Biomanufacturing of Tomato-Derived Nanovesicles
Abstract
:1. Introduction
2. Materials and Methods
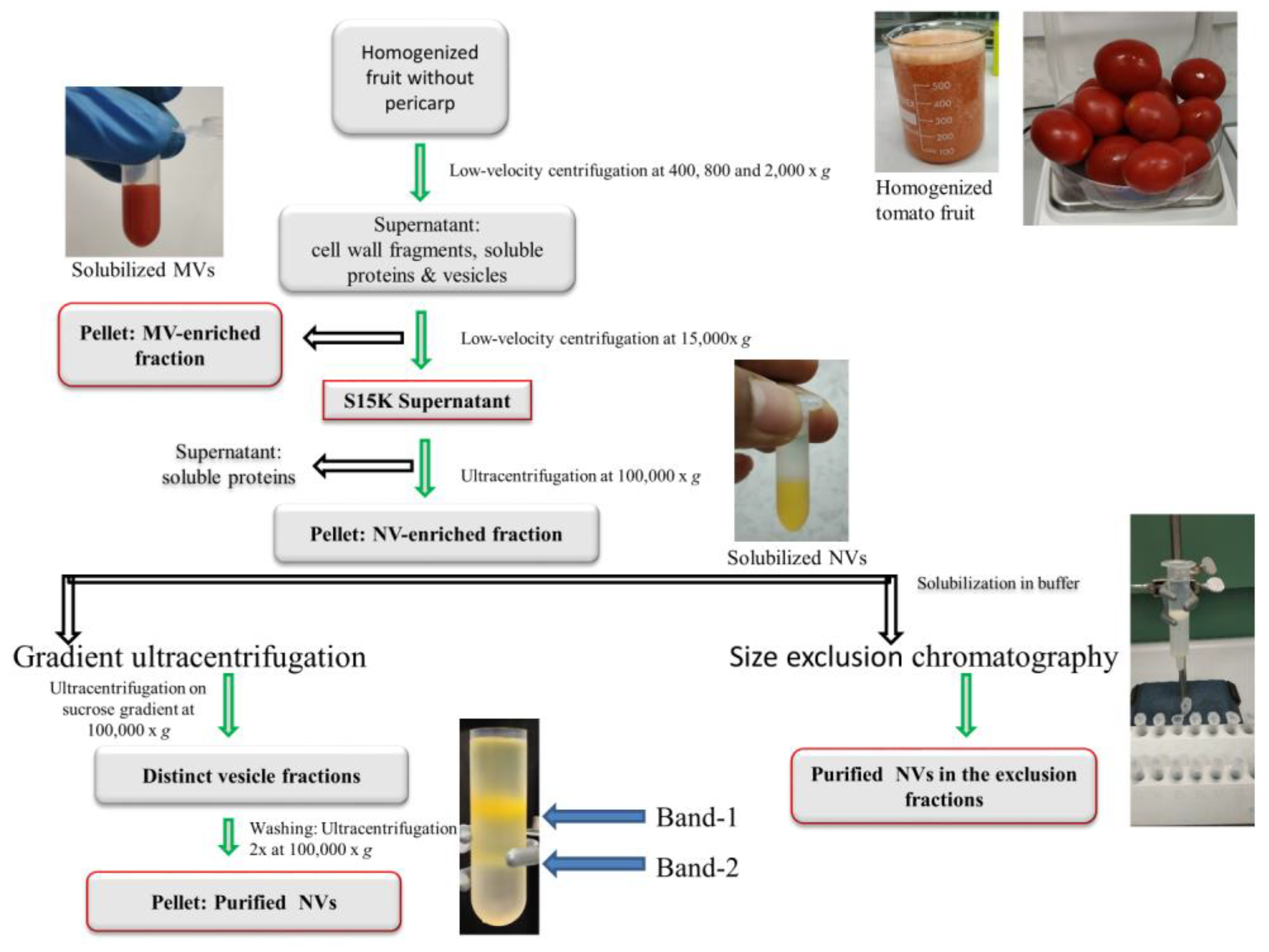
2.1. Plant Material and Isolation of Vesicles by Differential Ultracentrifugation
2.2. Gradient Ultracentrifugation
2.3. Size-Exclusion Chromatography
2.3.1. Column Packing
2.3.2. Size-Exclusion Chromatography of Tomato Fruit-Derived Vesicles
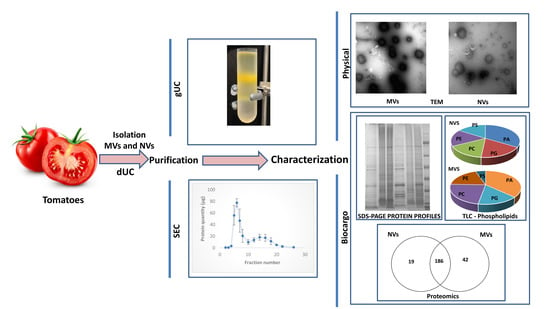
2.4. Characterization of MVs and NVs
2.4.1. Dynamic Light Scattering
2.4.2. Nanoparticle Tracking Analysis
2.4.3. Transmission Electron Microscope (TEM)
2.4.4. Protein Profiling by SDS-PAGE
2.4.5. Lipid Profiling by Thin-Layer Chromatography
2.4.6. Proteomic and Bioinformatics Analysis
3. Results and Discussion
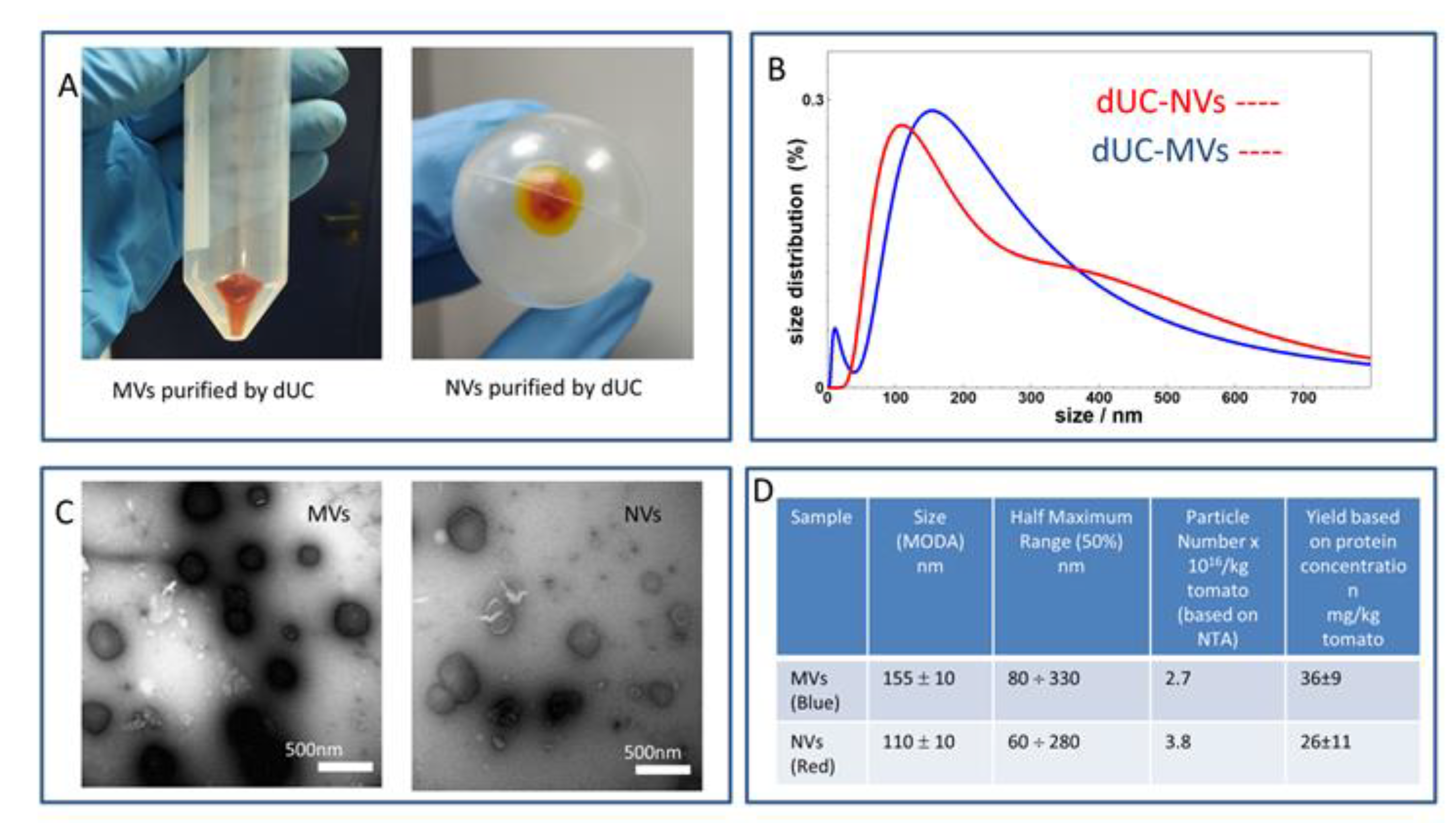
3.1. dUC Isolation and Characterization of Tomato-Derived MVs and NVs
3.2. Gradient Ultracentrifugation of NVs Isolated by dUC
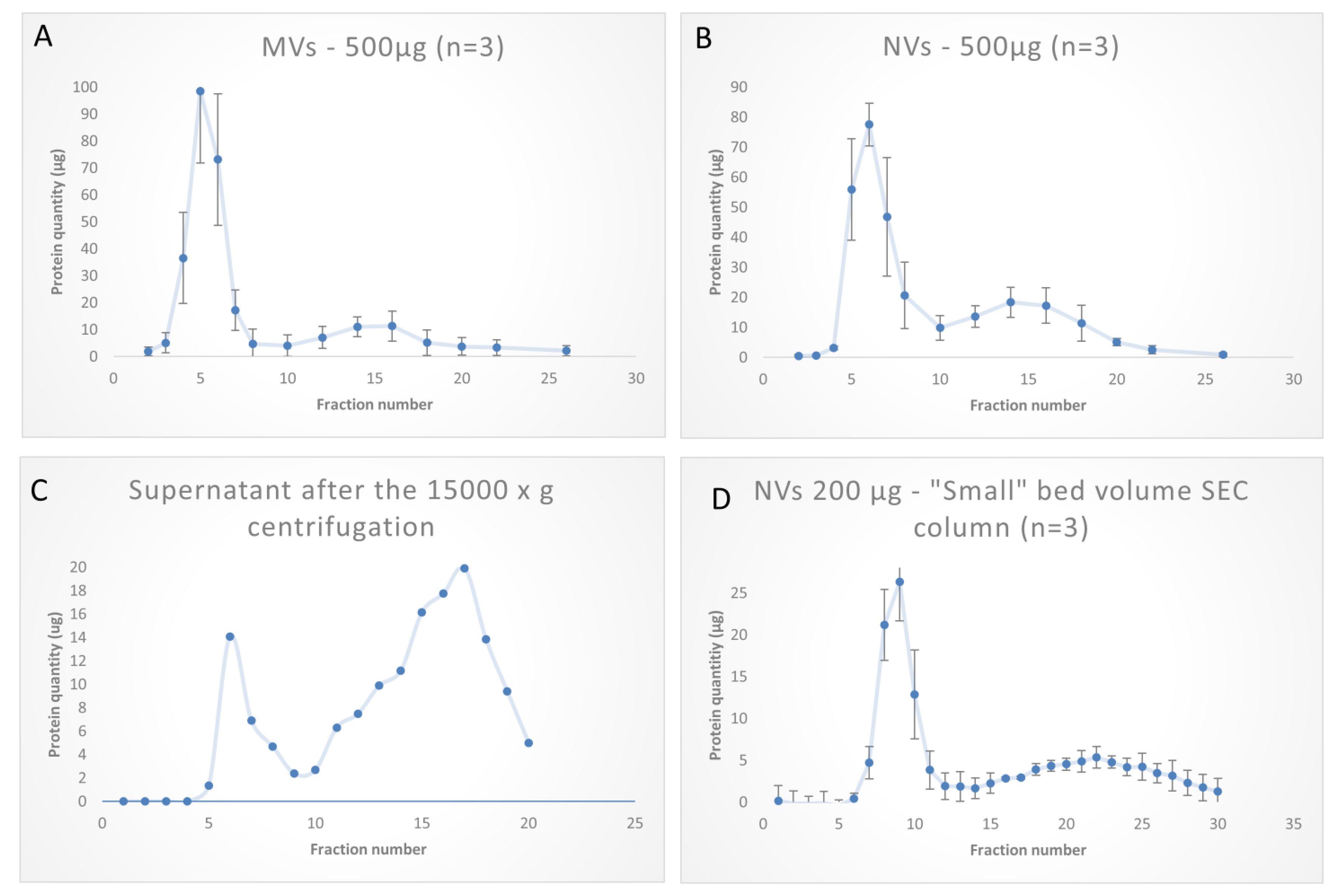
3.3. SEC Purification of MVs and NVs Isolated by dUC
3.4. Proteomic Characterization of dUC/SEC-Purified MVs and NVs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ding, Y.; Wang, J.; Wang, J.; Stierhof, Y.-D.; Robinson, D.G.; Jiang, L. Unconventional protein secretion. Trends Plant Sci. 2012, 17, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Rutter, B.D.; Innes, R. Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, Q.; Van Bel, A.J.E.; Hückelhoven, R. Do Plant Cells Secrete Exosomes Derived from Multivesicular Bodies? Plant Signal. Behav. 2007, 2, 4–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Feng, S.; Wang, X.; Long, K.; Luo, Y.; Wang, Y.; Ma, J.; Tang, Q.; Jin, L.; Li, X.; et al. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ 2018, 6, e5186. [Google Scholar] [CrossRef] [PubMed]
