; Xie, F.; Gong, Z.; Zhang, Y.; Stojkoska, A.; Xie, J. Transport mechanism of Mycobacterium tuberculosis MmpL/S family proteins and implications in pharmaceutical targeting. Biol. Chem. 2020, 401, 331–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalut, C. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberc. (Edinb. Scotl.) 2016, 100, 32–45. [Google Scholar] [CrossRef]
- Melly, G.; Purdy, G.E. MmpL Proteins in Physiology and Pathogenesis of M. tuberculosis. Microorganisms 2019, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arumugam, P.; Shankaran, D.; Bothra, A.; Gandotra, S.; Rao, V. The MmpS6-MmpL6 Operon Is an Oxidative Stress Response System Providing Selective Advantage to Mycobacterium tuberculosis in Stress. J. Infect. Dis. 2019, 219, 459–469. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Brennan, R.G. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 2002, 45, 885–893. [Google Scholar] [CrossRef]
- Grove, A. Regulation of Metabolic Pathways by MarR Family Transcription Factors. Comput. Struct. Biotechnol. J. 2017, 15, 366–371. [Google Scholar] [CrossRef]
- Bussmann, M.; Baumgart, M.; Bott, M. RosR (Cg1324), a hydrogen peroxide-sensitive MarR-type transcriptional regulator of Corynebacterium glutamicum. J. Biol. Chem. 2010, 285, 29305–29318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Chen, C.; Su, T.; Che, C.; Yao, S.; Liang, G.; Li, G.; Yang, G. CosR is an oxidative stress sensing a MarR-type transcriptional repressor in Corynebacterium glutamicum. Biochem. J. 2018, 475, 3979–3995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Su, T.; Chen, C.; Liu, J.; Gong, Z.; Che, C.; Li, G.; Yang, G. OhsR acts as an organic peroxide-sensing transcriptional activator using an S-mycothiolation mechanism in Corynebacterium glutamicum. Microb. Cell Fact. 2018, 17, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Su, T.; Chen, C.; Wei, Z.; Gong, Z.; Li, G. OsmC in Corynebacterium glutamicum was a thiol-dependent organic hydroperoxide reductase. Int. J. Boil. Macromol. 2019, 136, 642–652. [Google Scholar] [CrossRef]
- Si, M.; Chen, C.; Wei, Z.; Gong, Z.; Li, G.; Yao, S. CarR, a MarR-family regulator from Corynebacterium glutamicum, modulated antibiotic and aromatic compound resistance. Biochem. J. 2019, 476, 3141–3159. [Google Scholar] [CrossRef]
- Hünnefeld, M.; Persicke, M.; Kalinowski, J.; Frunzke, J. The MarR-Type Regulator MalR Is Involved in Stress-Responsive Cell Envelope Remodeling in Corynebacterium glutamicum. Front. Microbiol. 2019, 10, 1039. [Google Scholar] [CrossRef] [Green Version]
- Kallscheuer, N.; Vogt, M.; Kappelmann, J.; Krumbach, K.; Noack, S.; Bott, M.; Marienhagen, J. Identification of the phd gene cluster responsible for phenylpropanoid utilization in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2016, 100, 1871–1881. [Google Scholar] [CrossRef]
- Ng, C.Y.; Khodayari, A.; Chowdhury, A.; Maranas, C.D. Advances in de novo strain design using integrated systems and synthetic biology tools. Curr. Opin. Chem. Biol. 2015, 28, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.B.; Zeng, A.P. Engineering a Lysine-ON Riboswitch for Metabolic Control of Lysine Production in Corynebacterium glutamicum. ACS Synth. Biol. 2015, 4, 1335–1340. [Google Scholar] [CrossRef]
