Please note that, as of 21 September 2020, High-Throughput has been renamed to BioTech and is now published here.
Journal Description
High-Throughput
High-Throughput
- formerly Microarrays - is an international, scientific, peer-reviewed, open access journal published quarterly by MDPI
- Open Access—free for readers, with article processing charges (APC) paid by authors or their institutions.
- Rapid Publication: first decisions in 18 days; acceptance to publication in 4 days (median values for MDPI journals in the second half of 2024).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Latest Articles
Health Impact and Therapeutic Manipulation of the Gut Microbiome
High-Throughput 2020, 9(3), 17; https://doi.org/10.3390/ht9030017 - 29 Jul 2020
Cited by 15
Abstract
Recent advances in microbiome studies have revealed much information about how the gut virome, mycobiome, and gut bacteria influence health and disease. Over the years, many studies have reported associations between the gut microflora under different pathological conditions. However, information about the role
[...] Read more.
Recent advances in microbiome studies have revealed much information about how the gut virome, mycobiome, and gut bacteria influence health and disease. Over the years, many studies have reported associations between the gut microflora under different pathological conditions. However, information about the role of gut metabolites and the mechanisms by which the gut microbiota affect health and disease does not provide enough evidence. Recent advances in next-generation sequencing and metabolomics coupled with large, randomized clinical trials are helping scientists to understand whether gut dysbiosis precedes pathology or gut dysbiosis is secondary to pathology. In this review, we discuss our current knowledge on the impact of gut bacteria, virome, and mycobiome interactions with the host and how they could be manipulated to promote health.
Full article
(This article belongs to the Special Issue Human Microbiome and Diseases: Implications for Novel Therapies)
►
Show Figures
Open AccessArticle
Influence of the Ovine Genital Tract Microbiota on the Species Artificial Insemination Outcome. A Pilot Study in Commercial Sheep Farms
by
Malena Serrano, Eric Climent, Fernando Freire, Juan F. Martínez-Blanch, Carmen González, Luis Reyes, M. Carmen Solaz-Fuster, Jorge H. Calvo, M. Ángeles Jiménez and Francisco M. Codoñer
High-Throughput 2020, 9(3), 16; https://doi.org/10.3390/ht9030016 - 6 Jul 2020
Cited by 17
Abstract
►▼
Show Figures
To date, there is a lack of research into the vaginal and sperm microbiome and its bearing on artificial insemination (AI) success in the ovine species. Using hypervariable regions V3–V4 of the 16S rRNA, we describe, for the first time, the combined effect
[...] Read more.
To date, there is a lack of research into the vaginal and sperm microbiome and its bearing on artificial insemination (AI) success in the ovine species. Using hypervariable regions V3–V4 of the 16S rRNA, we describe, for the first time, the combined effect of the ovine microbiome of both females (50 ewes belonging to five herds) and males (five AI rams from an AI center) on AI outcome. Differences in microbiota abundance between pregnant and non-pregnant ewes and between ewes carrying progesterone-releasing intravaginal devices (PRID) with or without antibiotic were tested at different taxonomic levels. The antibiotic treatment applied with the PRID only altered Streptobacillus genus abundance, which was significantly lower in ewes carrying PRID with antibiotic. Mageebacillus, Histophilus, Actinobacilllus and Sneathia genera were significantly less abundant in pregnant ewes. In addition, these genera were more abundant in two farms with higher AI failure. Species of these genera such as Actinobacillus seminis and Histophilus somni have been associated with reproductive disorders in the ovine species. These genera were not present in the sperm samples of AI rams, but were found in the foreskin samples of rams belonging to herd 2 (with high AI failure rate) indicating that their presence in ewes’ vagina could be due to prior transmission by natural mating with rams reared in the herd.
Full article
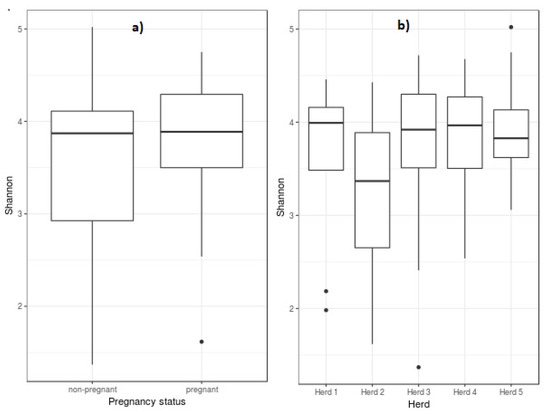
Figure 1
Open AccessArticle
Dark Proteome Database: Studies on Disorder
by
Nelson Perdigão, Pedro M. C. Pina, Cátia Rocha, João Manuel R. S. Tavares and Agostinho Rosa
High-Throughput 2020, 9(3), 15; https://doi.org/10.3390/ht9030015 - 30 Jun 2020
Cited by 1
Abstract
►▼
Show Figures
There is a misconception that intrinsic disorder in proteins is equivalent to darkness. The present study aims to establish, in the scope of the Swiss-Prot and Dark Proteome databases, the relationship between disorder and darkness. Three distinct predictors were used to calculate the
[...] Read more.
There is a misconception that intrinsic disorder in proteins is equivalent to darkness. The present study aims to establish, in the scope of the Swiss-Prot and Dark Proteome databases, the relationship between disorder and darkness. Three distinct predictors were used to calculate the disorder of Swiss-Prot proteins. The analysis of the results obtained with the used predictors and visualization paradigms resulted in the same conclusion that was reached before: disorder is mostly unrelated to darkness.
