A New Species of Spongilla (Porifera, Demospongiae) from a Karst Lake in Ha Long Bay (Vietnam)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Site and Sponge Collection
2.2. Morphological Study
2.3. Genotyping
2.4. Genetic Analysis
3. Results and Discussion
3.1. Taxonomical Account
3.1.1. Material Examined
3.1.2. Species Diagnosis
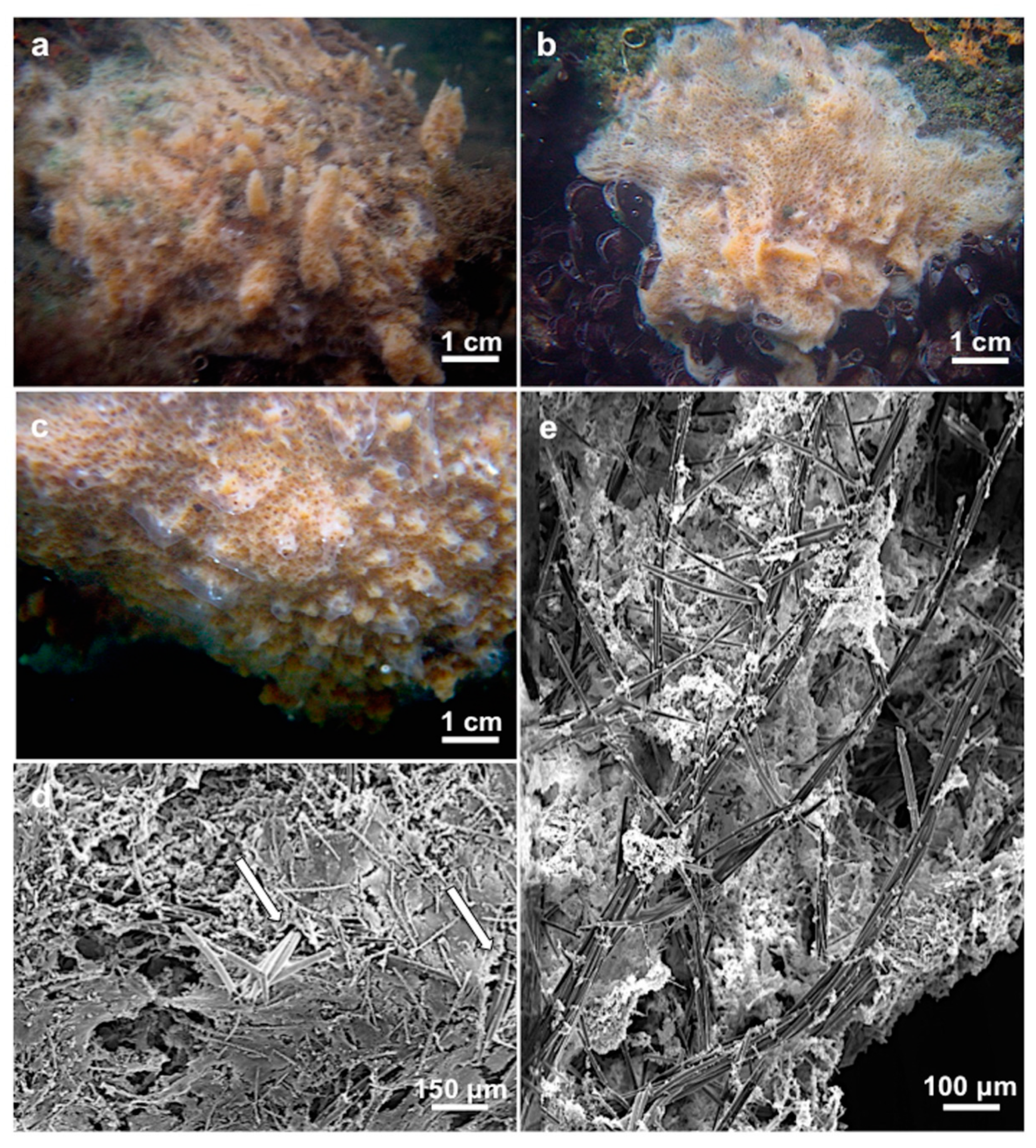
3.1.3. Description
3.1.4. Habitat and Distribution
3.1.5. Etymology
3.1.6. Remarks
3.2. Genotyping
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thung, D.G.; Ngai, N.D.; Sinh, N.V.; Quan, N.V.; Tien, D.D.; Thuoc, C.V.; Trang, C.T.T. Biodiversity of Limestone Island and Archipelagos Areas in Northeast Coast of Vietnam, Orientation of Sustainable Use Solutions; Publishing House of Natural Science and Technology: Ha Noi, Vietnam, 2018; pp. 56–57. (In Vietnamese) [Google Scholar]
- Vermeulen, J.J.; Anker, K. Outstanding global values in geology and environment in Cat Ba archipelago and Halong Bay. In Proceedings of the Biodiversity Conservation Solutions in Ha Long bay and Cat Ba Archipelago, IUCN, Haiphong City, Vietnam, 24 August 2017; pp. 16–23. [Google Scholar]
- Cerrano, C.; Azzini, F.; Bavestrello, G.; Calcinai, B.; Pansini, M.; Sarti, M.; Thung, D.C. Marine lakes of karst islands in the Ha Long Bay (Vietnam). Chem. Ecol. 2006, 22, 489–500. [Google Scholar] [CrossRef]
- Becking, L.E.; Renema, W.; Santodomingo, N.K.; Hoeksema, B.W.; Tuti, Y.; de Voogd, N.J. Recently discovered landlocked basins in Indonesia reveal high habitat diversity in anchialine systems. Hydrobiologia 2011, 677, 89–105. [Google Scholar] [CrossRef] [Green Version]
- Dawson, M.; Hamner, W.N. Rapid evolutionary radiation of marine zooplankton in peripheral environments. Proc. Natl. Acad. Sci. USA 2005, 102, 9235–9240. [Google Scholar] [CrossRef] [Green Version]
- Cerrano, C.; Bavestrello, G.; Calcinai, B.; Cattaneo-Vietti, V.; Chiantore, M.; Guidetti, M.; Sarà, A. Bioerosive processes in Antarctic seas. Pol. Biol. 2001, 24, 790–792. [Google Scholar] [CrossRef]
