Journal Description
LabMed
LabMed
is an international, peer-reviewed, open access journal devoted to laboratory medicine and clinical chemistry published quarterly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- Rapid Publication: first decisions in 19 days; acceptance to publication in 8 days (median values for MDPI journals in the second half of 2025).
- Recognition of Reviewers: APC discount vouchers, optional signed peer review, and reviewer names published annually in the journal.
- LabMed is a companion journal of Diagnostics.
subject
Imprint Information
Open Access
ISSN: 2813-9038
Latest Articles
Pre-Operational Validation of a Deviation-Ready QMS for Source Plasma Centers: Readiness Metrics and Hematology Supply Implications
LabMed 2026, 3(1), 2; https://doi.org/10.3390/labmed3010002 - 14 Jan 2026
Abstract
Source plasma centers sustain hematology therapeutics by safeguarding testing, traceability, and cold-chain integrity before fractionation. Despite regulatory requirements (21 CFR 606/640; EU Directive 2005/62/EC), published pre-operational validation frameworks demonstrating deviation-readiness before first collections remain sparse. We conducted a simulation-based pre-operational validation of an
[...] Read more.
Source plasma centers sustain hematology therapeutics by safeguarding testing, traceability, and cold-chain integrity before fractionation. Despite regulatory requirements (21 CFR 606/640; EU Directive 2005/62/EC), published pre-operational validation frameworks demonstrating deviation-readiness before first collections remain sparse. We conducted a simulation-based pre-operational validation of an electronic quality management system (eQMS) with an Incident → Deviation → Corrective Action and Preventive Action (CAPA) pathway at a new source plasma center, performing 20 chairside mock runs, 3 freezer-alarm drills, and a document-control stress test. Primary endpoints were anomaly rate, alarm-response time relative to a 15 min service-level agreement (SLA), and deviation-closure SLA compliance. Analyses were descriptive and designed to demonstrate system functionality, not long-term process stability. Minor anomalies occurred in 6/20 mock runs (30.0%; 95% CI 11.9–54.3); no major/critical events were observed (0/20; 95% CI 0–16.8). Deviation-closure SLAs were met in 6/6 tests (100%; 95% CI 54.1–100). Alarm-response times averaged 7.0 min (SD 1.0; range 6–8 min; 95% CI 4.5–9.5), and all drills met the 15 min vendor SLA, illustrating a preliminary readiness margin (Cpu ≈ 2.7) rather than a statistically stable capability estimate. Simulation-based pre-operational validation produced inspection-ready documentation and quantitative acceptance criteria aligned to U.S./EU expectations, supporting reproducible multi-site deployment. By protecting cold-chain integrity and traceability before first collections, the validated QMS helps preserve supply reliability for plasma-derived therapeutics central to hematology care and establishes the measurement infrastructure for post-operational performance validation.
Full article
(This article belongs to the Special Issue Laboratory Medicine in Hematology)
►
Show Figures
Open AccessArticle
Stewardship in Action: An Evaluation of Antibiotic De-Escalation Prescribing After Positive Pneumococcal Urinary Antigen Testing in a Safety Net Health System
by
Mehak Bhatia, Katherine Davanzo, Paul Kim, Jyothik Varun Inampudi, Mazhar Shapoo, Marco Scipione, Sorabh Dhar and Lea Monday
LabMed 2026, 3(1), 1; https://doi.org/10.3390/labmed3010001 - 31 Dec 2025
Abstract
►▼
Show Figures
The Infectious Diseases Society of America (IDSA) guidelines for community-acquired pneumonia (CAP) recommend pneumococcal urinary antigen testing (UAT) for a subset of inpatients admitted with pneumonia. Despite this, UAT testing is frequently performed on inpatients who do not meet the official IDSA criteria,
[...] Read more.
The Infectious Diseases Society of America (IDSA) guidelines for community-acquired pneumonia (CAP) recommend pneumococcal urinary antigen testing (UAT) for a subset of inpatients admitted with pneumonia. Despite this, UAT testing is frequently performed on inpatients who do not meet the official IDSA criteria, and current evidence regarding antibiotic de-escalation in UAT-positive cases remains inconclusive. To explore this further, we conducted a retrospective cohort study examining antibiotic de-escalation patterns among hospitalized CAP patients who underwent UAT over a 60-day period during peak respiratory illness season (November and December, 2023). Patients with positive UAT results were compared to those who had negative UAT; the primary outcome was whether a positive UAT impacted antibiotic de-escalation prescribing. A total of 268 patients were analyzed—235 UAT-negative and 33 UAT-positive. Both groups were comparable in terms of disease severity, underlying health conditions, and readmission rates. Empiric therapy targeting Pseudomonas aeruginosa (P. aeruginosa) and methicillin-resistant Staphylococcus aureus (MRSA) was used in 40% of patients (36% in the UAT-positive group and 46% of the UAG-negative group). The use of atypical coverage, MRSA coverage, or anti-pseudomonal β-lactams was frequently de-escalated in both cohorts (p < 0.05); however, the UAT-positive group had significantly shorter durations of anti-pseudomonal therapy (p = 0.03) and anti-MRSA therapy (p = 0.02). Despite this, the UAT-positive group was more commonly given fluoroquinolones, such as levofloxacin or moxifloxacin, over narrow-spectrum β-lactams for final antibiotic coverage (p = 0.021). Overall, positive UAT appeared to support earlier discontinuation of anti-MRSA and anti-pseudomonal antibiotics; however, it did not impact fluoroquinolone use. Future antimicrobial stewardship efforts may benefit from promoting greater use of narrow-spectrum β-lactams in these patients.