Full article
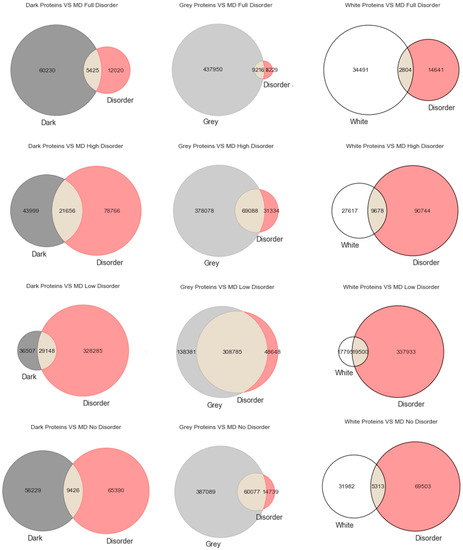
Figure 1
Open AccessArticle
Intra-Laboratory Evaluation of Luminescence Based High-Throughput Serum Bactericidal Assay (L-SBA) to Determine Bactericidal Activity of Human Sera against Shigella
by
Omar Rossi, Eleonora Molesti, Allan Saul, Carlo Giannelli, Francesca Micoli and Francesca Necchi
High-Throughput 2020, 9(2), 14; https://doi.org/10.3390/ht9020014 - 8 Jun 2020
Cited by 10
Abstract
►▼
Show Figures
Despite the huge decrease in deaths caused by Shigella worldwide in recent decades, shigellosis still causes over 200,000 deaths every year. No vaccine is currently available, and the morbidity of the disease coupled with the rise of antimicrobial resistance renders the introduction of
[...] Read more.
Despite the huge decrease in deaths caused by Shigella worldwide in recent decades, shigellosis still causes over 200,000 deaths every year. No vaccine is currently available, and the morbidity of the disease coupled with the rise of antimicrobial resistance renders the introduction of an effective vaccine extremely urgent. Although a clear immune correlate of protection against shigellosis has not yet been established, the demonstration of the bactericidal activity of antibodies induced upon vaccination may provide one means of the functionality of antibodies induced in protecting against Shigella. The method of choice to evaluate the complement-mediated functional activity of vaccine-induced antibodies is the Serum Bactericidal Assay (SBA). Here we present the development and intra-laboratory characterization of a high-throughput luminescence-based SBA (L-SBA) method, based on the detection of ATP as a proxy of surviving bacteria, to evaluate the complement-mediated killing of human sera. We demonstrated the high specificity of the assay against a homologous strain without any heterologous aspecificity detected against species-related and non-species-related strains. We assessed the linearity, repeatability and reproducibility of L-SBA on human sera. This work will guide the bactericidal activity assessment of clinical sera raised against S. sonnei. The method has the potential of being applicable with similar performances to determine the bactericidal activity of any non-clinical and clinical sera that rely on complement-mediated killing.
Full article
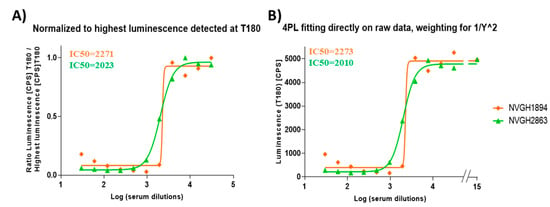
Figure 1
Open AccessCommunication
Genetic Counseling and NGS Screening for Recessive LGMD2A Families
by
Claudia Strafella, Valerio Caputo, Giulia Campoli, Rosaria Maria Galota, Julia Mela, Stefania Zampatti, Giulietta Minozzi, Cristina Sancricca, Serenella Servidei, Emiliano Giardina and Raffaella Cascella
High-Throughput 2020, 9(2), 13; https://doi.org/10.3390/ht9020013 - 10 May 2020
Cited by 2
Abstract
►▼
Show Figures
Genetic counseling applied to limb–girdle muscular dystrophies (LGMDs) can be very challenging due to their clinical and genetic heterogeneity and the availability of different molecular assays. Genetic counseling should therefore be addressed to select the most suitable approach to increase the diagnostic rate
[...] Read more.
Genetic counseling applied to limb–girdle muscular dystrophies (LGMDs) can be very challenging due to their clinical and genetic heterogeneity and the availability of different molecular assays. Genetic counseling should therefore be addressed to select the most suitable approach to increase the diagnostic rate and provide an accurate estimation of recurrence risk. This is particularly true for families with a positive history for recessive LGMD, in which the presence of a known pathogenetic mutation segregating within the family may not be enough to exclude the risk of having affected children without exploring the genetic background of phenotypically unaffected partners. In this work, we presented a family with a positive history for LGMD2A (OMIM #253600, also known as calpainopathy) characterized by compound heterozygosity for two CAPN3 mutations. The genetic specialist suggested the segregation analysis of both mutations within the family as a first-level analysis. Sequentially, next-generation sequencing (NGS) analysis was performed in the partners of healthy carriers to provide an accurate recurrence/reproductive risk estimation considering the genetic background of the couple. Finally, this work highlighted the importance of providing a genetic counseling/testing service even in unaffected individuals with a carrier partner. This approach can support genetic counselors in estimating the reproductive/recurrence risk and eventually, suggesting prenatal testing, early diagnosis or other medical surveillance strategies.