- Díaz, M.C.; Rützler, K. Sponges: An essential component of Caribbean coral reefs. Bull. Mar. Sci. 2001, 69, 535–546. [Google Scholar]
- Bertolino, M.; Cerrano, C.; Bavestrello, G.; Carella, M.; Pansini, M.; Calcinai, B. Diversity of Porifera in the Mediterranean coralligenous accretions, with description of a new species. Zookeys 2013, 336, 1–37. [Google Scholar] [CrossRef]
- Gerovasileiou, V.; Voultsiadou, E. Sponge diversity gradients in marine caves of the eastern Mediterranean. J. Mar. Biol. Assoc. UK 2016, 96, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Wulff, J.L. Ecological interactions of marine sponges. Can. J. Zool. 2006, 84, 146–166. [Google Scholar] [CrossRef]
- Becerro, M.A. Quantitative trends in sponge ecology research. Mar. Ecol. 2008, 29, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Azzini, F.; Calcinai, B.; Cerrano, C.; Bavestrello, G.; Pansini, M. Sponges of the marine karst lakes and of the coast of the islands of Ha Long Bay (North Vietnam). In Porifera Research: Biodiversity, Innovation and Sustainability; Custódio, M.R., Lôbo-Hajdu, G., Hajdu, E., Muricy, G., Eds.; Museu National Rio de Janeiro: Rio de Janeiro, Brazil, 2007; pp. 157–164. [Google Scholar]
- Quang, T.M. A review of the diversity of sponges (Porifera) in Vietnam. In Proceedings of the 2nd International Workshop on Marine Bioresources of Vietnam, Hanoi, Vietnam, 5–6 June 2013; pp. 109–115. [Google Scholar]
- Calcinai, B.; Azzini, F.; Bavestrello, G.; Cerrano, C.; Pansini, M.; Thung, D.C. Boring Sponges from Ha Long Bay, Tonkin Gulf, Vietnam. Zool. Stud. 2006, 45, 201–212. [Google Scholar]
- Cerrano, C.; Bavestrello, G.; Bertolino, M.; Pansini, M.; Núñez-Pons, L.; Sarti, M.; Thung, D.C.; Calcinai, C. The Ha Long Bay marine ecosystem. An unprecedented opportunity for evolutionary studies on marine taxa. In Innovations in Land, Water and Energy for Vietnam’s Sustainable Development; Anderle, M., Ed.; Unipa Springer Series; Springer: Cham, Switzerland, 2021; pp. 45–52. [Google Scholar] [CrossRef]