- Baldini, N.; Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Avnet, S. Exosome-like Nanovesicles Isolated from Citrus limon L. Exert Antioxidative Effect. Curr. Pharm. Biotechnol. 2018, 19, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Stanly, C.; Moubarak, M.; Fiume, I.; Turiák, L.; Pocsfalvi, G. Membrane Transporters in Citrus clementina Fruit Juice-Derived Nanovesicles. Int. J. Mol. Sci. 2019, 20, 6205. [Google Scholar] [CrossRef] [Green Version]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef]
- Kalarikkal, S.P.; Prasad, D.; Kasiappan, R.; Chaudhari, S.R.; Sundaram, G.M. A cost-effective polyethylene glycol-based method for the isolation of functional edible nanoparticles from ginger rhizomes. Sci. Rep. 2020, 10, 4456. [Google Scholar] [CrossRef]
- Pocsfalvi, G.; Turiák, L.; Ambrosone, A.; Del Gaudio, P.; Puska, G.; Fiume, I.; Silvestre, T.; Vékey, K. Physiochemical and protein datasets related to citrus juice sac cells-derived nanovesicles and microvesicles. Data Brief 2019, 22, 251–254. [Google Scholar] [CrossRef]
- Woith, E.; Melzig, M.F. Extracellular Vesicles from Fresh and Dried Plants—Simultaneous Purification and Visualization Using Gel Electrophoresis. Int. J. Mol. Sci. 2019, 20, 357. [Google Scholar] [CrossRef] [Green Version]
- Fujita, D.; Arai, T.; Komori, H.; Shirasaki, Y.; Wakayama, T.; Nakanishi, T.; Tamai, I. Apple-Derived Nanoparticles Modulate Expression of Organic-Anion-Transporting Polypeptide (OATP) 2B1 in Caco-2 Cells. Mol. Pharm. 2018, 15, 5772–5780. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Dico, A.L.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A.; et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514–19527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Yu, S.; Li, M.; Gui, X.; Li, P. Isolation of Exosome-Like Nanoparticles and Analysis of MicroRNAs Derived from Coconut Water Based on Small RNA High-Throughput Sequencing. J. Agric. Food Chem. 2018, 66, 2749–2757. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-S.; Yang, C.; Viennois, E.; Zhang, M.; Merlin, D. Isolation, Purification, and Characterization of Ginger-derived Nanoparticles (GDNPs) from Ginger, Rhizome of Zingiber officinale. Bio-protocol 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Viennois, E.; Xu, C.; Merlin, D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 2016, 4, e1134415. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [Green Version]