Full article
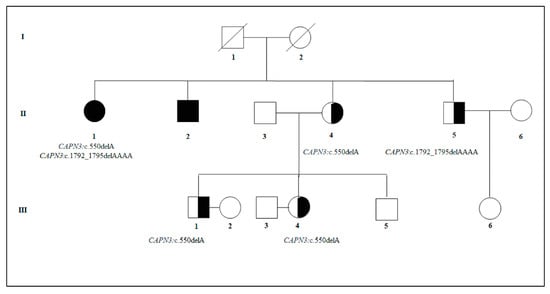
Figure 1
Open AccessReview
Microbiota and Human Reproduction: The Case of Female Infertility
by
Rossella Tomaiuolo, Iolanda Veneruso, Federica Cariati and Valeria D’Argenio
High-Throughput 2020, 9(2), 12; https://doi.org/10.3390/ht9020012 - 3 May 2020
Cited by 53
Abstract
During the last decade, the availability of next-generation sequencing-based approaches has revealed the presence of microbial communities in almost all the human body, including the reproductive tract. As for other body sites, this resident microbiota has been involved in the maintenance of a
[...] Read more.
During the last decade, the availability of next-generation sequencing-based approaches has revealed the presence of microbial communities in almost all the human body, including the reproductive tract. As for other body sites, this resident microbiota has been involved in the maintenance of a healthy status. As a consequence, alterations due to internal or external factors may lead to microbial dysbiosis and to the development of pathologies. Female reproductive microbiota has also been suggested to affect infertility, and it may play a key role in the success of assisted reproductive technologies, such as embryo implantation and pregnancy care. While the vaginal microbiota is well described, the uterine microbiota is underexplored. This could be due to technical issues, as the uterus is a low biomass environment. Here, we review the state of the art regarding the role of the female reproductive system microbiota in women’s health and human reproduction, highlighting its contribution to infertility.
Full article
(This article belongs to the Special Issue Human Microbiome and Diseases: Implications for Novel Therapies)
►▼
Show Figures

Graphical abstract
Open AccessFeature PaperArticle
Chimeric Virus Made from crTMV RNA and the Coat Protein of Potato Leafroll Virus is Targeted to the Nucleolus and Can Infect Nicotiana benthamiana Mechanically
by
Konstantin O. Butenko, Inna A. Chaban, Eugene V. Skurat, Olga A. Kondakova and Yuri F. Drygin
High-Throughput 2020, 9(2), 11; https://doi.org/10.3390/ht9020011 - 26 Apr 2020
Abstract
►▼
Show Figures
A genetically engineered chimeric virus crTMV-CP-PLRV composed of the crucifer-infecting tobacco mosaic virus (crTMV) RNA and the potato leafroll virus (PLRV) coat protein (CP) was obtained by agroinfiltration of Nicotiana benthamiana with the binary vector pCambia-crTMV-CPPLRV. The significant levels of the chimeric
[...] Read more.
A genetically engineered chimeric virus crTMV-CP-PLRV composed of the crucifer-infecting tobacco mosaic virus (crTMV) RNA and the potato leafroll virus (PLRV) coat protein (CP) was obtained by agroinfiltration of Nicotiana benthamiana with the binary vector pCambia-crTMV-CPPLRV. The significant levels of the chimeric virus enabled direct visualization of crTMV-CP-PLRV in the cell and to investigate the mechanism of the pathogenesis. Localization of the crTMV-CP-PLRV in plant cells was examined by immunoblot techniques, as well as light, and transmission electron microscopy. The chimera can transfer between vascular and nonvascular tissues. The chimeric virus inoculum is capable to infect N. benthamiana mechanically. The distinguishing feature of the chimeric virus, the RNA virus with the positive genome, was found to localize in the nucleolus. We also investigated the role of the N-terminal sequence of the PLRV P3 coat protein in the cellular localization of the virus. We believe that the gene of the PLRV CP can be substituted with genes from other challenging-to-study plant pathogens to produce other useful recombinant viruses.
Full article

Figure 1
Open AccessFeature PaperReview
Microbiota and Human Reproduction: The Case of Male Infertility
by
Rossella Tomaiuolo, Iolanda Veneruso, Federica Cariati and Valeria D’Argenio
High-Throughput 2020, 9(2), 10; https://doi.org/10.3390/ht9020010 - 13 Apr 2020
Cited by 38
Abstract
The increasing interest in metagenomics is enhancing our knowledge regarding the composition and role of the microbiota in human physiology and pathology. Indeed, microbes have been reported to play a role in several diseases, including infertility. In particular, the male seminal microbiota has
[...] Read more.
The increasing interest in metagenomics is enhancing our knowledge regarding the composition and role of the microbiota in human physiology and pathology. Indeed, microbes have been reported to play a role in several diseases, including infertility. In particular, the male seminal microbiota has been suggested as an important factor able to influence couple’s health and pregnancy outcomes, as well as offspring health. Nevertheless, few studies have been carried out to date to deeper investigate semen microbiome origins and functions, and its correlations with the partner’s reproductive tract microbiome. Here, we report the state of the art regarding the male reproductive system microbiome and its alterations in infertility.