Full article

Figure 1
Open AccessEditorial
Can Free AI Tools Replace Statistical Software in Data Analysis?
by
Giuseppe Lippi
LabMed 2025, 2(4), 27; https://doi.org/10.3390/labmed2040027 - 18 Dec 2025
Abstract
Artificial intelligence (AI) has become increasingly integrated into scientific publishing, performing a vast array of tasks for enhancing research efficiency, analysis, and dissemination [...]
Full article
Open AccessCommentary
Current Antibiotic Susceptibility Test Underestimates Minority Resistance: Implications for High-Risk Infections
by
Ivan Brukner and Matthew Oughton
LabMed 2025, 2(4), 26; https://doi.org/10.3390/labmed2040026 - 16 Dec 2025
Abstract
Antibiotic susceptibility testing (AST) reports classify isolates as “susceptible” despite potential undetected resistant subpopulations—a phenomenon termed susceptibility heterogeneity (SH). Found in 15–97% of clinical isolates of Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Klebsiella pneumoniae, SH arises from heteroresistance
[...] Read more.
Antibiotic susceptibility testing (AST) reports classify isolates as “susceptible” despite potential undetected resistant subpopulations—a phenomenon termed susceptibility heterogeneity (SH). Found in 15–97% of clinical isolates of Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Klebsiella pneumoniae, SH arises from heteroresistance or polyclonal diversity and may evade standard low-inoculum protocols. Clinically, this can lead to treatment failure, particularly in high-risk cases including immunocompromised patients, bloodstream infections, transplant recipients, and situations where minor resistant subpopulations significantly affect outcome. We argue that ethical principles of non-maleficence, transparency, and equity now compel laboratories to acknowledge this limitation. A simple annotation—“Limited susceptibility possible; resistant subpopulations may not be detected”—should accompany “susceptible” results in immunocompromised patients. High-risk cases warrant enhanced testing. This commentary calls for zone inspection, staff training, and Clinical and Laboratory Standards Institute (CLSI)/European Committee on Antimicrobial Susceptibility Testing (EUCAST) guideline updates to reflect SH. Transparency enhances clinical decision-making without implying diagnostic fault.
Full article
Open AccessCorrection
Correction: Barra et al. Impact of Tube Additives on Baseline Cell-Free DNA, Blood Nuclease Activity, and Cell-Free DNA Degradation in Serum and Plasma Samples: A Comparative Study. LabMed 2025, 2, 4
by
Gustavo Barcelos Barra, Ticiane Henriques Santa Rita, Rafael Henriques Jácomo and Lídia Freire Abdalla Nery
LabMed 2025, 2(4), 25; https://doi.org/10.3390/labmed2040025 - 1 Dec 2025
Abstract
In the original publication [...]
Full article
(This article belongs to the Collection Feature Papers in Laboratory Medicine)
Open AccessCase Report
Elevated Alpha-Fetoprotein in Hypothyroidism
by
Viola Ceconi, Valentina Kiren, Flora Maria Murru, Andrea Bon, Danica Dragovic, Lorenzo Zandonà, Alice Fachin, Gianluca Tamaro and Gianluca Tornese
LabMed 2025, 2(4), 24; https://doi.org/10.3390/labmed2040024 - 25 Nov 2025
Abstract
►▼
Show Figures
Alpha-fetoprotein (AFP) is a biomarker commonly used in the diagnosis of various malignancies but may also be elevated in non-neoplastic conditions, including hypothyroidism. We report the case of a 3-year-old girl with Down syndrome (DS) and newly diagnosed hypothyroidism, who presented with a
[...] Read more.
Alpha-fetoprotein (AFP) is a biomarker commonly used in the diagnosis of various malignancies but may also be elevated in non-neoplastic conditions, including hypothyroidism. We report the case of a 3-year-old girl with Down syndrome (DS) and newly diagnosed hypothyroidism, who presented with a hypoechoic oval lesion adjacent to the thymic parenchyma on ultrasound and markedly elevated AFP levels (169.2 ng/mL). Further investigations, including MRI, excluded the presence of germ cell tumors. Following initiation of levothyroxine therapy, AFP levels normalized in parallel with thyroid function. No evidence of malignancy was detected despite the initial suspicion. This case underscores the association between elevated AFP and hypothyroidism, highlighting the importance of evaluating thyroid status in patients with increased AFP to avoid unnecessary oncological investigations. In particular, elevated AFP in the context of hypothyroidism and DS warrants careful thyroid assessment and follow-up to prevent redundant diagnostic procedures and reduce patient and family anxiety. Thyroid function testing should be considered before extensive oncological evaluation in children with elevated AFP.
