Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Extract Preparation from Nigella sativa Seeds
2.2. Determination of Antioxidant Capacity Using Hydrogen Peroxide (H2O2)
2.3. DPPH Assay for the Evaluation of Antioxidant Activity
2.4. Evaluation of RBC Membrane Stability
2.4.1. Preparation of RBC Suspension
2.4.2. Inhibition of Heat-Induced Hemolysis
2.4.3. Inhibition of Hyposaline Induced Hemolysis
2.5. Screening of Anti-Glycating and AGEs Formation Inhibition Potentials
2.5.1. In Vitro System of Protein Glycation System
2.5.2. Assessment of Browning Intensity
2.5.3. Protein Aggregation Index
2.6. Biophysical Studies to Investigate AGEs Formation Inhibiting Properties of Extract
2.7. Molecular Docking Studies
2.7.1. The Receptors
2.7.2. The Ligands
2.7.3. In Silico Molecular Docking
2.8. Statistical Analysis
3. Results
3.1. H2O2 Reducing Activity of N. sativa Seed Extract
3.2. Scavenging of DPPH Radical by N. sativa Seed Extract
3.3. Effect of N. sativa on Membrane Stabilization
3.3.1. Protection against Hemolysis Caused by Heat
3.3.2. Protection against Hemolysis Caused by Hyposaline
3.4. Screening of Anti-Glycating and AGEs Formation Inhibition Potentials
3.4.1. Effect of N. sativa on Browning Intensity
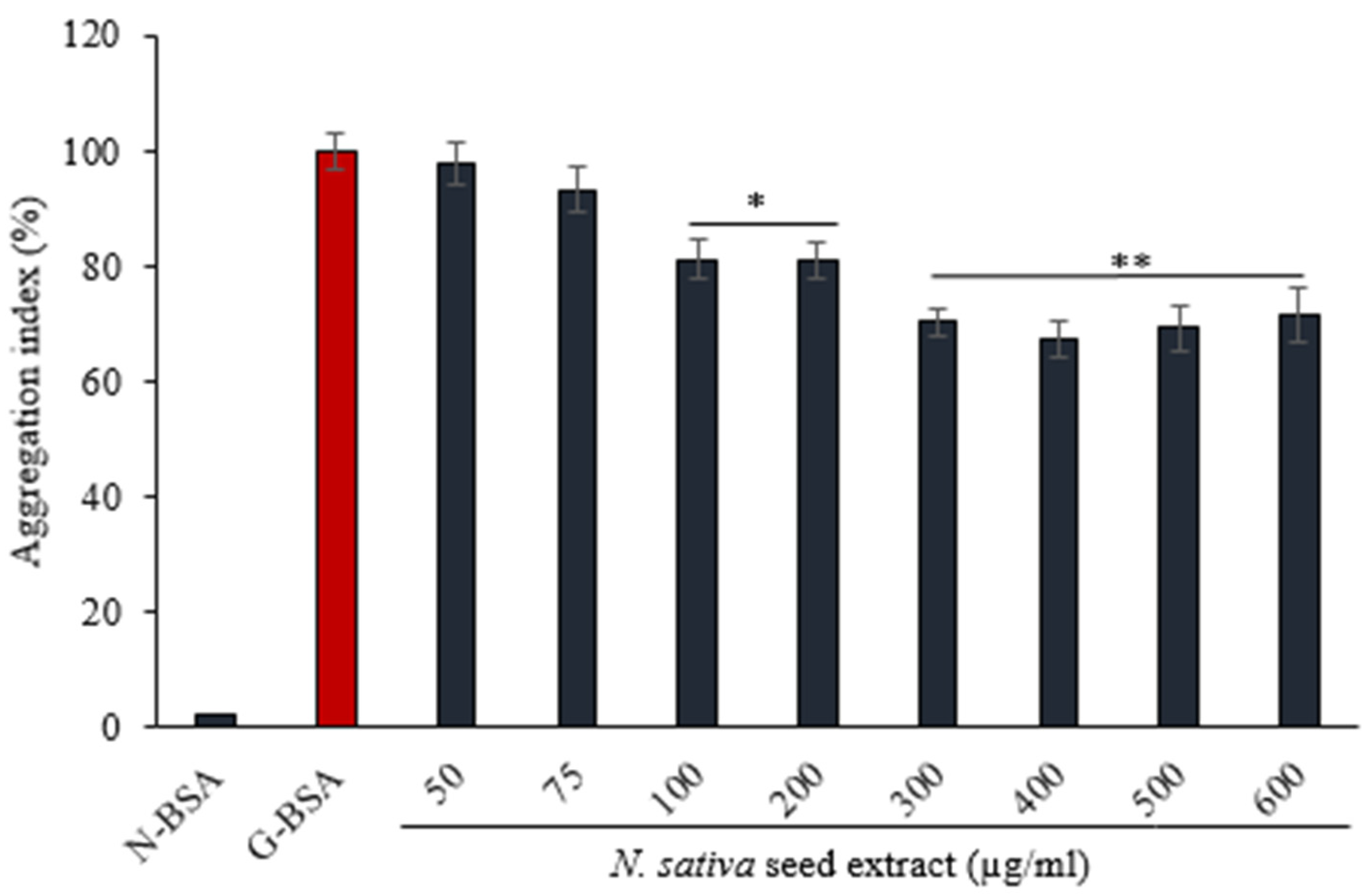
3.4.2. Effect of on N. sativa Seed Extract on Protein Aggregation Index
3.4.3. Fluorescence of AGE
3.5. Receptor-Ligand Interaction Study by Molecular Docking
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Qu, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Ragab, D.; Salah Eldin, H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef] [PubMed]
- Sherwani, S.; Khan, M.W.A. Cytokine Response in SARS-CoV-2 Infection in the Elderly. J. Inflamm. Res. 2020, 13, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Kubánková, M.; Hohberger, B.; Hoffmanns, J.; Fürst, J.; Herrmann, M.; Guck, J.; Kräter, M. Physical phenotype of blood cells is altered in COVID-19. Biophys. J. 2021, 120, 2838–2847. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salamanna, F.; Maglio, M.; Landini, M.P.; Fini, M. Body Localization of ACE-2: On the Trail of the Keyhole of SARS-CoV-2. Front. Med. (Lausanne) 2020, 7, 594495. [Google Scholar] [CrossRef] [PubMed]
- Jarlhelt, I.; Nielsen, S.K.; Jahn, C.X.H.; Hansen, C.B.; Pérez-Alós, L.; Rosbjerg, A.; Bayarri-Olmos, R.; Skjoedt, M.O.; Garred, P. SARS-CoV-2 Antibodies Mediate Complement and Cellular Driven Inflammation. Front. Immunol. 2021, 12, 767981. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Dzieciatkowska, M.; Issaian, A.; Nemkov, T.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Buehler, P.W.; Zimring, J.C.; et al. Evidence of Structural Protein Damage and Membrane Lipid Remodeling in Red Blood Cells from COVID-19 Patients. J. Proteome Res. 2020, 19, 4455–4469. [Google Scholar] [CrossRef]
- Roberts, J.; Pritchard, A.L.; Treweeke, A.T.; Rossi, A.G.; Brace, N.; Cahill, P.; MacRury, S.M.; Wei, J.; Megson, I.L. Why Is COVID-19 More Severe in Patients With Diabetes? The Role of Angiotensin-Converting Enzyme 2, Endothelial Dysfunction and the Immunoinflammatory System. Front. Cardiovasc. Med. 2021, 7, 629933. [Google Scholar] [CrossRef]