- Fenart, N.; Cat, N.; Drogue, C.; Van Canh, D.; Pistre, S. Influence of tectonics and neotectonics on the morphogenesis of the peak karst of Halong Bay, Vietnam. Geodin. Acta 1999, 12, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Egi, S.M.; Cousteau, P.Y.; Pieri, M.; Cerrano, C.; Özyigit, T.; Marroni, A. Designing a Diving Protocol for Thermocline Identification Using Dive Computers in Marine Citizen Science. Appl. Sci. 2018, 8, 2315. [Google Scholar] [CrossRef] [Green Version]
- Rützler, K. Sponges in coral reefs. In Coral Reefs: Research Methods. Monographs on Oceanografic Methodologies; Stoddart, D.R., Johannes, R.E., Eds.; UNESCO: Paris, France, 1978; Volume 5. [Google Scholar]
- Hooper, J.N.A. ‘Sponguide’. Guide to Sponge Collection and Identification. 2000. Available online: http://www.qm.qld.gov.au/organisation/sections/SessileMarineInvertebrates/spong.pdf (accessed on 1 May 2020).
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biot. 1994, 3, 294–299. [Google Scholar]
- Xavier, J.R.; Rachello-Dolmen, P.G.; Parra-Velandia, F.; Schönberg, C.H.L.; Breeuwer, J.J.; Van Soest, R.W.M. Molecular evidence of cryptic speciation in the “cosmopolitan” excavating sponge Cliona celata (Porifera, Clionaidae). Mol. Phylog. Evol. 2010, 56, 13–20. [Google Scholar] [CrossRef]
- Erpenbeck, D.; Hooper, J.N.A.; Wörheide, G. CO1 phylogenies in diploblasts and the ‘Barcoding of Life’—are we sequencing a suboptimal partition? Mol. Ecol. Notes 2006, 6, 550–553. [Google Scholar] [CrossRef]
- Wörheide, G.; Nichols, S.; Goldberg, J. Intragenomic variation of the rDNA internal transcribed spacers in sponges (Phylum Porifera): Implications for phylogenetic studies. Mol. Phylogenet. Evol. 2004, 33, 816–830. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, R.W.M.; Boury-Esnault, N.; Hooper, J.N.A.; Rützler, K.; de Voogd, N.J.; Alvarez, B.; Hajdu, E.A.; Pisera, B.; Manconi, R.; Schönberg, C.; et al. World Porifera Database. 2020. Available online: http://www.marinespecies.org/porifera (accessed on 20 November 2020).