Full article
(This article belongs to the Special Issue Human Microbiome and Diseases: Implications for Novel Therapies)
►▼
Show Figures
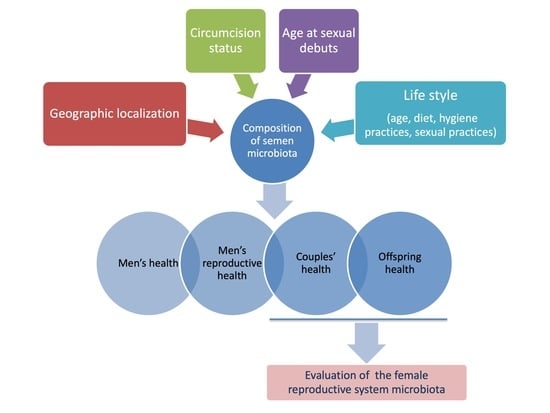
Graphical abstract
Open AccessArticle
A Simple, Label-Free, and High-Throughput Method to Evaluate the Epigallocatechin-3-Gallate Impact in Plasma Molecular Profile
by
Rúben Araújo, Luís Ramalhete, Helder Da Paz, Edna Ribeiro and Cecília R.C. Calado
High-Throughput 2020, 9(2), 9; https://doi.org/10.3390/ht9020009 - 9 Apr 2020
Cited by 7
Abstract
Epigallocatechin-3-gallate (EGCG), the major catechin present in green tea, presents diverse appealing biological activities, such as antioxidative, anti-inflammatory, antimicrobial, and antiviral activities, among others. The present work evaluated the impact in the molecular profile of human plasma from daily consumption of 225 mg
[...] Read more.
Epigallocatechin-3-gallate (EGCG), the major catechin present in green tea, presents diverse appealing biological activities, such as antioxidative, anti-inflammatory, antimicrobial, and antiviral activities, among others. The present work evaluated the impact in the molecular profile of human plasma from daily consumption of 225 mg of EGCG for 90 days. Plasma from peripheral blood was collected from 30 healthy human volunteers and analyzed by high-throughput Fourier transform infrared spectroscopy. To capture the biochemical information while minimizing the interference of physical phenomena, several combinations of spectra pre-processing methods were evaluated by principal component analysis. The pre-processing method that led to the best class separation, that is, between the plasma spectral data collected at the beginning and after the 90 days, was a combination of atmospheric correction with a second derivative spectra. A hierarchical cluster analysis of second derivative spectra also highlighted the fact that plasma acquired before EGCG consumption presented a distinct molecular profile after the 90 days of EGCG consumption. It was also possible by partial least squares regression discriminant analysis to correctly predict all unlabeled plasma samples (not used for model construction) at both timeframes. We observed that the similarity in composition among the plasma samples was higher in samples collected after EGCG consumption when compared with the samples taken prior to EGCG consumption. Diverse negative peaks of the normalized second derivative spectra, associated with lipid and protein regions, were significantly affected (p < 0.001) by EGCG consumption, according to the impact of EGCG consumption on the patients’ blood, low density and high density lipoproteins ratio. In conclusion, a single bolus dose of 225 mg of EGCG, ingested throughout a period of 90 days, drastically affected plasma molecular composition in all participants, which raises awareness regarding prolonged human exposure to EGCG. Because the analysis was conducted in a high-throughput, label-free, and economic analysis, it could be applied to high-dimension molecular epidemiological studies to further promote the understanding of the effect of bio-compound consumption mode and frequency.
Full article
(This article belongs to the Special Issue Shining the Light on High-Throughput—the Forgotten Role of Vibrational Spectroscopy)
►▼
Show Figures
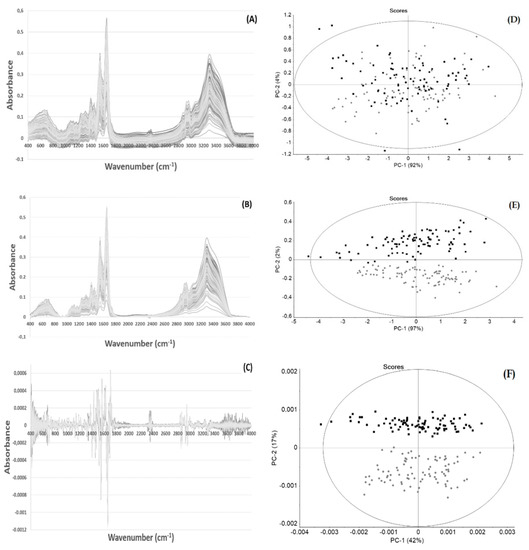
Figure 1
Open AccessFeature PaperReview
DMETTM Genotyping: Tools for Biomarkers Discovery in the Era of Precision Medicine
by
Giuseppe Agapito, Marzia Settino, Francesca Scionti, Emanuela Altomare, Pietro Hiram Guzzi, Pierfrancesco Tassone, Pierosandro Tagliaferri, Mario Cannataro, Mariamena Arbitrio and Maria Teresa Di Martino
High-Throughput 2020, 9(2), 8; https://doi.org/10.3390/ht9020008 - 29 Mar 2020
Cited by 8
Abstract
The knowledge of genetic variants in genes involved in drug metabolism may be translated into reduction of adverse drug reactions, increase of efficacy, healthcare outcomes improvement and economic benefits. Many high-throughput tools are available for the genotyping of Single Nucleotide Polymorphisms (SNPs) known
[...] Read more.