- Maiuolo, J.; Mollace, R.; Gliozzi, M.; Musolino, V.; Carresi, C.; Paone, S.; Scicchitano, M.; Macrì, R.; Nucera, S.; Bosco, F.; et al. The Contribution of Endothelial Dysfunction in Systemic Injury Subsequent to SARS-CoV-2 Infection. Int. J. Mol. Sci. 2020, 21, 9309. [Google Scholar] [CrossRef] [PubMed]
- Nabi, R.; Alvi, S.S.; Shah, A.; Chaturvedi, C.P.; Faisal, M.; Alatar, A.A.; Ahmad, S.; Khan, M.S. Ezetimibe attenuates experimental diabetes and renal pathologies via targeting the advanced glycation, oxidative stress and AGE-RAGE signalling in rats. Arch. Physiol. Biochem. 2021, 29, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Akhter, F.; Shahab, U.; Rafi, Z.; Khan, M.S.; Nabi, R.; Khan, M.S.; Ahmad, K.; Ashraf, J.M.; Moinuddin. Do all roads lead to the Rome? The glycation perspective! Semin. Cancer Biol. 2018, 49, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Uddin, M.; Habib, S.; Shahab, U.; Alam, K.; Ali, A. Autoimmune response to AGE modified human DNA: Implications in type 1 diabetes mellitus. J. Clin. Transl. Endocrinol. 2014, 1, 66–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Anwar, S.; Aljarbou, A.N.; Al-Orainy, M.; Aldebasi, Y.H.; Islam, S.; Younus, H. Protective effect of thymoquinone on glucose or methylglyoxal-induced glycation of superoxide dismutase. Int. J. Biol. Macromol. 2014, 65, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-García, J.; Osca-Verdegal, R.; Pallardó, F.V.; Ferreres, J.; Rodríguez, M.; Mulet, S.; Sanchis-Gomar, F.; Carbonell, N.; García-Giménez, J.L. Oxidative Stress and Inflammation in COVID-19-Associated Sepsis: The Potential Role of Anti-Oxidant Therapy in Avoiding Disease Progression. Antioxidants 2020, 9, 936. [Google Scholar] [CrossRef]
- De la Cruz-Enríquez, J.; Rojas-Morales, E.; Ruíz-García, M.G.; Tobón-Velasco, J.C.; Jiménez-Ortega, J.C. SARS-CoV-2 induces mitochondrial dysfunction and cell death by oxidative stress/inflammation in leukocytes of COVID-19 patients. Free Radic. Res. 2021, 55, 982–995. [Google Scholar] [CrossRef]
- Sartore, G.; Ragazzi, E.; Faccin, L.; Lapolla, A. A role of glycation and methylation for SARS-CoV-2 infection in diabetes? Med. Hypotheses 2020, 144, 110247. [Google Scholar] [CrossRef]
- Anwar, S.; Khan, S.; Almatroudi, A.; Khan, A.A.; Alsahli, M.A.; Almatroodi, S.A.; Rahmani, A.H. A review on mechanism of inhibition of advanced glycation end products formation by plant derived polyphenolic compounds. Mol. Biol. Rep. 2021, 48, 787–805. [Google Scholar] [CrossRef]
- Younus, H.; Anwar, S. Prevention of non-enzymatic glycosylation (glycation): Implication in the treatment of diabetic complication. Int. J. Health Sci. (Qassim) 2016, 10, 261–277. [Google Scholar] [CrossRef]
- Ahmad, A.; Husain, A.; Mujeeb, M.; Alam Khan, S.; Najmi, A.K.; Siddique, N.A.; Damanhouri, Z.A.; Anwar, F. A review on therapeutic potential of Nigella sativa: A miracle herb. Asian Pac. J. Trop. Biomed. 2013, 3, 337–352. [Google Scholar] [CrossRef] [Green Version]