- Rome, S. Biological properties of plant-derived extracellular vesicles. Food Funct. 2019, 10, 529–538. [Google Scholar] [CrossRef]
- Lee, R.; Ko, H.J.; Kim, K.; Sohn, Y.; Min, S.Y.; Kim, J.A.; Na, D.; Yeon, J.H. Anti-melanogenic effects of extracellular vesicles derived from plant leaves and stems in mouse melanoma cells and human healthy skin. J. Extracell. Vesicles 2020, 9, 1703480. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhang, M.; Merlin, D. Advances in plant-derived edible nanoparticle-based lipid nano-drug delivery systems as therapeutic nanomedicines. J. Mater. Chem. B 2018, 6, 1312–1321. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Feechan, A.; Böhlenius, H.; Ueda, T.; Thordal-Christensen, H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA 2012, 109, 11443–11448. [Google Scholar] [CrossRef] [Green Version]
- Takov, K.; Yellon, D.M.; Davidson, S.M. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: Yield, purity and functional potential. J. Extracell. Vesicles 2019, 8, 1560809. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S.; Varma, R.S. Plant-Derived Edible Nanoparticles and miRNAs: Emerging Frontier for Therapeutics and Targeted Drug-Delivery. ACS Sustain. Chem. Eng. 2019, 7, 8055–8069. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Res. Int. 2018, 2018, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Monguió-Tortajada, M.; Gálvez-Montón, C.; Bayes-Genis, A.; Roura, S.; Borràs, F.E. Extracellular vesicle isolation methods: Rising impact of size-exclusion chromatography. Cell. Mol. Life Sci. 2019, 76, 2369–2382. [Google Scholar] [CrossRef]