The knowledge of genetic variants in genes involved in drug metabolism may be translated into reduction of adverse drug reactions, increase of efficacy, healthcare outcomes improvement and economic benefits. Many high-throughput tools are available for the genotyping of Single Nucleotide Polymorphisms (SNPs) known to be related to drugs and xenobiotics metabolism. DMETTM platform represents an example of SNPs panel to discover biomarkers correlated to efficacy or toxicity in common and rare diseases. The difficulty in analyzing the mole of information generated by DMETTM platform led to the development and implementation of algorithms and tools for statistical and data mining analysis. These softwares allow efficient handling of the omics data to validate the explorative SNPs identified by DMET assay and to correlate them with drug efficacy, toxicity and/or cancer susceptibility. In this review we present a suite of bioinformatic frameworks for the preprocessing and analysis of DMET-SNPs data. In particular, we introduce a workflow that uses the GenoMetric Query Language, a high-level query language specifically designed for genomics, able to query public datasets (such as ENCODE, TCGA, GENCODE annotation dataset, etc.) as well as to combine them with private datasets (e.g., output from Affymetrix® DMETTM Platform).
Full article
(This article belongs to the Special Issue Selected Papers from 1st International and 32nd Annual Conference of Italian Association of Cell Culture)
►▼
Show Figures
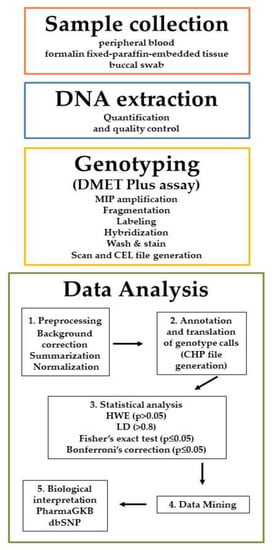
Figure 1
Open AccessFeature PaperReview
Colonization Resistance in the Infant Gut: The Role of B. infantis in Reducing pH and Preventing Pathogen Growth
by
Rebbeca M. Duar, David Kyle and Giorgio Casaburi
High-Throughput 2020, 9(2), 7; https://doi.org/10.3390/ht9020007 - 27 Mar 2020
Cited by 43
Abstract
Over the past century, there has been a steady increase in the stool pH of infants from industrialized countries. Analysis of historical data revealed a strong association between abundance of Bifidobacterium in the gut microbiome of breasted infants and stool pH, suggesting that
[...] Read more.
Over the past century, there has been a steady increase in the stool pH of infants from industrialized countries. Analysis of historical data revealed a strong association between abundance of Bifidobacterium in the gut microbiome of breasted infants and stool pH, suggesting that this taxon plays a key role in determining the pH in the gut. Bifidobacterium longum subsp. infantis is uniquely equipped to metabolize human milk oligosaccharides (HMO) from breastmilk into acidic end products, mainly lactate and acetate. The presence of these acidic compounds in the infant gut is linked to a lower stool pH. Conversely, infants lacking B. infantis have a significantly higher stool pH, carry a higher abundance of potential pathogens and mucus-eroding bacteria in their gut microbiomes, and have signs of chronic enteric inflammation. This suggests the presence of B. infantis and low intestinal pH may be critical to maintaining a protective environment in the infant gut. Here, we summarize recent studies demonstrating that feeding B. infantis EVC001 to breastfed infants results in significantly lower fecal pH compared to controls and propose that low pH is one critical factor in preventing the invasion and overgrowth of harmful bacteria in the infant gut, a process known as colonization resistance.
Full article
(This article belongs to the Special Issue Human Microbiome and Diseases: Implications for Novel Therapies)
►▼
Show Figures
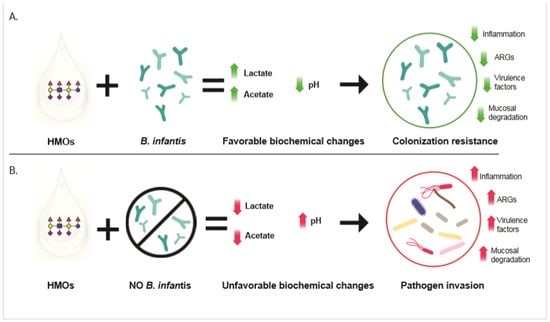
Figure 1
Open AccessReview
Factors Influencing Epigenetic Mechanisms: Is There A Role for Bariatric Surgery?
by
Alessio Metere and Claire E. Graves
High-Throughput 2020, 9(1), 6; https://doi.org/10.3390/ht9010006 - 20 Mar 2020
Cited by 11
Abstract
Epigenetics is the interaction between the genome and environmental stimuli capable of influencing gene expression during development and aging. A large number of studies have shown that metabolic diseases are highly associated with epigenetic alterations, suggesting that epigenetic factors may play a central
[...] Read more.
Epigenetics is the interaction between the genome and environmental stimuli capable of influencing gene expression during development and aging. A large number of studies have shown that metabolic diseases are highly associated with epigenetic alterations, suggesting that epigenetic factors may play a central role in obesity. To investigate these relationships, we focus our attention on the most common epigenetic modifications that occur in obesity, including DNA methylation and post-translational modifications of histones. We also consider bariatric surgery as an epigenetic factor, evaluating how the anatomic and physiologic modifications induced by these surgical techniques can change gene expression. Here we discuss the importance of epigenetic mechanisms in chronic disease and cancer, and the role of epigenetic disturbances in obesity, with a focus on the role of bariatric surgery.