Full article

Graphical abstract
Open AccessSystematic Review
Systematic Review and Meta-Analysis of Early Detection of Myocardial Injury: Advances in Biomarker-Based Risk Stratification and Diagnostic Precision
by
Diana Gabriela Ilaș, Sebastian Ciurescu, Raluca Ibănescu, Diana-Alexandra Mîțu and Daniel Florin Lighezan
LabMed 2025, 2(4), 23; https://doi.org/10.3390/labmed2040023 - 10 Nov 2025
Abstract
►▼
Show Figures
Chronic heart failure (CHF) carries high morbidity and mortality. Circulating biomarkers of myocardial stretch, injury, and remodelling aids diagnosis and prognosis, but utility varies, especially in HFpEF, where natriuretic peptide (NP) values may be lower or normal in obesity. We systematically searched PubMed,
[...] Read more.
Chronic heart failure (CHF) carries high morbidity and mortality. Circulating biomarkers of myocardial stretch, injury, and remodelling aids diagnosis and prognosis, but utility varies, especially in HFpEF, where natriuretic peptide (NP) values may be lower or normal in obesity. We systematically searched PubMed, Scopus, and Web of Science (2010–2025) for primary adult chronic-HF studies evaluating blood-based biomarkers: NPs, high-sensitivity troponins (hs-cTn), galectin-3, soluble ST2 (sST2), and microRNAs. Secondary sources (reviews/meta-analyses/guidelines) informed context only. Acute-HF studies were not pooled with chronic-HF analyses. Where appropriate, log hazard ratios were meta-analysed with random effects models. Twenty-nine studies met criteria. NT-proBNP remained the diagnostic/prognostic reference; across five prognostic cohorts, the pooled HR was 1.68 (95% CI 1.54–1.82; I2 ≈ 55%). hs-cTn consistently improved risk stratification. Galectin-3 and sST2 were associated with adverse outcomes but typically provided modest incremental value beyond NPs/hs-cTn; galectin-3 is influenced by renal function, and sST2 is commonly interpreted around ~28–35 ng/mL. MicroRNAs (e.g., miR-21, miR-210-3p, miR-22-3p) showed promising yet heterogeneous signals across platforms and preanalytical workflows; therefore, findings were synthesised narratively without pooling. NT-proBNP and hs-cTn form the evidence-based backbone for biomarker-guided assessment in chronic HF. Galectin-3 and sST2 add adjunct prognostic information, while microRNAs remain investigational, pending standardised methods and external validation. Overall, evidence supports a multimarker, phenotype-tailored approach, with core NPs + hs-cTn and selective adjunct use of sST2/galectin-3 in context (HFrEF vs. HFpEF, obesity, renal function) to refine risk stratification and guide clinical decision-making.
Full article

Figure 1
Open AccessArticle
Usefulness of Dried Blood Spot Samples for Syphilis Screening
by
Victoria González Soler, Gema Fernández-Rivas, Héctor Martínez Riveros, Pablo Pillado Alonso, Yesika Díaz Rodríguez, Marcos Montoro Fernández, Miquel Saña Miralles, Pere Joan Cardona Iglesias, Jordi Casabona Barbarà and C. Agusti
LabMed 2025, 2(4), 22; https://doi.org/10.3390/labmed2040022 - 4 Nov 2025
Abstract
►▼
Show Figures
Dried blood spots (DBSs) are a practical tool for diagnosing infectious diseases, especially in remote or resource-limited settings. This study assessed the efficacy of DBS-based serological assays for syphilis screening. EDTA blood samples from 171 syphilis-seropositive and 122 seronegative individuals were used to
[...] Read more.
Dried blood spots (DBSs) are a practical tool for diagnosing infectious diseases, especially in remote or resource-limited settings. This study assessed the efficacy of DBS-based serological assays for syphilis screening. EDTA blood samples from 171 syphilis-seropositive and 122 seronegative individuals were used to prepare DBSs by spotting whole blood onto filter paper. After drying, 12 mm disks were punched, incubated overnight in buffered solution, and centrifuged. Syphilis serological screening was conducted using the Liaison® Treponema Screen assay, Macro-Vue™ Reagin Plasma Rapid (RPR) card test, and Dual Path Platform (DPP) Syphilis Screen and Confirm test. The Liaison® assay demonstrated 100% sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) with an optimized cut-off. The nontreponemal RPR test showed very low sensitivity (2.9%) on DBS but perfect specificity (100%). The DPP test for treponemal antibodies achieved high sensitivity (92.1%) and specificity (98.2%) with microreader adjustment. Visual reading of the DPP test had variable accuracy, with sensitivity reaching 100% but lower specificity (42.1%). Nontreponemal antibody detection by DPP showed moderate sensitivity and specificity. Although nontreponemal testing requires refinement, DBS testing combined with point-of-care tests like DPP holds promise for expanding syphilis screening accessibility and decentralization globally, particularly in resource-constrained environments.