- Wang, Q.; Zhuang, X.; Mu, J.; Deng, Z.-B.; Jiang, H.; Zhang, L.; Xiang, X.; Wang, B.; Yan, J.; Miller, D.L.; et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013, 4, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Akuma, P.; Okagu, O.D.; Udenigwe, C.C. Naturally Occurring Exosome Vesicles as Potential Delivery Vehicle for Bioactive Compounds. Front. Sustain. Food Syst. 2019, 3. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652.e8. [Google Scholar] [CrossRef] [Green Version]
- Pocsfalvi, G.; Turiák, L.; Ambrosone, A.; Del Gaudio, P.; Puska, G.; Fiume, I.; Silvestre, T.; Vékey, K. Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. J. Plant Physiol. 2018, 229, 111–121. [Google Scholar] [CrossRef]
- Baldrich, P.; Rutter, B.D.; Zandkarimi, H.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 2019, 31, 315–324. [Google Scholar] [CrossRef] [Green Version]
- Regente, M.; Corti-Monzón, G.; Maldonado, A.M.; Pinedo, M.; Jorrín, J.; De La Canal, L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009, 583, 3363–3366. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, B.M.; Rico, A.; McCraw, S.; Fones, H.N.; Preston, G.M. The Infiltration-centrifugation Technique for Extraction of Apoplastic Fluid from Plant Leaves Using Phaseolus vulgaris as an Example. J. Vis. Exp. 2014, e52113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouchi, I.; Hayashi, K.; Hiradate, S.; Ishikawa, S.; Fukuoka, M.; Chen, C.P.; Kobayashi, K. Overcoming the Difficulties in Collecting Apoplastic Fluid from Rice Leaves by the Infiltration–Centrifugation method. Plant Cell Physiol. 2012, 53, 1659–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, K.S.; Phillies, G.D.J. An Introduction to Dynamic Light Scattering by Macromolecules. Phys. Today 1991, 44, 66. [Google Scholar] [CrossRef]
- Webber, J.; Clayton, A. How pure are your vesicles? J. Extracell. Vesicles 2013, 2, 19861. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Hibino, H.; Omura, T. Release of Rice dwarf virusfrom insect vector cells involves secretory exosomes derived from multivesicular bodies. Commun. Integr. Biol. 2009, 2, 324–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Viennois, E.; Prasad, M.; Zhang, Y.; Wang, L.; Zhang, Z.; Han, M.K.; Xiao, B.; Xu, C.; Srinivasan, S.; et al. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials 2016, 101, 321–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Rong, Y.; Teng, Y.; Mu, J.; Zhuang, X.; Tseng, M.; Samykutty, A.; Zhang, L.; Yan, J.; Miller, D.; et al. Broccoli-Derived Nanoparticle Inhibits Mouse Colitis by Activating Dendritic Cell AMP-Activated Protein Kinase. Mol. Ther. 2017, 25, 1641–1654. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Merlin, D. Curcuma Longa-Derived Nanoparticles Reduce Colitis and Promote Intestinal Wound Repair by Inactivating the NF-ΚB Pathway. Gastroenterology 2017, 152, S567. [Google Scholar] [CrossRef]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.-B.; Wang, B.; Zhang, L.; et al. Grape Exosome-like Nanoparticles Induce Intestinal Stem Cells and Protect Mice From DSS-Induced Colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhou, Y.; Yu, J. Exosome-like Nanoparticles from Ginger Rhizomes Inhibited NLRP3 Inflammasome Activation. Mol. Pharm. 2019, 16, 2690–2699. [Google Scholar] [CrossRef]
- Zhang, M.; Xiao, B.; Wang, H.; Han, M.K.; Zhang, Z.; Viennois, E.; Xu, C.; Merlin, D. Edible Ginger-derived Nano-lipids Loaded with Doxorubicin as a Novel Drug-delivery Approach for Colon Cancer Therapy. Mol. Ther. 2016, 24, 1783–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baysal, T.; Demirdöven, A. Lipoxygenase in fruits and vegetables: A review. Enzym. Microb. Technol. 2007, 40, 491–496. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Yang, Y.; Fei, Z.; Yuan, H.; Fish, T.; Thannhauser, T.W.; Mazourek, M.; Kochian, L.V.; Wang, X.; Li, L. Proteomic analysis of chromoplasts from six crop species reveals insights into chromoplast function and development. J. Exp. Bot. 2013, 64, 949–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, A.M.; De Clerck, Y.A.; Langley, K.E.; Reisfeld, R.A.; Mueller, B.M. Melanoma-mediated dissolution of extracellular matrix: Contribution of urokinase-dependent and metalloproteinase-dependent proteolytic pathways. Cancer Res. 1993, 53, 693–700. [Google Scholar]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef] [PubMed]