- Hadi, V.; Pahlavani, N.; Malekahmadi, M.; Alam Khan, S.; Najmi, A.K.; Siddique, N.A.; Damanhouri, Z.A.; Anwar, F. Nigella sativa in controlling Type 2 diabetes, cardiovascular, and rheumatoid arthritis diseases: Molecular aspects. J. Res. Med. Sci. 2021, 26, 20. [Google Scholar] [PubMed]
- Norman, G.A.V. Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC Basic Transl. Sci. 2016, 1, 170–179. [Google Scholar]
- Singh, R.; Bhardwaj, V.K.; Purohit, R. Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: An in-silico approach. Comp. Biol. Med. 2021, 139, 104965. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Bhardwaj, V.K.; Das, P.; Purohit, R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comp. Biol. Med. 2021, 135, 104555. [Google Scholar] [CrossRef]
- Singh, R.; Bhardwaj, V.K.; Das, P.; Bhattacherjee, D.; Zyryanov, G.V.; Purohit, R. Benchmarking the ability of novel compounds to inhibit SARS-CoV-2 main protease using steered molecular dynamics simulations. Comp. Biol. Med. 2022, 146, 105572. [Google Scholar] [CrossRef]
- Anwar, S.; Almatroudi, A.; Allemailem, K.S.; Jacob-Joseph, R.; Khan, A.A.; Rahmani, A.H. Protective Effects of Ginger Extract against Glycation and Oxidative Stress-Induced Health Complications: An In Vitro Study. Processes 2020, 8, 468. [Google Scholar] [CrossRef]
- Khan, M.W.A.; Otaibi, A.A.; Alhumaid, A.F.M.; Alsukaibi, A.K.D.; Alshamari, A.K.; Alshammari, E.M.; Al-Zahrani, S.A.; Almudyani, A.Y.M.; Sherwani, S. Garlic Extract: Inhibition of Biochemical and Biophysical Changes in Glycated HSA. Appl. Sci. 2021, 11, 11028. [Google Scholar] [CrossRef]
- Khan, M.W.A.; Otaibi, A.A.; Alsukaibi, A.K.D.; Alshammari, E.M.; Al-Zahrani, S.A.; Sherwani, S.; Khan, W.A.; Saha, R.; Verma, S.R.; Ahmed, N. Biophysical, Biochemical, and Molecular Docking Investigations of Anti-Glycating, Antioxidant, and Protein Structural Stability Potential of Garlic. Molecules 2022, 27, 1868. [Google Scholar] [CrossRef]
- Anwar, S.; Almatroudi, A.; Alsahli, M.A.; Khan, M.A.; Khan, A.A.; Rahmani, A.H. Natural Products: Implication in Cancer Prevention and Treatment through Modulating Various Biological Activities. Anticancer Agents Med. Chem. 2020, 20, 2025–2040. [Google Scholar] [CrossRef]
- Chanda, S.; Juvekar, A. In vitro anti-inflammatory activity of syringic acid. Int. J. Pharm. Pharm. Sci. 2019, 11, 71–73. [Google Scholar] [CrossRef]
- Brownlee, M.; Vlassara, H.; Kooney, A.; Ulrich, P.; Cerami, A. Aminoguanidine prevents diabetes-induced arterial wall protein cross-linking. Science 1986, 232, 1629–1632. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Ali, A. Antiglycation and antiaggregation potential of thymoquinone. Nat. Volatiles Essent. Oils 2019, 6, 25–33. [Google Scholar]
- Khan, M.W.A.; Rasheed, Z.; Khan, W.A.; Ali, R. Biochemical, biophysical, and thermodynamic analysis of in vitro glycated human serum albumin. Biochemistry 2007, 72, 146–152. [Google Scholar] [PubMed]