Full article
Open AccessFeature PaperArticle
Rational Engineering of the Substrate Specificity of a Thermostable D-Hydantoinase (Dihydropyrimidinase)
by
Hovsep Aganyants, Pierre Weigel, Yeranuhi Hovhannisyan, Michèle Lecocq, Haykanush Koloyan, Artur Hambardzumyan, Anichka Hovsepyan, Jean-Noël Hallet and Vehary Sakanyan
High-Throughput 2020, 9(1), 5; https://doi.org/10.3390/ht9010005 - 12 Feb 2020
Cited by 5
Abstract
►▼
Show Figures
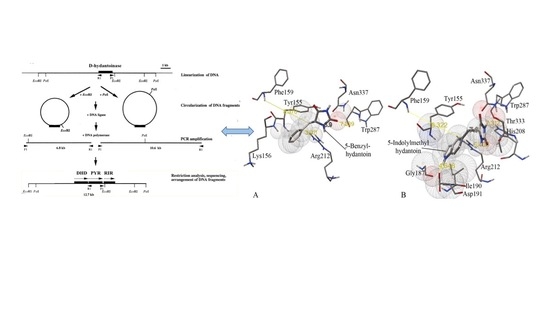
D-hydantoinases catalyze an enantioselective opening of 5- and 6-membered cyclic structures and therefore can be used for the production of optically pure precursors for biomedical applications. The thermostable D-hydantoinase from Geobacillus stearothermophilus ATCC 31783 is a manganese-dependent enzyme and exhibits low activity towards
[...] Read more.
D-hydantoinases catalyze an enantioselective opening of 5- and 6-membered cyclic structures and therefore can be used for the production of optically pure precursors for biomedical applications. The thermostable D-hydantoinase from Geobacillus stearothermophilus ATCC 31783 is a manganese-dependent enzyme and exhibits low activity towards bulky hydantoin derivatives. Homology modeling with a known 3D structure (PDB code: 1K1D) allowed us to identify the amino acids to be mutated at the substrate binding site and in its immediate vicinity to modulate the substrate specificity. Both single and double substituted mutants were generated by site-directed mutagenesis at appropriate sites located inside and outside of the stereochemistry gate loops (SGL) involved in the substrate binding. Substrate specificity and kinetic constant data demonstrate that the replacement of Phe159 and Trp287 with alanine leads to an increase in the enzyme activity towards D,L-5-benzyl and D,L-5-indolylmethyl hydantoins. The length of the side chain and the hydrophobicity of substrates are essential parameters to consider when designing the substrate binding pocket for bulky hydantoins. Our data highlight that D-hydantoinase is the authentic dihydropyrimidinase involved in the pyrimidine reductive catabolic pathway in moderate thermophiles.
Full article
Graphical abstract
Open AccessFeature PaperArticle
Comparative Study of NGS Platform Ion Torrent Personal Genome Machine and Therascreen Rotor-Gene Q for the Detection of Somatic Variants in Cancer
by
Angela Lombardi, Margherita Russo, Amalia Luce, Floriana Morgillo, Virginia Tirino, Gabriella Misso, Erika Martinelli, Teresa Troiani, Vincenzo Desiderio, Gianpaolo Papaccio, Francesco Iovino, Giuseppe Argenziano, Elvira Moscarella, Pasquale Sperlongano, Gennaro Galizia, Raffaele Addeo, Alois Necas, Andrea Necasova, Fortunato Ciardiello, Andrea Ronchi, Michele Caraglia and Anna Grimaldiadd
Show full author list
remove
Hide full author list
High-Throughput 2020, 9(1), 4; https://doi.org/10.3390/ht9010004 - 11 Feb 2020
Cited by 2
Abstract
Molecular profiling of a tumor allows the opportunity to design specific therapies which are able to interact only with cancer cells characterized by the accumulation of several genomic aberrations. This study investigates the usefulness of next-generation sequencing (NGS) and mutation-specific analysis methods for
[...] Read more.
Molecular profiling of a tumor allows the opportunity to design specific therapies which are able to interact only with cancer cells characterized by the accumulation of several genomic aberrations. This study investigates the usefulness of next-generation sequencing (NGS) and mutation-specific analysis methods for the detection of target genes for current therapies in non-small-cell lung cancer (NSCLC), metastatic colorectal cancer (mCRC), and melanoma patients. We focused our attention on EGFR, BRAF, KRAS, and BRAF genes for NSCLC, melanoma, and mCRC samples, respectively. Our study demonstrated that in about 2% of analyzed cases, the two techniques did not show the same or overlapping results. Two patients affected by mCRC resulted in wild-type (WT) for BRAF and two cases with NSCLC were WT for EGFR according to PGM analysis. In contrast, these samples were mutated for the evaluated genes using the therascreen test on Rotor-Gene Q. In conclusion, our experience suggests that it would be appropriate to confirm the WT status of the genes of interest with a more sensitive analysis method to avoid the presence of a small neoplastic clone and drive the clinician to correct patient monitoring.