Full article

Figure 1
Open AccessArticle
Laboratory Science Students’ Reflections on Clinical Educators and Clinical Training Experiences
by
Shelley Robin Latchem, Benedict K. Jikong, Heather L. Phillips and Eleanor K. Jator
LabMed 2025, 2(4), 21; https://doi.org/10.3390/labmed2040021 - 30 Oct 2025
Abstract
A 39-question survey targeting recent graduates was deployed by the American Society for Clinical Pathology (ASCP) to its membership nationwide by email. Participants were prompted to reflect on clinical educators from whom they had learned the most and least. This survey was open
[...] Read more.
A 39-question survey targeting recent graduates was deployed by the American Society for Clinical Pathology (ASCP) to its membership nationwide by email. Participants were prompted to reflect on clinical educators from whom they had learned the most and least. This survey was open for approximately six weeks with 177 respondents. Participants included medical laboratory scientists (71.8%), medical laboratory technicians (21.8%), phlebotomists (4.5%), blood bank specialists (0.9%) and laboratory administration (0.9%). This paper focuses on three survey questions. The first question asked participants to reflect on clinical educators from whom they had learned the most and explain why. Themes included teaching ability (37.2%), engagement (25.6%), passion (18.6%) and knowledge (16.3%). The second question asked participants to reflect on educators they had learned the least from and explain why. Themes included teaching challenges (48.8%), disengagement (29.3%) and unprofessionalism (19.5%). The third question asked about barriers to clinical training. Main themes included staffing shortages (25.8%), COVID-19-related issues (12.9%) and work culture (12.9%). Little research has been published on the student perspective of clinical training in laboratory sciences. This research provides insight into what students consider helpful in their training and what hinders their learning.
Full article
Open AccessArticle
Improving Turnaround Time in Pediatric Clinical Microbiology Results: Implementation of the Kaizen Method in a Chilean Hospital Laboratory
by
Dona Benadof, Agustin Zamorano, Judith Aguirre, Abigail Veas, Esteban Araneda and Gustavo Saint-Pierre
LabMed 2025, 2(4), 20; https://doi.org/10.3390/labmed2040020 - 25 Oct 2025
Abstract
►▼
Show Figures
Timely reporting of microbiological results is critical for clinical decision-making, particularly in pediatric hospitals where delays can significantly impact outcomes. Despite advances in laboratory automation, workflow inefficiencies and resistance to change remain barriers to improvement in Latin America. This study aimed to evaluate
[...] Read more.
Timely reporting of microbiological results is critical for clinical decision-making, particularly in pediatric hospitals where delays can significantly impact outcomes. Despite advances in laboratory automation, workflow inefficiencies and resistance to change remain barriers to improvement in Latin America. This study aimed to evaluate the effect of implementing a Kaizen-based change management strategy on reducing turnaround time (TAT) in the microbiology laboratory of Hospital Roberto del Río, Santiago, Chile. We conducted a prospective, pre–post intervention study focusing on blood culture processing. The baseline period (July 2022) included 961 cultures processed with the BacT/ALERT® 3D system. A Kaizen/LEAN intervention was designed, comprising workflow redesign, staff training, and installation of the BACT/ALERT® Virtuo® (bioMerieux, Marcy l’Etoile, France) continuous-loading blood culture system. The intervention engaged all technical and professional staff in a five-day Kaizen immersion, followed by eight months of monitoring. Outcomes were assessed by comparing TAT for positive blood cultures before and after implementation (June 2023, 496 samples). Statistical analysis was performed using the Mann–Whitney U test, with p < 0.05 considered significant. The intervention achieved a median reduction in TAT from 68.22 h (IQR 56.14–88.59) pre-intervention to 51.52 h (IQR 41.17–66.57) post-intervention, corresponding to a 24.48% improvement (p < 0.001), surpassing the 20% target. Time to preliminary Gram reporting also decreased, and workflow standardization enhanced staff productivity and culture validation frequency. Implementation of Kaizen principles in a pediatric microbiology laboratory significantly reduced blood culture TAT and improved workflow efficiency. Beyond technological upgrades, active staff engagement and structured change management were key to success. These findings support the applicability of Kaizen-based interventions to optimize laboratory performance in resource-constrained public healthcare systems.