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipmann, D.J. Gapped BLAST and PSI- BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, S.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. Model Finder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 1–8. [Google Scholar]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- De Lamarck, J.B.D.M. Histoire Naturelle Des Animaux Sans Vertèbres; Verdière: Paris, France, 1816; tome second; p. 568. [Google Scholar]
- Núñez-Pons, L.; Calcinai, B.; Gates, R.D. Who’s there?—First morphological and DNA barcoding catalogue of the shallow Hawai’ian sponge fauna. PLoS ONE 2017, 12, e0189357. [Google Scholar] [CrossRef] [Green Version]
- Itskovich, V.; Gontcharov, A.; Masuda, Y.; Nohno, T.; Belikov, S.; Efremova, S.; Meixner, M.; Janussen, D. Ribosomal ITS sequences allow resolution of freshwater sponge phylogeny with alignments guided by secondary structure prediction. J. Mol. Evol. 2008, 67, 608–620. [Google Scholar] [CrossRef]
- Huang, D.; Meier, R.; Todd, P.A.; Chou, M. Slow Mitochondrial COI Sequence Evolution at the Base of the Metazoan Tree and Its Implications for DNA Barcoding. J. Mol. Evol. 2008, 66, 167–174. [Google Scholar] [CrossRef]
- Erpenbeck, D.; Steiner, M.; Schuster, A.; Genner, M.; Manconi, R.; Pronzato, R.; Ruthensteiner, B.; Spiegel, D.; Van Soest, R.W.M.; Wörheide, G. Minimalist barcodes for sponges—A case study classifying African freshwater Spongillida. Genome 2018, 62, 1–10. [Google Scholar] [CrossRef]
- Jakhalekar, S.S.; Ghate, H.V. Taxonomy of freshwater sponges of Maharashtra, India, with illustrated descriptions and notes on ecology and habitats (Porifera: Spongillida: Spongillidae). Zootaxa 2016, 4173, 501–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| 7/12/2017 | 21/08/2018 |
|---|---|
| S‰ = 12 | S‰ = 8 |
| pH = 7.2 | pH = 7.65 |
| DO mg/L (Dissolved Oxygen) = 6.7 | DO mg/L (Dissolved Oxygen) = 5.09 |
| T = 20.7 °C At the surface | T = 33.4 °C At the surface |
| Specimens | Oxeas (µm) | Microxeas (µm) | Gemmuloscleres (µm) |
|---|---|---|---|
| MSNG 61501 holotype | 205 (272.5 ± 28.4) 332.1 × 8.2 (11.8 ± 3.2) 16.4 | 80.6 (103.59 ± 18.5) 161.2 × 1.3 (2.08 ± 0.6) 2.6 | 78 (97.85 ± 8.8) 111.8 × 7.8 |
| MSNG 61502 paratype | 255 (291.1 ± 23.8) 350 × 10 | 75 (94.3 ± 12.7) 112.5 × 2.5 | 87.5 (100.4 ± 11.9) 124 × 5 |
| HL 108 topotype | 255 (283.5 ± 19.1) 310 × 7.5 (8.9 ± 1.5) 11.25 | 80 (103.6 ± 11.4) 120 × 2.5 | 82.5 (101.8 ± 9) 115 × 5 |
| HL108A topotype | 260 (308 ± 26.3) 345 × 6.2 (8.9 ±1.5) 10 | 82.5 (107.1 ± 111.4) 122.5 × 2.5 | * |
| HL108B topotype | 275 (306 ± 16.4) 340 × 7.5 | 67.5 (94.8 ± 13.3) 110 × 2.5 | 100–105 * |
| HL108C topotype | 260 (282 ± 17.2) 320 × 6.2 (8.5 ± 1.6) 10 | 90 (103.2 ± 9.1) 122.5 × 2.5 | * |
| HL108D topotype | 280 (311 ± 14.4) 340 × 7.5 (8.2 ± 1.1) 10 | 77.5 (101.5 ± 15.3) 125 × 2.5 | 75 (96.9 ± 11) 120 × 5 |