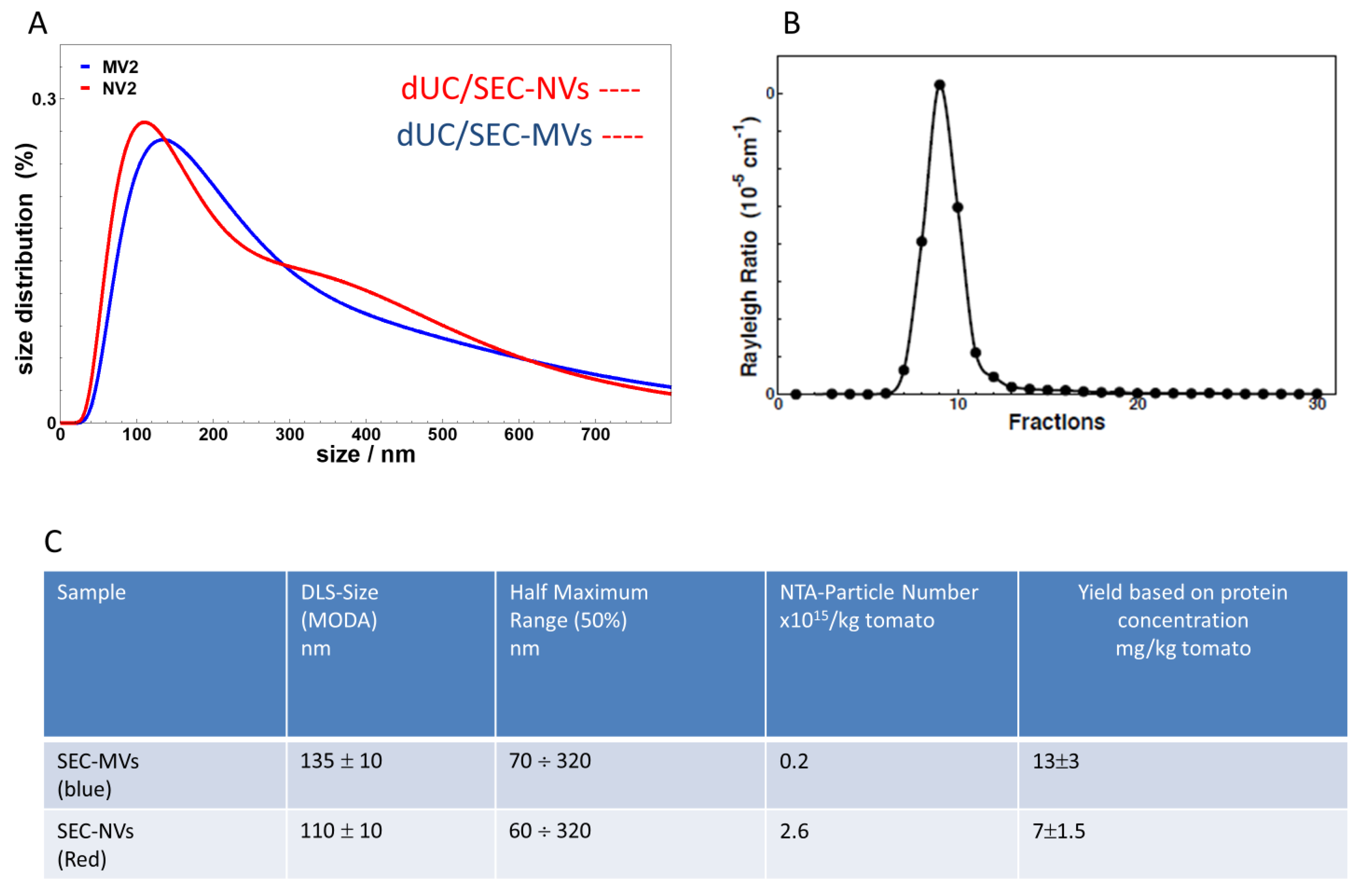
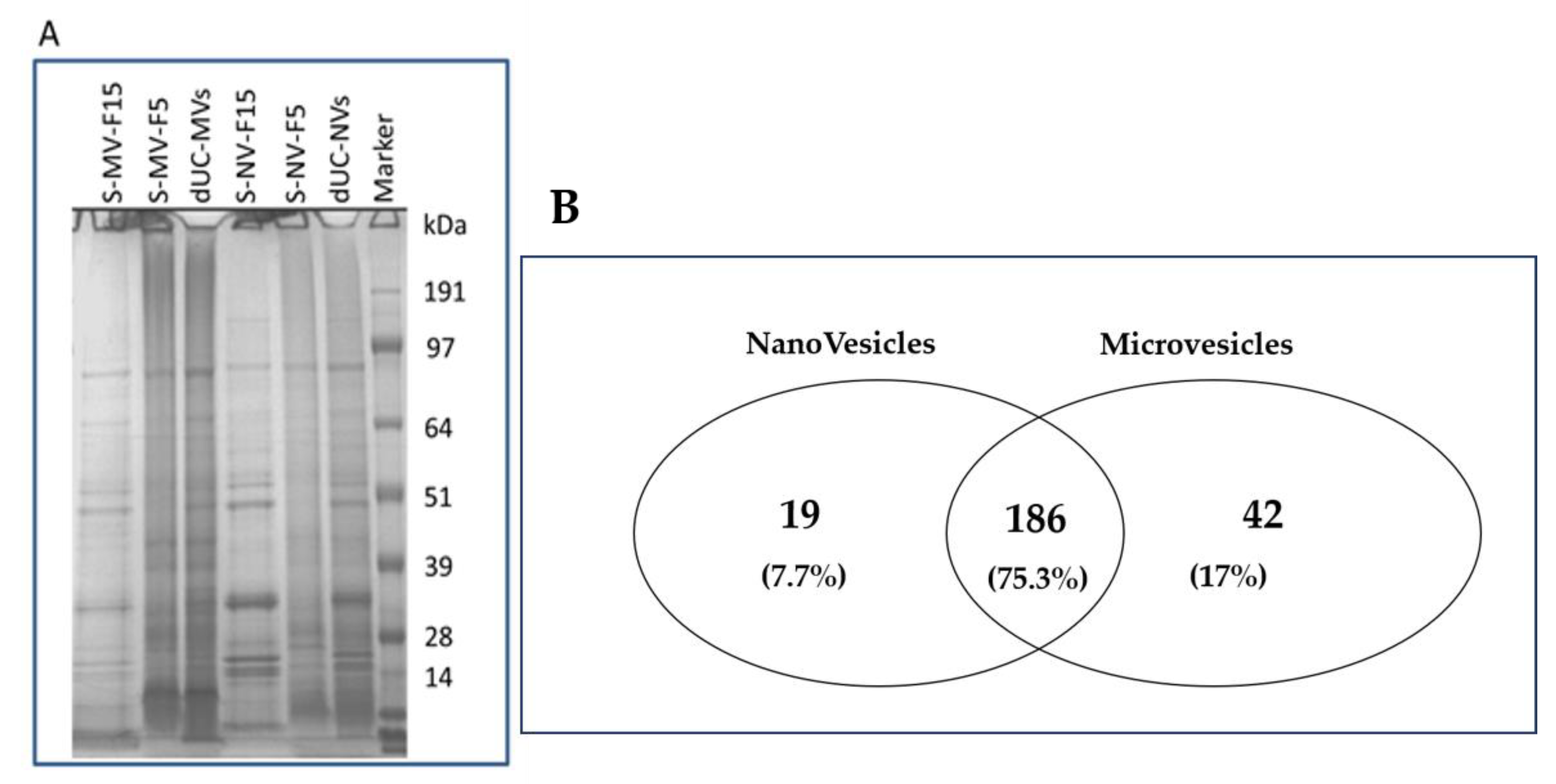
| Resource | Organ | Isolation Method | Vesicle Type(s) Isolated | Yield (g/L or g/kg of Starting Plant Material) | Particle Number (Particles/kg or Particles/L of Starting Plant Material) | Ref. |
|---|---|---|---|---|---|---|
| Ginger | rhizome | dUC/gUC | NVs | 0.25–1.25 g/L | n.r. | [21] |
| Ginger | rhizome | dUC/precipitation | NVs | 2-3.8 | n.r. | [22] |
| Ginger | rhizome | dUC | NVs | 4 | n.r. | [22] |
| Ginger | rhizome | dUC | NVs | 48.5 ± 4.8 × 10−3 | n.r. | [20] |
| Ginger | rhizome | dUC/gUC | NVs | 0.890 | 4.2 × 1012 | [23] |
| Ginger | rhizome | dUC | NVs | n.r. | 0.5 − 2 × 1014 | [17] |
| Ginger | rhizome | dUC/gUC | NVs | Three bands each containing ≅0.05 | n.r. | [13] |
| Grape | fruit | dUC/gUC | NVs | 1.76 ± 0.15 | n.r. | [19] |
| Grapefruit | fruit | dUC/gUC | NVs | 2.21 ± 0.044 | n.r. | [19] |
| Tomatoes | fruit | dUC/gUC | NVs | 0.44 ± 0.02 | n.r. | [19] |
| Grape | fruit | dUC/gUC | NVs | n.r. | n.r. | [16] |
| Broccoli | flower | dUC/gUC | NVs, MVs | n.r. | n.r. | [14] |
| Apple | fruit | dUC | NVs | n.r. | 1.6 × 1013 particles/L | [24] |
| Coconut | fruit | dUC/MF | NVs | n.r. | n.r. | [25] |
| Citrus clementina | fruit | dUC/gUC | NVs | 1.67 × 10−3 g/L (protein) | 1.16 × 1012 particles/L | [26] |
| Citrus sinensis | fruit | dUC | NVs | 0.178 g/L (protein) | n.r. | [27] |
| Citrus paradisi | fruit | dUC | NVs | 0.134 g/L (protein) | n.r. | [27] |
| Citrus aurantium | fruit | dUC | NVs | 0.161 g/L (protein) | n.r. | [27] |
| Citrus limon | fruit | dUC | NVs | 0.409 g/L (protein) | n.r. | [27] |
| Citrus limon | fruit | dUC/MF/gUC | NVs | 2.5 × 10−3 g/L | n.r. | [12] |
| Carrot | root | dUC/gUC | NVs | 0.298 | n.r. | [28] |
| Blueberry | fruit | dUC/MF | NVs | n.r. | n.r. | [29] |