Full article

Figure 1
Open AccessArticle
Adapting the Illumina COVIDSeq for Whole Genome Sequencing of Other Respiratory Viruses in Multiple Workflows and a Single Rapid Workflow
by
Nqobile Mthembu, Sureshnee Pillay, Hastings Twalie Musopole, Shirelle Janine Naidoo, Nokukhanya Msomi, Bertha Cinthia Baye, Derek Tshiabuila, Nokulunga Zamagambu Memela, Thembelihle Tombo, Tulio de Oliveira and Jennifer Giandhari
LabMed 2025, 2(4), 19; https://doi.org/10.3390/labmed2040019 - 4 Oct 2025
Abstract
Acute respiratory infections (ARIs) continue to pose a major global health threat, particularly among vulnerable populations. These infections often present with similar clinical symptoms, complicating accurate diagnosis and facilitating unmonitored transmissions. Genomic surveillance has emerged as an invaluable tool for pathogen identification and
[...] Read more.
Acute respiratory infections (ARIs) continue to pose a major global health threat, particularly among vulnerable populations. These infections often present with similar clinical symptoms, complicating accurate diagnosis and facilitating unmonitored transmissions. Genomic surveillance has emerged as an invaluable tool for pathogen identification and monitoring of such infectious pathogens; however, its implementation is frequently limited by high costs. The widespread use of high-throughput sequencing during the COVID-19 pandemic has created an opportunity to repurpose existing genomic platforms for broader respiratory virus surveillance. In this study, we evaluated the feasibility of adapting the Illumina COVIDSeq assay—initially designed for SARS-CoV-2 whole-genome sequencing—for use with Influenza A/B, Respiratory Syncytial Virus (RSV), and Rhinovirus. Positive control samples were processed using two approaches for library preparation: four virus-specific multiple workflows and a combined rapid workflow. Both workflows incorporated pathogen-specific primers for amplification and followed the Illumina COVIDSeq protocol for library preparation and sequencing. Sequencing quality metrics were analysed, including Phred scores, read length distribution, and coverage depth. The study did not identify significant differences in genome coverage and genetic diversity metrics between workflows. Genome Detective consistently identified the correct species across both methods. The findings of this study demonstrate that the COVIDSeq assay can be effectively adapted for multi-pathogen genomic surveillance and that the combined rapid workflow can offer a cost- and labour-efficient alternative with minimal compromise to data quality.
Full article
(This article belongs to the Special Issue Rapid Diagnostic Methods for Infectious Diseases)
►▼
Show Figures

Figure 1
Open AccessArticle
SNP rs3737883 in PPFIA4 Gene Associated with Atrial Fibrillation Risk: A Case–Control Study in a Chinese Population
by
Jiahui Zhuo, Pengyun Wang and Chengqi Xu
LabMed 2025, 2(4), 18; https://doi.org/10.3390/labmed2040018 - 25 Sep 2025
Abstract
►▼
Show Figures
Atrial fibrillation (AF), the most prevalent cardiac arrhythmia, significantly elevates the risk of stroke and heart failure. The etiology of AF is complex and multifactorial, involving genetic predisposition, environmental risk factors, and their potential interactions. A previous genome-wide association study (GWAS) of AF
[...] Read more.
Atrial fibrillation (AF), the most prevalent cardiac arrhythmia, significantly elevates the risk of stroke and heart failure. The etiology of AF is complex and multifactorial, involving genetic predisposition, environmental risk factors, and their potential interactions. A previous genome-wide association study (GWAS) of AF in a Korean population has identified an association between the rs3737883 single-nucleotide polymorphism (SNP) in the PPFIA4 gene and an increased risk of AF. However, the association needs to be replicated in other populations. In this paper, we conducted a case–control association study including 724 AF cases and 1475 controls, and successfully validated the association between SNP rs3737883 with the risk of AF in a Chinese population (OR = 1.33 with an adjusted p was 2.83 × 10−11). Given that the PPFIA4 variant has been reported to influence high-sensitivity cardiac troponin T (hs-cTnT) levels, we further investigated the relationship between rs3737883 and hs-cTnT in 48 AF patients. Notably, we observed that the risk allele was also associated with elevated hs-cTnT levels. Our findings provide further genetic substantiation for the association of rs3737883 with AF. These results suggest a potential association between the PPFIA4 gene variant, hs-cTnT levels, and AF risk, although further studies are needed to clarify the underlying mechanisms.
Full article
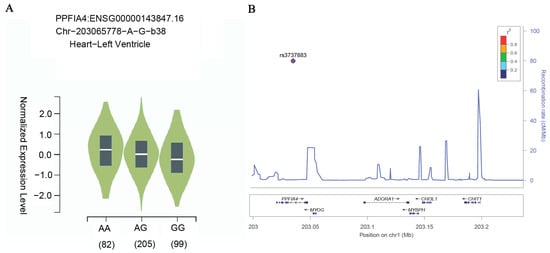
Figure 1
Open AccessReview
Non-Coding RNAs in Health and Disease: From Biomarkers to Therapeutic Targets
by
Marios A. Diamantopoulos, Michaela A. Boti, Triantafyllia Sarri and Andreas Scorilas
LabMed 2025, 2(3), 17; https://doi.org/10.3390/labmed2030017 - 17 Sep 2025
Cited by 4
Abstract
►▼
Show Figures
Non-coding RNAs (ncRNAs) are critical regulators of gene expression, taking part in the modulation of multiple biological functions across a range of cell types. Initially dismissed as transcriptional noise, ncRNAs are now recognized for their significant roles in key cellular mechanisms, including differentiation,
[...] Read more.