| Species | Oxeas (µm) | Microxeas (µm) | Gemmuloscleres (µm) | Distribution |
|---|---|---|---|---|
| S. alba Carter, 1849 | 277–356 × 16–26 | 55–142 × 2–3 | 90–143 × 6–10 | Indonesia, Australia, South America, India, Caribbean |
| S. arctica Annandale, 1915 | 154–280 × 4–14 | 32–70 × 2–4 | 80–140 × 4–6 | Arctic, Russia |
| S. cenota Penney and Racek, 1968 | 310–410 × 14–22 | 68–123 × 2–3 | 65–86 × 9–13 | Yucatan |
| S. friabilis Lamarck, 1816 | taxon inquirendum | – | – | Asia ? |
| S. chaohuensis Cheng, 1991 | 208–280 × 8–15.2 | – | 80–112 × 2.4–8.8 | China |
| S. helvetica Annandale, 1909 | 148–200 × 4–9 | 24–55 × 1–2 | 32–68 × 2–5 | Switzerland |
| S. jiujiangensis Cheng, 1991 | 204–296 × 6.4–16 | - | 72–152 × 3.2–9.6 | China |
| S. lacustris (Linnaeus, 1759) | 90–350 × 2–18 | 25–178 × 2–8 | 21–130 × 1–10 | Northern Polar Circle, Ireland, England, France, Italy, California, |
| S. mucronata Topsent, 1932 | 245–275 × 10–11 | 35–90 × 1.2–2 | 95–120 × 8–9 | Africa |
| S. permixta Weltner, 1895 | – | – | 320–390 × 3–5 | Egypt |
| S. prespensis Hadzische, 1953 | 170–200 × 6–13 | 157.4 × 2–4 | 50–120 × 3–7 | Macedonia |
| S. sarasinorum Weltner, 1901 | up to 250 | 180 × 8 | 160 (190) 210 × 13 (18) 20 | Indonesia |
| S. shikaribensis Sasaki, 1934 | 240 (293) 350 × 13 (13.9) 15 | 61 (80.1) 100 × 3 (4.1) 5 | 50 (75.7) 101 | Japan |
| S. stankovici Arndt, 1938 | 176–235 × 3–6 | 180 × 8 | 160–210 × 8 | Albania |
| S. wagneri Potts, 1889 | 144–270 × 7–12 | 49–62 × 2–4 | 48–75 × 6–8 | United States |
| Spongilla manconiae sp. nov. | 250–350 × 6.2–16.4 | 67.5–161.2 × 1.3–2.6 | 75–124 × 5–7.8 | Vietnam |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calcinai, B.; Cerrano, C.; Núñez-Pons, L.; Pansini, M.; Thung, D.C.; Bertolino, M. A New Species of Spongilla (Porifera, Demospongiae) from a Karst Lake in Ha Long Bay (Vietnam). J. Mar. Sci. Eng. 2020, 8, 1008. https://doi.org/10.3390/jmse8121008
Calcinai B, Cerrano C, Núñez-Pons L, Pansini M, Thung DC, Bertolino M. A New Species of Spongilla (Porifera, Demospongiae) from a Karst Lake in Ha Long Bay (Vietnam). Journal of Marine Science and Engineering. 2020; 8(12):1008. https://doi.org/10.3390/jmse8121008
Chicago/Turabian StyleCalcinai, Barbara, Carlo Cerrano, Laura Núñez-Pons, Maurizio Pansini, Do Cong Thung, and Marco Bertolino. 2020. "A New Species of Spongilla (Porifera, Demospongiae) from a Karst Lake in Ha Long Bay (Vietnam)" Journal of Marine Science and Engineering 8, no. 12: 1008. https://doi.org/10.3390/jmse8121008
APA StyleCalcinai, B., Cerrano, C., Núñez-Pons, L., Pansini, M., Thung, D. C., & Bertolino, M. (2020). A New Species of Spongilla (Porifera, Demospongiae) from a Karst Lake in Ha Long Bay (Vietnam). Journal of Marine Science and Engineering, 8(12), 1008. https://doi.org/10.3390/jmse8121008