| Hami melon | fruit | dUC/MF | NVs | n.r. | n.r. | [29] |
| Pea | seed | dUC/MF | NVs | n.r. | n.r. | [29] |
| Pear | fruit | dUC/MF | NVs | n.r. | n.r. | [29] |
| Soybean | seed | dUC/MF | NVs | n.r. | n.r. | [29] |
| Orange | fruit | dUC/MF | NVs | n.r. | n.r. | [29] |
| Kiwifruit | fruit | dUC/MF | NVs | n.r. | n.r. | [29] |
| Arabidopsis thaliana L. | leaf | dUC/gUC | EVs | n.r. | n.r. | [30] |
| Sunflower | seed | MF/dUC | AVs | n.r. | n.r. | [31] |
| Nicotiana tabacum L. | leaf | dUC | AVs | n.r. | n.r. | [32] |
| Vinca minor L. | leaf | dUC | AVs | n.r. | n.r. | [32] |
| Viscum album L. | leaf | dUC | AVs | n.r. | n.r. | [32] |
| Phaseolus vulgaris L. | leaf | dUC | EVs | 0.081 ± 0.03 | n.r. | [33] |
| Oryza sativa L. (Rice) | leaf | dUC | AVs | n.r. | n.r. | [34] |
| UniProtKB | Protein Names | Typical Subcellular Location | NV Intesnsity | MV Intensity | |
|---|---|---|---|---|---|
| 1 | Q42873_SOLLC | Lipoxygenase | cytoplasm | 2.94 × 108 | 3.62 × 108 |
| 2 | ADH2_SOLLC | Alcohol dehydrogenase 2 | cytoplasm | 1.45 × 108 | 1.47 × 108 |
| 3 | ACCH3_SOLLC | 1-aminocyclopropane-1-carboxylate oxidase homolog | 1.23 × 108 | 1.16 × 108 | |
| 4 | ATPB_SOLLC | ATP synthase subunit beta, chloroplastic | chloroplast | 1.14 × 108 | 6.02 × 107 |
| 5 | ASR1_SOLLC | Abscisic stress-ripening protein 1 | nucleus | 1.04 × 108 | 1.48 × 108 |
| 6 | PGLR_SOLLC | Polygalacturonase-2 (Pectinase) | cell wall, apoplast | 1.04 × 108 | 2.04 × 108 |
| 7 | ATPA_SOLLC | ATP synthase subunit alpha, chloroplastic | chloroplast | 9.11 × 107 | 4.44 × 107 |
| 8 | EF1A_SOLLC | Elongation factor 1-alpha | cytoplasm | 8.20 × 107 | 9.24 × 107 |
| 9 | P93767_SOLLC | ADP/ATP translocator | membrane | 7.28 × 107 | 5.78 × 107 |
| 10 | Q40140_SOLLC | Aspartic protease | 6.38 × 107 | 3.58 × 107 | |
| 11 | Q9XEX8_SOLLC | Remorin 1 | 6.14 × 107 | 4.06 × 107 | |
| 12 | H1ZXA9_SOLLC | Heat shock protein 70 isoform 3 | cytoplasm | 6.09 × 107 | 9.74 × 107 |
| 13 | HSP80_SOLLC | Heat shock cognate protein 80 | cytoplasm | 6.06 × 107 | 7.05 × 107 |
| 14 | G5DGD4_SOLLC | Class I small heat shock protein | 6.01 × 107 | 4.57 × 107 | |
| 15 | K4CJ46_SOLLC | 2-Isopropylmalate synthase | chloroplast, cytoplasm | 5.88 × 107 | 5.40 × 107 |
| 16 | B0JEU3_SOLLC | Vicilin | 5.81 × 107 | 4.65 × 107 | |
| 17 | Q4W5U7_SOLLC | Calnexin-like protein | ER | 5.74 × 107 | 4.24 × 107 |
| 18 | Q6IV07_SOLLC | UDP-arabinopyranose mutase | cytosol/Golgi | 5.27 × 107 | 1.50 × 107 |
| 19 | G8Z279_SOLLC | Hop-interacting protein THI113 | 4.54 × 107 | 4.69 × 107 | |
| 20 | Q38JD4_SOLLC | Temperature-induced lipocalin | cytoplasm | 4.50 × 107 | 3.70 × 107 |
| 21 | FSPM_SOLLC | Fruit-specific protein | 6.51× 106 | 5.78 × 107 | |
| 22 | O81536_SOLLC | Annexin | cytoplasm | 2.92 × 107 | 5.76 × 107 |
| 23 | CATA1_SOLLC | Catalase isozyme 1 | peroxisome | 2.16 × 107 | 4.42 × 107 |
| 24 | RS27A_SOLLC | Ubiquitin-40S ribosomal protein S27a | nucleus | 3.64 × 107 | 4.10 × 107 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokka, R.; Ramos, A.P.; Fiume, I.; Manno, M.; Raccosta, S.; Turiák, L.; Sugár, S.; Adamo, G.; Csizmadia, T.; Pocsfalvi, G. Biomanufacturing of Tomato-Derived Nanovesicles. Foods 2020, 9, 1852. https://doi.org/10.3390/foods9121852
Bokka R, Ramos AP, Fiume I, Manno M, Raccosta S, Turiák L, Sugár S, Adamo G, Csizmadia T, Pocsfalvi G. Biomanufacturing of Tomato-Derived Nanovesicles. Foods. 2020; 9(12):1852. https://doi.org/10.3390/foods9121852
Chicago/Turabian StyleBokka, Ramesh, Anna Paulina Ramos, Immacolata Fiume, Mauro Manno, Samuele Raccosta, Lilla Turiák, Simon Sugár, Giorgia Adamo, Tamás Csizmadia, and Gabriella Pocsfalvi. 2020. "Biomanufacturing of Tomato-Derived Nanovesicles" Foods 9, no. 12: 1852. https://doi.org/10.3390/foods9121852
APA StyleBokka, R., Ramos, A. P., Fiume, I., Manno, M., Raccosta, S., Turiák, L., Sugár, S., Adamo, G., Csizmadia, T., & Pocsfalvi, G. (2020). Biomanufacturing of Tomato-Derived Nanovesicles. Foods, 9(12), 1852. https://doi.org/10.3390/foods9121852