- Zhou, L.B.; Zeng, A.P. Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum. ACS Synth. Biol. 2015, 4, 729–734. [Google Scholar] [CrossRef]
- Tetali, S.D. Terpenes and isoprenoids: A wealth of compounds for global use. Planta 2019, 249, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Handbook of Corynebacterium Glutamicum; Eggeling, L.; Bott, M. (Eds.) CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2005. [Google Scholar]
- Sambrook, J.; Russell, D. Molecular Cloning. A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratoy Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.P.; Polen, T.; Youn, J.W.; Emer, D.; Eikmanns, B.J.; Wendisch, V.F. Regulation of the malic enzyme gene malE by the transcriptional regulator MalR in Corynebacterium glutamicum. J. Biotechnol. 2012, 159, 204–215. [Google Scholar] [CrossRef] [PubMed]
| Strain, gDNA or Plasmid | Relevant Characteristics or Sequence | Reference |
|---|---|---|
| E. coli strains | ||
| E.coli DH5α | F-thi-1 endA1 hsdr17(r-, m-) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 | [42] |
| S17-1 | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 integrated into the chromosome | [43] |
| E.coli BL21 (DE3) | F– ompT gal dcm lon hsdSB(rB–mB–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) | [44] |
| E.coli BL21 (DE3) (pLysS) | F– ompT gal dcm lon hsdSB(rB–mB–)λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) pLysS[T7p20 orip15A](CmR) | Promega |
| C. glutamicum strains | ||
| C. glutamicum WT | ATCC 13032, wild type | [45] |
| WTΔcrtR | ATCC 13,032 with deletion of crtR (cg0725) | [23] |
| WTΔcrtBΔcrtB2I’I2 | ATCC 13,032 with deletion of crtB (cg0721) and crtB2I’I2 (OP_cg2672) | this work |
| GGPPA | WTΔcrtBΔcrtB2I’I2 derivative with plasmid-driven IPTG-inducible expression of MEP pathway genes dxs (cg2083) and idi (cg2531) from pEKEx3 and the GGPP synthase gene idsA (cg2384) from pEC-XT. | this work |
| GGPPB | WTΔcrtBΔcrtB2I’I2 derivative with plasmid-driven IPTG-inducible expression of MEP pathway genes dxs (cg2083) and idi (cg2531) from pEKEx3 and the GGPP synthase gene idsA (2384) and primary sigma factor gene sigA (cg2092) from pEC-XT. | this work |
| Genomic DNA | ||
| Acidipropionibacterium jensenii | Wild type, DSM 20535, ATCC 4868 | [46], DSMZ |
| Corynebacterium callunae | Wild type, DSM 20147, ATCC 15991 | [47] |
| Micrococcus luteus | Wild type, DSM 20030, ATCC 4698 | [48], DSMZ |
| Paenarthrobacter nicotinovorans | Wild type, DSM 420, ATCC 49919 | [49], DSMZ |
| Pseudarthrobacter chlorophenolicus | Wild type, DSM 12829, ATCC 700700 | [50], DSMZ |
| Plasmids | ||
| pEPR1 | KmR, pCG1 oriVCG, gfpuv, promoterless, C. glutamicum/E.coli shuttle promoter-probe vector | [37] |
| pEPR1_PcrtE | pEPR1 derivate containing the promoter of crtE (PcrtE) | [23] |
| pTEST | pEPR1_PcrtE derivate containing an additional expression cassette for expression of crtR orthologs from the gap promoter | this work |
| pTEST_crtRCg | pTEST derivate for expression of the crtR from C. glutamicum | this work |
| pTEST_crtRCc | pTEST derivate for heterologous expression of the crtR orthologs from C. callunae | this work |
| pTEST_crtRAj | pTEST derivate for heterologous expression of the crtR ortholog from A. jensenii | this work |
| pTEST_crtRPn | pTEST derivate for heterologous expression of the crtR ortholog from P. nicotinovorans | this work |