Non-coding RNAs (ncRNAs) are critical regulators of gene expression, taking part in the modulation of multiple biological functions across a range of cell types. Initially dismissed as transcriptional noise, ncRNAs are now recognized for their significant roles in key cellular mechanisms, including differentiation, apoptosis, and proliferation, as well as their profound implications for the pathogenesis of numerous human diseases. Due to their remarkable stability, tissue-specific expression patterns, and abundance in body fluids, ncRNAs hold significant promise as non-invasive biomarkers for diagnosis, prognosis, and therapeutic monitoring. Furthermore, advances in RNA-targeted therapeutics have introduced novel strategies to modulate ncRNA activity, although challenges related to delivery efficiency, specificity, and clinical validation remain. This review comprehensively summarizes the classification, biogenesis, and molecular functions of ncRNAs, elucidates their involvement in health and disease, and evaluates their potential as clinical biomarkers and therapeutic targets. Additionally, it discusses the emerging technologies for RNA manipulation, including CRISPR-based RNA editing, that can advance ncRNA research and revolutionize ncRNA-based therapeutics.
Full article

Figure 1
Open AccessReview
Quality Management in a Hemostasis Laboratory
by
Mayukh K. Sarkar
LabMed 2025, 2(3), 16; https://doi.org/10.3390/labmed2030016 - 1 Sep 2025
Abstract
Quality assurance in a clinical laboratory is essential to ensure reliable, accurate and precise laboratory test results all the time. A hemostasis laboratory is an important part of a clinical laboratory setting in a hospital or a healthcare center, and clinical laboratory tests
[...] Read more.
Quality assurance in a clinical laboratory is essential to ensure reliable, accurate and precise laboratory test results all the time. A hemostasis laboratory is an important part of a clinical laboratory setting in a hospital or a healthcare center, and clinical laboratory tests play a crucial role in diagnosis and management of conditions related to bleeding or clotting of diseased individuals. This review discusses all aspects of coagulation laboratory testing from pre-analytical, analytical, and post-analytical variables as part of daily quality assurance processes undertaken as well as the quality management process of assay validation and implementation in a laboratory prior to patient testing. The internal and external quality processes that drive a hemostasis laboratory will be discussed that shows a rigorous process in assurance of testing that is reliable and accurate every time, at all times.
Full article
(This article belongs to the Collection Feature Papers in Laboratory Medicine)
►▼
Show Figures

Figure 1
Open AccessArticle
Preliminary Evaluation of Cell Population Data Parameters in Different Blood Collection Tubes on Sysmex XN-Series Analysers
by
James V. Harte, Ciara Hoy and Vitaliy Mykytiv
LabMed 2025, 2(3), 15; https://doi.org/10.3390/labmed2030015 - 26 Aug 2025
Abstract
►▼
Show Figures
Cell population data (CPD) parameters generated by Sysmex XN-series analysers are promising biomarkers for a variety of disease states. Routine use of CPD parameters, however, will require extensive evaluation of potential pre-analytical variables that may affect reliability. At present, no information on the
[...] Read more.
Cell population data (CPD) parameters generated by Sysmex XN-series analysers are promising biomarkers for a variety of disease states. Routine use of CPD parameters, however, will require extensive evaluation of potential pre-analytical variables that may affect reliability. At present, no information on the comparability of CPD parameters generated using different blood collection tubes is available. In this preliminary study, we evaluated the impact of four commonly used blood collection tubes—dipotassium (K2) EDTA, tripotassium (K3) EDTA, trisodium citrate, and lithium heparin—on the generation of CPD parameters in whole blood from a cohort of 10 healthy donors. We also evaluated the stability of the CPD parameters generated at 4 h post-collection. Statistically significant differences in the CPD were observed across all blood collection tubes: whole blood anticoagulated with K3EDTA induced minimal biases and was comparable to whole blood anticoagulated with K2EDTA at collection; however, whole blood anticoagulated with citrate and heparin were associated with more substantial and more widespread biases in several parameters with potential clinical relevance. Notably, the biases observed in whole blood anticoagulated with K3EDTA increased in both number and magnitude at 4 h post-collection, whilst the CPD parameters generated with whole blood anticoagulated with K2EDTA remained stable. Although further confirmatory investigations are required, these findings highlight the importance of anticoagulant selection, as well as the need for further pre-analytical research, to support the integration of CPD parameters generated by Sysmex XN-series analysers into routine diagnostic workflows.