- Khan, M.W.A.; Otaibi, A.A.; Al-Zahrani, S.A.; Alshammari, E.M.; Haque, A.; Alouffi, S.; Khan, W.A.; Khan, S.N. Experimental and theoretical insight into resistance to glycation of bovine serum albumin. J. Mol. Struc. 2021, 1230, 129645. [Google Scholar] [CrossRef]
- Ahmad, S.; Abbasi, H.W.; Shahid, S.; Gul, S.; Abbasi, S.W. Molecular docking, simulation and MM-PBSA studies of nigella sativa compounds: A computational quest to identify potential natural antiviral for COVID-19 treatment. J. Biomol. Struct. Dyn. 2021, 39, 4225–4233. [Google Scholar] [CrossRef] [PubMed]
- Tiruppur Venkatachallam, S.K.; Pattekhan, H.; Divakar, S.; Kadimi, U.S. Chemical composition of Nigella sativa L. seed extracts obtained by supercritical carbon dioxide. J. Food Sci. Technol. 2010, 47, 598–605. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.L.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, G.N.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Swargiary, A.; Mahmud, S.; Saleh, M.A. Screening of phytochemicals as potent inhibitor of 3-chymotrypsin and papain-like proteases of SARS-CoV2: An in silico approach to combat COVID-19. J. Biomol. Struct. Dyn. 2022, 40, 2067–2081. [Google Scholar] [CrossRef] [PubMed]
- Alouffi, S.; Sherwani, S.; Al-Mogbel, M.S.; Sherwani, M.K.A.; Khan, M.W.A. Depression and Smoking Augment the Production of Circulating Autoantibodies against Glycated HSA in Rheumatoid Arthritis Patients. Int. Arch. Allergy Immunol. 2018, 177, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Bouma, B.; Kroon-Batenburg, L.M.; Wu, Y.P.; Brünjes, B.; Posthuma, G.; Kranenburg, O.; de Groot, P.G.; Voest, E.E.; Gebbink, M.F. Glycation induces formation of amyloid cross-beta structure in albumin. J. Biol. Chem. 2003, 278, 41810–41819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, D.; Ramasamy, R.; Schmidt, A.M. Journey to a Receptor for Advanced Glycation End Products Connection in Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Akhter, N.; Ahmad, S.; Alzahrani, F.A.; Dar, S.A.; Wahid, M.; Haque, S.; Bhatia, K.; Sr Almalki, S.; Alharbi, R.A.; Sindi, A.A.A. Impact of COVID-19 on the cerebrovascular system and the prevention of RBC lysis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10267–10278. [Google Scholar] [PubMed]
- Gerber, G.F.; Yuan, X.; Yu, J.; Cher, B.A.Y.; Braunstein, E.M.; Chaturvedi, S.; Brodsky, R.A. COVID-19 vaccines induce severe hemolysis in paroxysmal nocturnal hemoglobinuria. Blood 2021, 137, 3670–3673. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6, Erratum in: 2020, 183, 1735. [Google Scholar] [CrossRef]
- Xia, X. Domains and Functions of Spike Protein in SARS-CoV-2 in the Context of Vaccine Design. Viruses 2021, 13, 109. [Google Scholar] [CrossRef]
- Shaikh, Y.I.; Shaikh, V.S.; Ahmed, K.; Nazeruddin, G.M.; Pathan, H.M. The revelation of various compounds found in Nigella sativa L.(Black Cumin) and their possibility to inhibit COVID-19 infection based on the molecular docking and physical properties. Eng. Sci. 2020, 11, 31–35. [Google Scholar] [CrossRef]