| pTEST_crtRPc | pTEST derivate for heterologous expression of the crtR ortholog from P. chlorophenolicus | this work |
| pTEST_crtRMl | pTEST derivate for heterologous expression of the crtR ortholog from M. luteus | this work |
| pET16b | Expression plasmid for production of His-tagged proteins | Novagen |
| pET16b_crtRCg | pET16b derivate for production of His-tagged CrtR from C. glutamicum | this work |
| pET16b_crtRCc | pET16b derivate for production of His-tagged CrtR C. callunae | this work |
| pET16b_crtRAj | pET16b derivate for production of His-tagged CrtR A. jensenii | this work |
| pET16b_crtRPn | pET16b derivate for expression of the crtR from P. nicotinovorans | this work |
| pET16b_crtRPc | pET16b derivate for production of His-tagged CrtR P. chlorophenolicus | this work |
| pET16b_crtRMl | pET16b derivate for production of His-tagged CrtR M. luteus | this work |
| pEKEx3 | SpecR, Ptac lacIq, pBL1 oriVCg, C. glutamicum/E. coli expression shuttle vector | [51] |
| pEKEx3_dxs_idi | pEKEx3 derivate for IPTG-inducible expression of dxs and idi from C. glutamicum containing an artificial ribosome binding site | this work |
| pEC-XT99A | TetR, Ptrc lacIq, pGA1 oriVCg, C. glutamicum/E. coli expression shuttle vector | [52] |
| pEC-XT_idsA | pEC-XT99A derivate for IPTG-inducible expression of idsA from C. glutamicum containing an artificial ribosome binding site | this work |
| pEC-XT_idsA_sigA | pEC-XT99A derivate for IPTG-inducible expression of idsA and sigA from C. glutamicum containing an artificial ribosome binding site | this work |
| Oligonucleotide (5′→3′) | |
|---|---|
| NH45 | CATGCCTGCAGGTCGACTCTAGAGGAAAGGAGGCCCTTCAGATGGGAATTCTGAACAGTATTTCAA |
| NH46 | GTTCGTGTGGCAGTTTTATTCCCCGAACAGGGAATC |
| NH47 | AACTGCCACACGAACGAAAGGAGGCCCTTCAGATGTCTAAGCTTAGGGGCATG |
| NH48 | ATTCGAGCTCGGTACCCGGGGATCTTACTCTGCGTCAAACGCTTC |
| NH49 | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGGCTTACTCCGCTATGGCTA |
| NH50 | GCATGCCTGCAGGTCGACTCTAGAGGATCTTAGTTCTGGCGGAAAGCAA |
| NH51 | GTTCGTGTGGCAGTTTTAGTTCTGGCGGAAAGCAA |
| NH52 | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGGACTTTCCGCAGCAACTCG |
| NH53 | GCATGCCTGCAGGTCGACTCTAGAGGATCTTATTTATTACGCTGGATGATGTAGTCC |
| NH54 | GTTCGTGTGGCAGTTTTATTTATTACGCTGGATGATGTAGTCC |
| NH55 | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGAGCAGTTTCGATGCCCA |
| NH56 | GCATGCCTGCAGGTCGACTCTAGAGGATCTTACATCCGACGTTCGGTTGA |
| NH57 | GTTCGTGTGGCAGTTTTACATCCGACGTTCGGTTGA |
| NH58 | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGGTAGAAAACAACGTAGCAA |
| NH59 | GCATGCCTGCAGGTCGACTCTAGAGGATCTTAGTCCAGGTAGTCGCGAAG |
| NH60 | AACTGCCACACGAACGAAAGGAGGCCCTTCAGATGGTAGAAAACAACGTAGCAA |
| NH63 | GCAAAGTTGTTGTCGTAGTC |
| NH64 | ATGAAAACGTTGTTGCCAT |
| NH65 | ATGAAGACGCCACTGAC |
| NH66 | CGGTGAGCTCGGCATCT |
| NH67 | GTGCCTTGCGAGCTGTCT |
| TH17 | CTGTTGATGACGACGAGGAG |
| pE-CXT fw | AATACGCAAACCGCCTCTCC |
| pE-CXT rv | TACTGCCGCCAGGCAAATTC |
| crtE-E | GTGACCATGAGGGCGAAAGC |
| crtE-F | TCACATAGTCCGGCGTTTGC |
| idsA-E | GCAGCTTCGCCAGAGTGTAT |
| idsA-F | CAATGCGGACAATGCTCCAG |
| 581 | CATCATAACGGTTCTGGC |
| 582 | ATCTTCTCTCATCCGCCA |
| Pgap fw | TGGCCTTTTGCTGGCCTTTTGCTCACTGCGAAATCTTTGTTTCCCCG |
| Pgap rv | GGATCCGTTGTGTCTCCTCTAAAGATT |
| term fw | AATCTTTAGAGGAGACACAACGGATCCTTTTGGCGGATGAGAGAA |