Full article

Figure 1
Open AccessArticle
Advancing Drug Resistance Detection: Comparative Analysis Using Short-Read and Long-Read Next-Generation Sequencing Technologies
by
Julie Martinez, Rezak Drali, Amira Doudou, Chalom Sayada, Ronan Boulmé, Dimitri Gonzalez, Laurent Deblir, Matthieu Barralon, Jérome Wautrin, Jonathan Porzio, Arnaud Reffay, Mohamed Errafyqy, Jonathan Kolsch, Jonathan Léonard, Giuseppina Zuco, Aitor Modol and Sofiane Mohamed
LabMed 2025, 2(3), 14; https://doi.org/10.3390/labmed2030014 - 20 Aug 2025
Abstract
In recent years, antiviral therapy has proved crucial in the treatment of infectious diseases, particularly infections by highly variable viruses such as human immunodeficiency virus, hepatitis B, hepatitis C, SARS-CoV-2 or bacteria such as Mycobacterium tuberculosis. Under the effect of selection pressure,
[...] Read more.
In recent years, antiviral therapy has proved crucial in the treatment of infectious diseases, particularly infections by highly variable viruses such as human immunodeficiency virus, hepatitis B, hepatitis C, SARS-CoV-2 or bacteria such as Mycobacterium tuberculosis. Under the effect of selection pressure, this variability induces mutations that lead to resistance to antiviral and antibacterial drugs, and thus to escape from treatment. The use of Advanced Biological Laboratories (ABL) assays technology combined with next-generation sequencing (NGS) and automatized software to detect majority and minority variants involved in treatment resistance has become a mainstay for establishing therapeutic strategies. The present study demonstrated high concordance between majority and minority subtypes and mutations identified in 15 samples across four NGS platforms: ISeq100 (Illumina (San Diego, CA, USA)), MiSeq (Illumina), DNBSEQ-G400 (MGI (Santa Clara, CA, USA)) and Mk1C MinION (Oxford Nanopore (Oxford Science Park, UK)). However, nanopore technology showed a higher number of minority mutations (<20%). The analysis also validated the pooling of microbiological samples as a method for detecting mutations and genotypes in viral and bacterial organisms, using the easy-to-use DeepChek® bioinformatics software, compatible with all four sequencing platforms. This study underlines the constant evolution of microbiological diagnostic research and the need to adapt rapidly to improve patient care.
Full article
(This article belongs to the Special Issue Rapid Diagnostic Methods for Infectious Diseases)
►▼
Show Figures

Figure 1
Open AccessReview
Monoclonal Protein Evaluation in the Diagnostic Algorithm for Cardiac Amyloidosis
by
Syed Bukhari
LabMed 2025, 2(3), 13; https://doi.org/10.3390/labmed2030013 - 28 Jul 2025
Abstract
►▼
Show Figures
Cardiac amyloidosis (CA) results from the deposition of either immunoglobulin light chain (AL) or transthyretin (ATTR) amyloid fibrils in the myocardium, causing restrictive cardiomyopathy and, if left untreated, can lead to early death. Advancements in non-invasive diagnostic modalities have led to an increased
[...] Read more.
Cardiac amyloidosis (CA) results from the deposition of either immunoglobulin light chain (AL) or transthyretin (ATTR) amyloid fibrils in the myocardium, causing restrictive cardiomyopathy and, if left untreated, can lead to early death. Advancements in non-invasive diagnostic modalities have led to an increased recognition of the disease. Monoclonal gammopathy plays a pivotal role in the diagnostic algorithm for CA, particularly in differentiating AL from ATTR. This review highlights the importance of monoclonal protein detection through serum protein electrophoresis, immunofixation electrophoresis, and serum free light chain assays as initial screening tools. However, these tests alone are insufficient for a definitive diagnosis due to the complexities associated with coexisting monoclonal gammopathies and the potential for false negative and positive results. Advanced imaging modalities, such as echocardiography, cardiac magnetic resonance, and nuclear scintigraphy, along with tissue biopsy, are crucial for confirming CA and accurately determining the CA subtype.
Full article

Figure 1
Open AccessArticle
Direct PCR for Rapid and Safe Pathogen Detection: Laboratory Evaluation Supporting Field Use in Infectious Disease Outbreak
by
Ivan Brukner and Matthew Oughton
LabMed 2025, 2(3), 12; https://doi.org/10.3390/labmed2030012 - 11 Jul 2025
Cited by 1
Abstract
Rapid, safe, and field-deployable molecular diagnostics are crucial for the effective management of infectious disease outbreaks, particularly those involving highly infectious pathogens, which can produce clinical symptoms similar to less infectious pathogens, thus raising potential biosafety concerns. In this study, we evaluated DNA/RNA
[...] Read more.