Full article
(This article belongs to the Special Issue Selected Papers from 1st International and 32nd Annual Conference of Italian Association of Cell Culture)
►▼
Show Figures
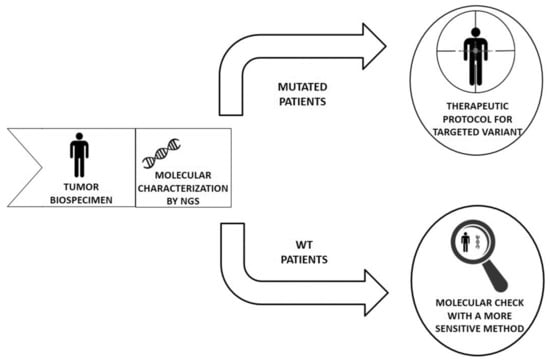
Figure 1
Open AccessFeature PaperCommentary
Precision Medicine in Non-Communicable Diseases
by
Giuseppe Novelli, Michela Biancolella, Andrea Latini, Aldo Spallone, Paola Borgiani and Marisa Papaluca
High-Throughput 2020, 9(1), 3; https://doi.org/10.3390/ht9010003 - 7 Feb 2020
Cited by 7
Abstract
The increase in life expectancy during the 20th century ranks as one of society’s greatest achievements, with massive growth in the numbers and proportion of the elderly, virtually occurring in every country of the world. The burden of chronic diseases is one of
[...] Read more.
The increase in life expectancy during the 20th century ranks as one of society’s greatest achievements, with massive growth in the numbers and proportion of the elderly, virtually occurring in every country of the world. The burden of chronic diseases is one of the main consequences of this phenomenon, severely hampering the quality of life of elderly people and challenging the efficiency and sustainability of healthcare systems. Non-communicable diseases (NCDs) are considered a global emergency responsible for over 70% of deaths worldwide. NCDs are also the basis for complex and multifactorial diseases such as hypertension, diabetes, and obesity. The epidemics of NCDs are a consequence of a complex interaction between health, economic growth, and development. This interaction includes the individual genome, the microbiome, the metabolome, the immune status, and environmental factors such as nutritional and chemical exposure. To counteract NCDs, it is therefore essential to develop an innovative, personalized, preventative, early care model through the integration of different molecular profiles of individuals to identify both the critical biomarkers of NCD susceptibility and to discover novel therapeutic targets.
Full article
(This article belongs to the Special Issue Selected Papers from 1st International and 32nd Annual Conference of Italian Association of Cell Culture)
►▼
Show Figures
Figure 1
Open AccessEditorial
Acknowledgement to Reviewers of High-Throughput in 2019
by
High-Throughput Editorial Office
High-Throughput 2020, 9(1), 2; https://doi.org/10.3390/ht9010002 - 16 Jan 2020
Abstract
The editorial team greatly appreciates the reviewers who have dedicated their considerable time and expertise to the journal’s rigorous editorial process over the past 12 months, regardless of whether the papers are finally published or not [...]
Full article
Open AccessFeature PaperReview
Applications of Next Generation Sequencing to the Analysis of Familial Breast/Ovarian Cancer
by
Veronica Zelli, Chiara Compagnoni, Katia Cannita, Roberta Capelli, Carlo Capalbo, Mauro Di Vito Nolfi, Edoardo Alesse, Francesca Zazzeroni and Alessandra Tessitore
High-Throughput 2020, 9(1), 1; https://doi.org/10.3390/ht9010001 - 10 Jan 2020
Cited by 24
Abstract
Next generation sequencing (NGS) provides a powerful tool in the field of medical genetics, allowing one to perform multi-gene analysis and to sequence entire exomes (WES), transcriptomes or genomes (WGS). The generated high-throughput data are particularly suitable for enhancing the understanding of the
[...] Read more.
Next generation sequencing (NGS) provides a powerful tool in the field of medical genetics, allowing one to perform multi-gene analysis and to sequence entire exomes (WES), transcriptomes or genomes (WGS). The generated high-throughput data are particularly suitable for enhancing the understanding of the genetic bases of complex, multi-gene diseases, such as cancer. Among the various types of tumors, those with a familial predisposition are of great interest for the isolation of novel genes or gene variants, detectable at the germline level and involved in cancer pathogenesis. The identification of novel genetic factors would have great translational value, helping clinicians in defining risk and prevention strategies. In this regard, it is known that the majority of breast/ovarian cases with familial predisposition, lacking variants in the highly penetrant BRCA1 and BRCA2 genes (non-BRCA), remains unexplained, although several less penetrant genes (e.g., ATM, PALB2) have been identified. In this scenario, NGS technologies offer a powerful tool for the discovery of novel factors involved in familial breast/ovarian cancer. In this review, we summarize and discuss the state of the art applications of NGS gene panels, WES and WGS in the context of familial breast/ovarian cancer.
Full article
(This article belongs to the Special Issue Selected Papers from 1st International and 32nd Annual Conference of Italian Association of Cell Culture)
Open AccessFeature PaperArticle
Descriptors for High Throughput in Structural Materials Development
by
Matthias Steinbacher, Gabriela Alexe, Michael Baune, Ilya Bobrov, Ingmar Bösing, Brigitte Clausen, Tobias Czotscher, Jérémy Epp, Andreas Fischer, Lasse Langstädtler, Daniel Meyer, Sachin Raj Menon, Oltmann Riemer, Heike Sonnenberg, Arne Thomann, Anastasiya Toenjes, Frank Vollertsen, Nicole Wielki and Nils Ellendt
High-Throughput 2019, 8(4), 22; https://doi.org/10.3390/ht8040022 - 5 Dec 2019
Cited by 17
Abstract
►▼
Show Figures
The development of novel structural materials with increasing mechanical requirements is a very resource-intense process if conventional methods are used. While there are high-throughput methods for the development of functional materials, this is not the case for structural materials. Their mechanical properties are
[...] Read more.