| term rv | AATCAGGGGATAACGCAGGAAAGAACAAAAGAGTTTGTAGAA |
| NA25- Cg fw | TACAATCTTTAGAGGAGACACAACGGAAAGGAGGCCCTTCAGATGCTGAATATGCAGGAACCA |
| NA26- Cg rv | AAAATCTTCTCTCATCCGCCAAAAGTTACTCCGTGTTGAGCCATGG |
| NA27- Cc fw | TACAATCTTTAGAGGAGACACAACGGAAAGGAGGCCCTTCAGATGTCCGATCCGCAAGAACC |
| NA28- Cc rv | AAAATCTTCTCTCATCCGCCAAAAGTTAATGTGAGGAAGACTCGAAC |
| NA31- Aj fw | TACAATCTTTAGAGGAGACACAACGGAAAGGAGGCCCTTCAGATGAGTGAAGACCGCGATG |
| NA32- Aj rv | AAAATCTTCTCTCATCCGCCAAAAGTTACCGCGGGTGGCGC |
| NA33-An fw | TACAATCTTTAGAGGAGACACAACGGAAAGGAGGCCCTTCAGATGTCCAGTCTTGAAGAAATGC |
| NA34-An rv | AAAATCTTCTCTCATCCGCCAAAAGTTAGCGTGGAGCCGCAG |
| NA39- Ml fw | TACAATCTTTAGAGGAGACACAACGGAAAGGAGGCCCTTCAGATGACCACGCAGCCCC |
| NA40- Ml rv | AAAATCTTCTCTCATCCGCCAAAAGTTACGGGTCCTCCGGGG |
| NA41- Pc fw | TACAATCTTTAGAGGAGACACAACGGAAAGGAGGCCCTTCAGATGAACGGCAACAATCCG |
| NA42- Pc rv | AAAATCTTCTCTCATCCGCCAAAAGTTACCCGGCTGGACGC |
| HN83-Cg-fw | GCGGCCATATCGAAGGTCGTCATCTGAATATGCAGGAACCAG |
| HN84-Cg-rv | TAGCAGCCGGATCCTCGAGCATTACTCCGTGTTGAGCCATG |
| HN85-Cc-fw | GCGGCCATATCGAAGGTCGTCATTCCGATCCGCAAGAACCCC |
| HN86-Cc-rv | TAGCAGCCGGATCCTCGAGCATTAATGTGAGGAAGACTCGAAC |
| HN87-Aj-fw | GCGGCCATATCGAAGGTCGTCATAGTGAAGACCGCGATGC |
| HN88-Aj-rv | TAGCAGCCGGATCCTCGAGCATTACCGCGGGTGGCGC |
| HN89-Pn-fw | GCGGCCATATCGAAGGTCGTCATTCCAGTCTTGAAGAAATGCC |
| HN90-Pn-rv | TAGCAGCCGGATCCTCGAGCATTAGCGTGGAGCCGCAG |
| HN93-Ml-fw | GCGGCCATATCGAAGGTCGTCATACCACGCAGCCCCCC |
| HN94-Ml-rv | TAGCAGCCGGATCCTCGAGCATTACGGGTCCTCCGGGG |
| HN95-Pc-fw | GCGGCCATATCGAAGGTCGTCATAACGGCAACAATCCGGGC |
| HN96-Pc-rv | TAGCAGCCGGATCCTCGAGCATTACCCGGCTGGACGC |
| Pc-PcrtR-fw | TGCCTTCCATGCGGATGGTC |
| Pc-PcrtR-rv | TGCCCGGATTGTTGCCGTTC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henke, N.A.; Austermeier, S.; Grothaus, I.L.; Götker, S.; Persicke, M.; Peters-Wendisch, P.; Wendisch, V.F. Corynebacterium glutamicum CrtR and Its Orthologs in Actinobacteria: Conserved Function and Application as Genetically Encoded Biosensor for Detection of Geranylgeranyl Pyrophosphate. Int. J. Mol. Sci. 2020, 21, 5482. https://doi.org/10.3390/ijms21155482
Henke NA, Austermeier S, Grothaus IL, Götker S, Persicke M, Peters-Wendisch P, Wendisch VF. Corynebacterium glutamicum CrtR and Its Orthologs in Actinobacteria: Conserved Function and Application as Genetically Encoded Biosensor for Detection of Geranylgeranyl Pyrophosphate. International Journal of Molecular Sciences. 2020; 21(15):5482. https://doi.org/10.3390/ijms21155482
Chicago/Turabian StyleHenke, Nadja A., Sophie Austermeier, Isabell L. Grothaus, Susanne Götker, Marcus Persicke, Petra Peters-Wendisch, and Volker F. Wendisch. 2020. "Corynebacterium glutamicum CrtR and Its Orthologs in Actinobacteria: Conserved Function and Application as Genetically Encoded Biosensor for Detection of Geranylgeranyl Pyrophosphate" International Journal of Molecular Sciences 21, no. 15: 5482. https://doi.org/10.3390/ijms21155482
APA StyleHenke, N. A., Austermeier, S., Grothaus, I. L., Götker, S., Persicke, M., Peters-Wendisch, P., & Wendisch, V. F. (2020). Corynebacterium glutamicum CrtR and Its Orthologs in Actinobacteria: Conserved Function and Application as Genetically Encoded Biosensor for Detection of Geranylgeranyl Pyrophosphate. International Journal of Molecular Sciences, 21(15), 5482. https://doi.org/10.3390/ijms21155482