Rapid, safe, and field-deployable molecular diagnostics are crucial for the effective management of infectious disease outbreaks, particularly those involving highly infectious pathogens, which can produce clinical symptoms similar to less infectious pathogens, thus raising potential biosafety concerns. In this study, we evaluated DNA/RNA Defend Pro (DRDP) buffer, a novel viral-inactivating transport medium designed to stabilize nucleic acids and allow direct PCR without nucleic acid extraction. To ensure critical qPCR parameters were not compromised by using DRDP, we conducted serial dilution tests using herpes simplex viruses 1 and 2 (HSV-1, HSV-2) and varicella-zoster virus (VZV), comparing DRDP to standard universal transport medium (UTM). Detection sensitivity, determined by cycle quantification (Cq) values, favored DRDP, as UTM samples required a 2–3-fold dilution to mitigate PCR inhibition. DRDP maintained reliable PCR compatibility at reaction volumes containing up to 25% buffer. At higher DRDP concentrations (30–35%), PCR inhibition occurred due to EDTA content but was fully reversible by adding supplemental magnesium. Furthermore, DRDP samples did not require an initial 95 °C thermal lysis step, thus simplifying the procedure without reducing PCR sensitivity or efficiency.
Full article
(This article belongs to the Special Issue Rapid Diagnostic Methods for Infectious Diseases)
►▼
Show Figures

Figure 1
Open AccessArticle
Role of Windowing Image Technique to Decipher Soft Tissue Pathologies
by
Saavi Reddy Pellakuru, Neha Nischal, Hasaam Uldin, Nathan Jenko, Anshu Firake, David Beale, Karthikeyan P. Iyengar and Rajesh Botchu
LabMed 2025, 2(3), 11; https://doi.org/10.3390/labmed2030011 - 30 Jun 2025
Abstract
►▼
Show Figures
Fluid-sensitive sequences on MRI [Magnetic Resonance Imaging] have widely been used to assess soft tissue oedema. Windowing techniques play a significant role in adjusting the contrast to highlight the pathology. The purpose of this study is to establish the impact of modified MRI
[...] Read more.
Fluid-sensitive sequences on MRI [Magnetic Resonance Imaging] have widely been used to assess soft tissue oedema. Windowing techniques play a significant role in adjusting the contrast to highlight the pathology. The purpose of this study is to establish the impact of modified MRI window parameters, with a narrower window width than window level, in assessing soft tissue oedema in a plethora of musculoskeletal pathologies. Fifty randomly selected patients with a range of musculoskeletal pathologies resulting in soft tissue oedema on MRI were included in the study. Two separate images of each MRI study were taken on a PD fat suppressed sequence, one with default windowing range and another with window width lower than that of window level. Both images were reviewed by two radiologists and were assessed for diagnostic effectiveness in terms of image resolution and depiction of pathology. Assessment was semi-quantitatively compared and graded on the Likert scale, from 1 to 5, with 1 indicating poor quality and 5 indicating excellent quality. Friedman’s test was then conducted to compare the scores of both images. In most of the cases, the image with the modified window/level setting was significantly better in terms of depicting pathology and having better resolution, though some cases showed no clear preference. Friedman’s test showed that the score for images with modified window settings was significantly higher. Images with modified windowing in conjunction with standard imaging protocols help to assess soft tissue oedema.
Full article

Figure 1
Open AccessArticle
Residual Direct Oral Anticoagulant Activity in the Preoperative Setting: Review of the Literature and a Pilot Study Regarding Direct Oral Anticoagulant Preoperative Interruption (Based on Guidelines) and Its Correlation with Patient Characteristics and Blood Product Transfusion
by
Eleni C. Georgiadi, Apostolos Nousias and Paraskevi Kotsi
LabMed 2025, 2(2), 10; https://doi.org/10.3390/labmed2020010 - 13 Jun 2025
Abstract
►▼
Show Figures
Direct oral anticoagulants (DOACs) have been licensed worldwide for several years for various indications. Each year, 10–15% of patients receiving oral anticoagulants will undergo an interventional procedure, and expert groups have issued several guidelines for perioperative management in such situations. According to the
[...] Read more.
Direct oral anticoagulants (DOACs) have been licensed worldwide for several years for various indications. Each year, 10–15% of patients receiving oral anticoagulants will undergo an interventional procedure, and expert groups have issued several guidelines for perioperative management in such situations. According to the PAUSE study, the proposed randomized strategy of stopping DOACs without bridging therapy in patients with atrial fibrillation was associated with low rates of major bleeding and arterial thromboembolism so that its implementation is increasingly safe. The present study was carried out in order to investigate the efficacy and safety of the standardized perioperative DOAC management strategy by measuring the residual activity of oral anticoagulants when stopping them preoperatively in daily practice in a regional hospital. Thirty-two patients were included in the present study. They were patients who suffered from atrial fibrillation or deep vein thrombosis and were receiving an oral anticoagulant, rivaroxaban or apixaban at the indicated dose. These patients underwent an elective surgery or invasive procedure at the Karditsa General Hospital between May 2022 and April 2023. The results showed that in a percentage of >90% of the patients on the day of surgery they had a residual anti-Xa activity below 0.5 U/mL. This rate is considered high and confirms the safety and efficacy of the guideline-recommended protocol for perioperative discontinuation of DOACs.
Full article

Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
LabMed
Rapid Diagnostic Methods for Infectious Diseases
Guest Editor: Emmanouil MagiorkinisDeadline: 30 April 2026