The development of novel structural materials with increasing mechanical requirements is a very resource-intense process if conventional methods are used. While there are high-throughput methods for the development of functional materials, this is not the case for structural materials. Their mechanical properties are determined by their microstructure, so that increased sample volumes are needed. Furthermore, new short-time characterization techniques are required for individual samples which do not necessarily measure the desired material properties, but descriptors which can later be mapped on material properties. While universal micro-hardness testing is being commonly used, it is limited in its capability to measure sample volumes which contain a characteristic microstructure. We propose to use alternative and fast deformation techniques for spherical micro-samples in combination with classical characterization techniques such as XRD, DSC or micro magnetic methods, which deliver descriptors for the microstructural state.
Full article
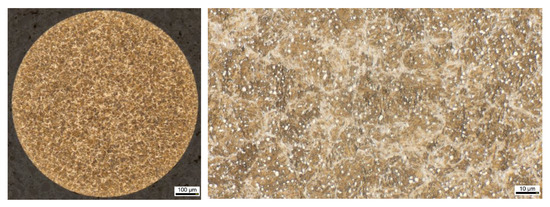
Figure 1
Open AccessFeature PaperReview
From the Laboratory to The Vineyard—Evolution of The Measurement of Grape Composition using NIR Spectroscopy towards High-Throughput Analysis
by
Aoife Power, Vi Khanh Truong, James Chapman and Daniel Cozzolino
High-Throughput 2019, 8(4), 21; https://doi.org/10.3390/ht8040021 - 30 Nov 2019
Cited by 25
Abstract
Compared to traditional laboratory methods, spectroscopic techniques (e.g., near infrared, hyperspectral imaging) provide analysts with an innovative and improved understanding of complex issues by determining several chemical compounds and metabolites at once, allowing for the collection of the sample “fingerprint”. These techniques have
[...] Read more.
Compared to traditional laboratory methods, spectroscopic techniques (e.g., near infrared, hyperspectral imaging) provide analysts with an innovative and improved understanding of complex issues by determining several chemical compounds and metabolites at once, allowing for the collection of the sample “fingerprint”. These techniques have the potential to deliver high-throughput options for the analysis of the chemical composition of grapes in the laboratory, the vineyard and before or during harvest, to provide better insights of the chemistry, nutrition and physiology of grapes. Faster computers, the development of software and portable easy to use spectrophotometers and data analytical methods allow for the development of innovative applications of these techniques for the analyses of grape composition.
Full article
(This article belongs to the Special Issue Shining the Light on High-Throughput—the Forgotten Role of Vibrational Spectroscopy)
►▼
Show Figures
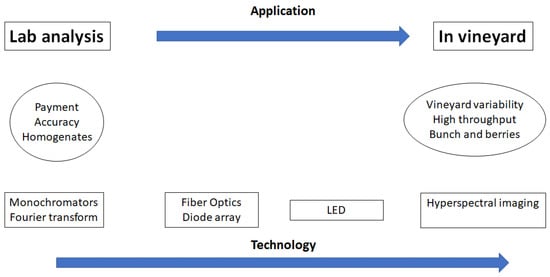
Figure 1
Open AccessArticle
Comparative Analysis of Strategies for De Novo Transcriptome Assembly in Prokaryotes: Streptomyces clavuligerus as a Case Study
by
Carlos Caicedo-Montoya, Laura Pinilla, León F. Toro, Jeferyd Yepes-García and Rigoberto Ríos-Estepa
High-Throughput 2019, 8(4), 20; https://doi.org/10.3390/ht8040020 - 30 Nov 2019
Cited by 1
Abstract
►▼
Show Figures
The performance of software tools for de novo transcriptome assembly greatly depends on the selection of software parameters. Up to now, the development of de novo transcriptome assembly for prokaryotes has not been as remarkable as that for eukaryotes. In this contribution, Rockhopper2
[...] Read more.
The performance of software tools for de novo transcriptome assembly greatly depends on the selection of software parameters. Up to now, the development of de novo transcriptome assembly for prokaryotes has not been as remarkable as that for eukaryotes. In this contribution, Rockhopper2 was used to perform a comparative transcriptome analysis of Streptomyces clavuligerus exposed to diverse environmental conditions. The study focused on assessing the incidence of software parameters on software performance for the identification of differentially expressed genes as a final goal. For this, a statistical optimization was performed using the Transrate Assembly Score (TAS). TAS was also used for evaluating the software performance and for comparing it with related tools, e.g., Trinity. Transcriptome redundancy and completeness were also considered for this analysis. Rockhopper2 and Trinity reached a TAS value of 0.55092 and 0.58337, respectively. Trinity assembles transcriptomes with high redundancy, with 55.6% of transcripts having some duplicates. Additionally, we observed that the total number of differentially expressed genes (DEG) and their annotation greatly depends on the method used for removing redundancy and the tools used for transcript quantification. To our knowledge, this is the first work aimed at assessing de novo assembly software for prokaryotic organisms.
Full article

Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics