1. Introduction
Nowadays, body sensors monitor heart signals with a high precision and negligible noise. Smart sensors are considered an alternative solution to physical sensors. In heart disease applications, smart sensors are used to predict a biomedical signal, such as the ECG, based on the history of that signal or to predict a signal using other available means.
ECG electrodes are sensitive to several distortions and may be damaged by certain environmental factors. The displacement of electrodes from the desired measurement site or the detachment of the transmission cable could change the temporal and frequential characteristics of ECG signals, which may lead to a false analysis and poor measurement quality. Innovative biosensors are considered an alternative solution to reduce noise and signal cuts in case of external or internal perturbation. The intelligent sensors are also used to reduce the number of electrodes placed on the patient’s chest to ensure a proper interpretation and diagnosis of the heart health condition.
Smart sensors are generally based on machine learning (ML) methods. Among the most popular and widely used ML techniques is the ANN. Indeed, this technique has been successfully used in various application areas such as climate [
1], energy [
2], and communication technology [
3]. The literature presents different techniques of ECG biosensors. In [
4], the authors proposed an ECG biosensor based on the autoregressive integrated moving average (ARIMA) model with a discrete wavelet transform. Despite the good prediction of the amplitude, it should be noted that the assumption of a local linearity limits such a method. To avoid this drawback, other ECG biosensors based on the ANN model are also used to predict ECG signals [
5,
6]. In [
5], the authors applied the ANN to detect the ECG abnormalities by using a multilayer perceptron network trained by three different algorithms such as the Bayesian regularization (BR), the Levenberg–Marquardt (LM) and backpropagation (BP). The obtained results are satisfactory. Other biosensors, such as [
7,
8] can also be cited. In [
7], Gutta et al. proposed a biosensor allowing the extraction features and classification of ECG signals using a recurrent neural network. [
8] proposed another ECG identification algorithm based on the support vector machine.
Despite the multitude of existing ECG biosensors, the one based on the ANN is of great interest. However, despite its success in various applications, ANN methods have different limitations. The two most known issues that have a substantial impact on the estimation quality are catastrophic forgetting (CF) and the choice of hyperparameters [
9,
10]. Indeed, CF is the loss of information related to a first task after training for a second task. This information declination could almost be catastrophic in some cases [
11,
12,
13,
14,
15]. For example, the problem of using many hidden layers with a nonlinear activation function is that the proposed ANN model can stop learning, leading to poor results. Indeed, the so-called vanishing gradient problem is highlighted in this case. It is caused by the information expression that takes place at each iteration. From a practical standpoint, rectified linear units (ReLU) present an excellent function. Many models also rely heavily on ReLU to give good results since it preserves the information. However, this function has its disadvantages; for example, it runs into a dying ReLU problem in rare cases and can block the negative values that prevent the propagation of certain information.
Hyperparameter optimization is another issue that can affect the ANN prediction efficiency. In this perspective, many researchers suggested using the random search method [
16,
17]. This technique consists of generating architectures with different parameter values. The grid search method also relies on checking all possible architectures with a limited set of parameters defined values [
18]. However, this will require a lot of resources and can be inefficient and time-consuming, even impossible when the problem presents a high complexity. The output is a nonlinear combination of the input variables. Indeed, the ANN method finds the best neural network architecture and determines the weight using retro-propagation. The retro-propagation algorithm is applied at each iteration to update the weights. Decreasing the number of epochs will thus not provide good results but increase the number of epochs which might lead to over-fitting. Therefore, it is advised to try different combinations to determine the adequate values.
To solve the ANN hyperparameter problem, several optimization techniques were proposed in the literature. Conventional methods such as gradient descent [
19] and graph theory [
20] have some constraints mainly defined by the local optimization and unknown search space problems. Moreover, heuristic and metaheuristic methods have been used. These techniques are divided into four subclasses: Algorithms based on swarm intelligence such as the Harris Hawks optimization (HHO) [
21] and the genetic algorithm (GA) [
22], evolutionary algorithms, natural phenomena-based algorithms, and natural sciences-based algorithms such as the Henry gas solubility optimization (HGSO) [
23]. In the latter, the authors adapted the HGSO algorithm (based on Henry’s law for the imitation of gas behavior) to tune the random forest’s hyperparameters in the prediction of caesarean births. In [
24], the authors proposed an improved prediction algorithm based on the HHO method with the aim of assessing early the severity of COVID-19. Similarly, the authors in [
25] proposed an optimized machine learning approach by combining the Harris Hawk method with the feature analysis based on the SHapely adaptive exPlanations (SHAP) method for a better COVID-19 prediction. Over the last decade, nature-inspired algorithms and swarm intelligence have been increasingly used and applied in different disciplines due to their efficiencies and flexibility. The major advantages they have are black-box and gradient-free optimizers. However, there are some key issues regarding these algorithms concerning their analysis in terms of their stability, convergence, convergence rate, and robustness. In [
26], an in-depth review of some recent nature-inspired algorithms emphasizes five open issues regarding the analysis of algorithmic convergence, parameter tuning, role of calibration, etc.
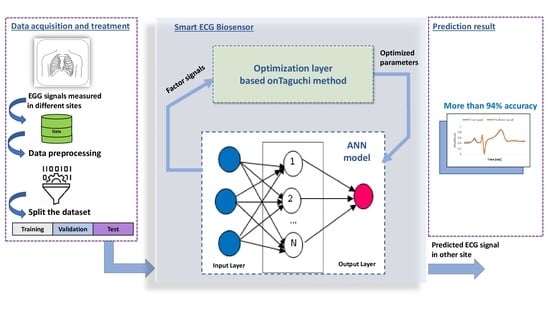
From this perspective, the main idea of this paper is to propose a smart ECG biosensor defined by a hybrid architecture based on ANNs and the Taguchi optimizer. This optimizer is a powerful and efficient statistical tool for the design of high-quality systems [
27,
28]. It is used for different kinds of applications [
29,
30,
31,
32,
33]. The Taguchi has also proved its effectiveness as a robust tool for deep learning applications [
34,
35]. It is well-known for its robustness-based orthogonal array design. The Taguchi optimizer allows reducing the time needed to study the influence of individual factors and determine which factor has more influence by applying the simplest technique with any mathematical complexity. According to [
36], a comparative study between the Taguchi method and some genetic algorithms (GA) was performed. It is proven that it is preferred to use the Taguchi method rather than GA, given the cost of the experiment design, the number of experiments, quality, and performance. Such an approach consists of narrowing the search field after each iteration by checking the statistics of the result on each iteration and readjusting the search parameters to predict the measured ECG signals in different locations. In summary, this optimizer reduces the computational time, avoiding the problems associated with the hyperparameters and the computing time related to the ANN predator.
The contributions of this work are multifold. The Taguchi optimizer layer has been applied on ECG signal prediction for the first time with a high accuracy, enabling the optimization of the number of body ECG sensors. It is well known that the most common method to reconstruct the 12-lead ECG from a limited lead set is a linear regression. Despite the use of ANNs in the literature [
37], the enhancement of ANNs through the Taguchi optimizer has never been attempted before. While many papers [
38,
39] have proposed an ECG signal reconstruction of missing samples, their methods are mainly based on multiple ECG leads that are linearly connected. To the best of our knowledge, no other paper has attempted the prediction of ECG using independent traces. This novel approach is significant and challenging as the reconstruction is based on single leads measured at different auscultation sites. The clinical applications of the proposed approach include designing and implementing miniaturized devices for measuring and tracking heart diseases. Such devices are suggested to make use of ECGs to drive the segmentation of phonocardiograms [
39]. Furthermore, the field of monitoring and tracking utilizes telemedicine to transmit data through various connectionless channels. These channels are often affected by the loss of data packages (occurring during transmission) that need to be recovered. While reconstructed from multiple leads, the ECG has received much attention [
40], and novel methods are required for independent channels. The importance of data reconstruction has proven to be very relevant in the remote monitoring of patients in rural areas [
41].
This paper is structured as follows.
Section 2 introduces the ECG biosensor design with an overview on the prediction approach. Then, the Taguchi-based ANN model is discussed in detail. For that, the basics of this method are recalled and applied to a study case. The simulation results with a comprehensive performance comparison between the basic ANN algorithm and the proposed Taguchi-based ANN are presented in
Section 3. Last,
Section 4 concludes the paper.
3. Results and Discussion
This section tackles the numerical validation of the ECG biosensor based on the performance of the optimized ANN model using the technical and software requirements described in
Table 6.
With five signals available, one signal will be the output while the remaining signals are combined to form the input data. Due to the data collection method and in order to reduce the search field, it is advisable to lower the amount of content with three signals, Lead1, ECGA, and either ECGP or ECGT or ECGM, narrowing the possibilities to only 27.
Figure 7 shows a high
R2 score for all of the possible combinations of ECG signals measured in different sites; for example, ECGP, ECGA, and LEAD1, using LEAD1 as an input and ECGA as an output. As for the signals ECGT and ECGM, the R2 score was between 50% and 80%, which is due to the low correlation factor, unlike the case with ECGP, which makes this site a good location to rely on for predicting other signals demonstrated in
Figure 7. We note that the best efficiency estimation is shown in
Figure 8. It is the result of the use of ECGP and ECGA to predict LEAD1 of subject nine and of a neural network’s architecture based on seven hidden layers 100–100–10–10–200–10–200, respectively, with activation functions: Selu-Selu-Relu-Relu-Elu-Selu-Sigmoid.
The predicted signal’s standard deviation (SD) with the optimized method rounds up to 0.1448. The actual signal’s SD rounds up to 0.1450, whereas the signal obtained from the random search only method results in an SD of 0.1313, which is far from the actual value. As shown in
Figure 9, both the actual signal and the predicted signals using the Taguchi method show many similarities in the median values with a 0.57% error, upper quartile, lower quartile, maximum, and minimum values with a 1.3% error between the interval of points.
The signal predicted by the random search only presents a 4.7% error in the median value and an 18% error in the maximum and minimum values compared to the actual signal. Consequently, the properties of the signal are entirely changed. ECG signals are very sensitive and delicate, and such errors can change their characteristics.
Via the proposed prediction approach, it is also possible to know how many model inputs and which ECG signal sites to use to obtain the best result. Indeed, the prediction with an optimized search proves that estimating an ECG signal via two ECG inputs instead of one is more accurate with respect to the imposed performance criteria.
To verify the efficiency of the Taguchi method, we reran the search randomly using the random search function on the Keras tuner, and the difference was noticeable in
Figure 10.
The percentage of the trials in the search field is less than 0.1%, which means that the number of tests for each combination is minimal. That is why the random search method gives poor results. Running all of the input combinations for the random search method took more than 15 h for each type of signal (ECGP, ECGT, or ECGM), whereas in the optimized method, it took almost six hours and gave better and more consistent results.
As shown in
Figure 10a, the searches with an
R2 score of 94% were the best results, with ECGP or Lead1 as an output signal. This method was accurate because of the small sample taken, and ECGP and Lead1 have a high correlation factor with the other signals.
In conclusion, the ANN model optimized with the Taguchi method gives better results than those obtained via a random search. In addition, if the data presents a medium-low correlation coefficient, this method is preferred since it cuts a lot of research time and eliminates unnecessary parameter values.
For the same database, a comparative study between different estimators (Linear Regression (LR), K-nearest neighbors (KNN), random forest regression (RFR) and ANNs) was proposed in [
54] to predict the ECG signals measured in different sites. It was shown that the RFR and KNN models perform better in terms of prediction efficiency than the LR model in direct and cross validation.
Table 7 summarizes the MSE, MAE, and
R2 values that represent the average values obtained for all of subjects for the proposed optimized approach and for the LR, KNN, RFR, and ANN algorithms. These results show the interest of the proposed estimator compared with the other.