- Koshak, A.E.; Koshak, E.A.; Mobeireek, A.F.; Badawi, M.A.; Wali, S.O.; Malibary, H.M.; Atwah, A.F.; Alhamdan, M.M.; Almalki, R.A.; Madani, T.A. Nigella sativa for the treatment of COVID-19: An open-label randomized controlled clinical trial. Complement. Ther. Med. 2021, 61, 102769. [Google Scholar] [CrossRef]
- Anwar, S.; Raut, R.; Alsahli, M.A.; Almatroudi, A.; Alfheeaid, H.; Alzahrani, F.M.; Khan, A.A.; Allemailem, K.S.; Almatroodi, S.A.; Rahmani, A.H. Role of Ajwa Date Fruit Pulp and Seed in the Management of Diseases through In Vitro and In Silico Analysis. Biology 2022, 11, 78. [Google Scholar] [CrossRef]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef] [PubMed]
- Du, A.; Zheng, R.; Disoma, C.; Li, S.; Chen, Z.; Li, S.; Liu, P.; Zhou, Y.; Shen, Y.; Liu, S.; et al. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int. J. Biol. Macromol. 2021, 176, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Minie, M.; Chopra, G.; Sethi, G.; Horst, J.; White, G.; Roy, A.; Hatti, K.; Samudrala, R. CANDO and the infinite drug discovery frontier. Drug Discov. Today 2014, 9, 1353–1363. [Google Scholar]








| Ligands | PubChem CID | Spike Protein S (6VXX) | |
|---|---|---|---|
| Binding Energy (kcal/mol) | Interacting Amino Acids | ||
| Pentadecanoic acid | 13849 | −3.4 | Asp467, Thr108, Thr114, Ile468, Asn234, Gln155, Glu465, Arg466, Gly232, Ile233 |
| Octadecadienoic acid | 5312457 | −3.9 | The108, Ile233, Asn234, Gly232, Glu465, Arg466, Thr114, Asp467, Ile468, Gln115, Lys113 |
| (Z)6-Pentadecen-1-ol | 5365626 | −3.7 | Ile468, Arg466, Gln115, Gly232, Ile233, Gle465, Lys462, Thr114, Thr108, Asn234, Ile235 |
| 9,12-Octadecadien-1-ol | 5462912 | −3.6 | Ile468, Arg466, Gln115, Gly232, Ile233, Thr114, Thr108, Asn234, Ile233, Asp467, Glu465, Gly107 |
| Dioctyl phthalate | 8346 | −4.2 | Asp467, Glu465, Ile233, Gly232, Arg466, Asn234, Thr114, Thr109, Thr108, Gln115, Ile468, Lys113 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sherwani, S.; Rajendrasozhan, S.; Khan, M.W.A.; Saleem, M.; Khan, M.; Khan, S.; Raafat, M.; Othman Alqahtani, F. Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies. Processes 2022, 10, 1346. https://doi.org/10.3390/pr10071346
Sherwani S, Rajendrasozhan S, Khan MWA, Saleem M, Khan M, Khan S, Raafat M, Othman Alqahtani F. Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies. Processes. 2022; 10(7):1346. https://doi.org/10.3390/pr10071346
Chicago/Turabian StyleSherwani, Subuhi, Saravanan Rajendrasozhan, Mohd Wajid Ali Khan, Mohd Saleem, Mahvish Khan, Saif Khan, Mohamed Raafat, and Fatimah Othman Alqahtani. 2022. "Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies" Processes 10, no. 7: 1346. https://doi.org/10.3390/pr10071346
APA StyleSherwani, S., Rajendrasozhan, S., Khan, M. W. A., Saleem, M., Khan, M., Khan, S., Raafat, M., & Othman Alqahtani, F. (2022). Pharmacological Profile of Nigella sativa Seeds in Combating COVID-19 through In-Vitro and Molecular Docking Studies. Processes, 10(7), 1346. https://doi.org/10.3390/pr10071346






