Influence of Adiposity on the Gut Microbiota Composition of Arab Women: A Case-Control Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Study Tools
2.2.1. Anthropometric Measures
2.2.2. Clinical Data and Questionnaire
2.2.3. Stool Sample and DNA Extraction
2.3. Bioinformatics and Statistical Analysis
3. Results
3.1. Descriptive Characteristics
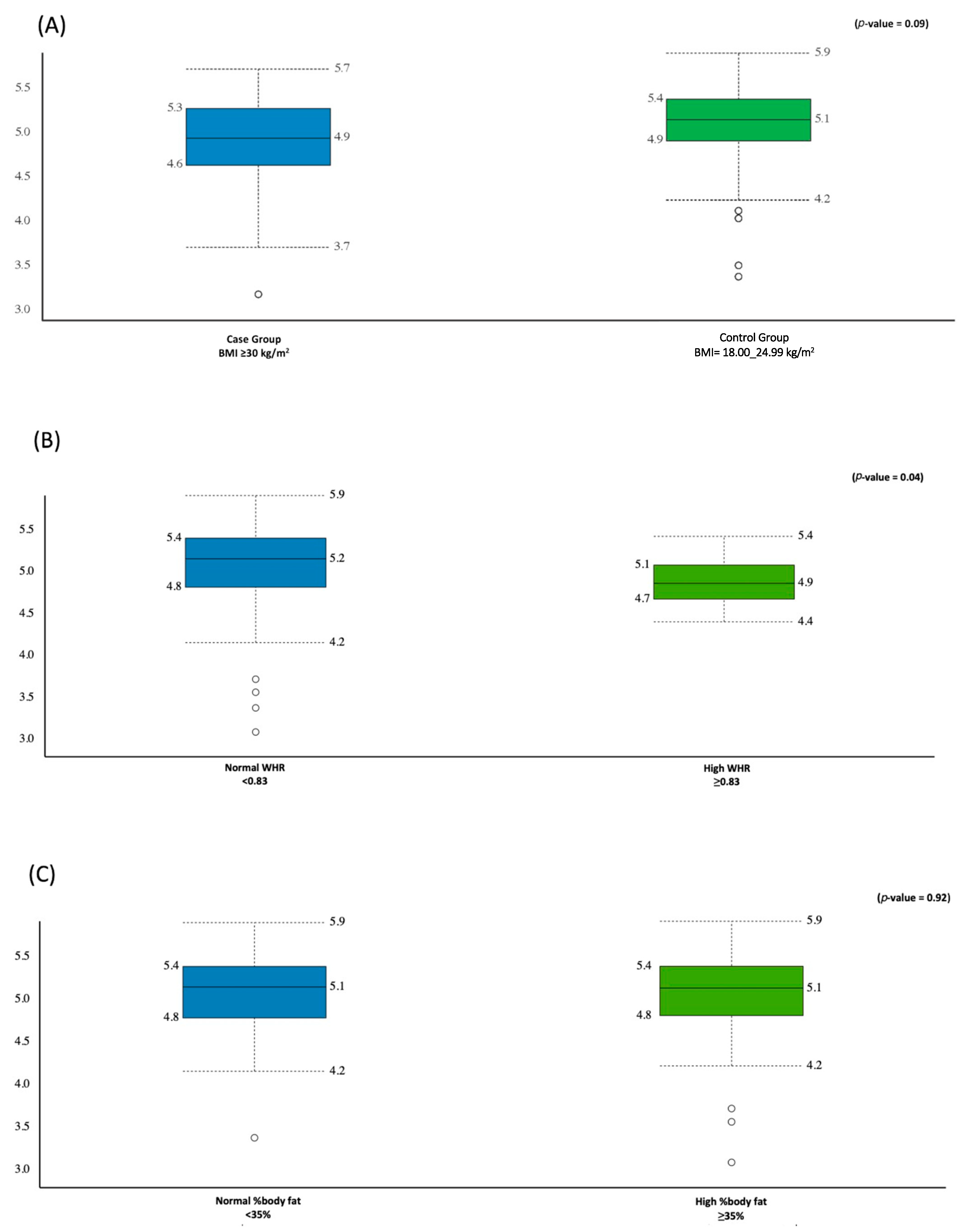
3.2. Alpha Diversity Analysis
3.3. Beta Diversity Analysis
3.4. Gut Microbiota Signatures in the Case and Control Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdelaal, M.; le Roux, C.W.; Docherty, N.G. Morbidity and mortality associated with obesity. Ann. Transl. Med. 2017, 5, 161. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Obesity and Overweight. Available online: http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 20 February 2019).
- World Health Organization. Prevalence of Obesity among Adults, BMI ≥ 30 (Age-Standardized Estimate) (%). Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/prevalence-of-obesity-among-adults-bmi-=-30-(age-standardized-estimate)-(-) (accessed on 4 March 2019).
- Barker, M.; Dombrowski, S.U.; Colbourn, T.; Fall, C.H.D.; Kriznik, N.M.; Lawrence, W.T.; Norris, S.A.; Ngaiza, G.; Patel, D.; Skordis-Worrall, J.; et al. Intervention strategies to improve nutrition and health behaviours before conception. Lancet 2018, 391, 1853–1864. [Google Scholar] [CrossRef] [Green Version]
- Archer, E.; Lavie, C.J. Obesity Subtyping: The Etiology, Prevention, and Management of Acquired versus Inherited Obese Phenotypes. Nutrients 2022, 14, 2286. [Google Scholar] [CrossRef] [PubMed]
- Castaner, O.; Goday, A.; Park, Y.-M.; Lee, S.-H.; Magkos, F.; Shiow, S.-A.T.E.; Schröder, H. The Gut Microbiome Profile in Obesity: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 9. [Google Scholar] [CrossRef]
- Shanahan, F.; Ghosh, T.S.; O’Toole, P.W. The Healthy Microbiome—What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, Q.; Hu, L.; Xing, S.; Gong, H.; Liu, Z.; Qin, P.; Xu, J.; Du, J.; Ai, W.; et al. Dynamic Alteration of the Gut Microbiota Associated with Obesity and Intestinal Inflammation in Ovariectomy C57BL/6 Mice. Int. J. Endocrinol. 2022, 2022, 6600158. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Ma, Y.; Zhang, F.; Zhao, C.; Nie, Y. Insights into the role of gut microbiota in obesity: Pathogenesis, mechanisms, and therapeutic perspectives. Protein Cell 2018, 9, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.N.; Yao, Y.; Ju, S.Y. Short Chain Fatty Acids and Fecal Microbiota Abundance in Humans with Obesity: A Systematic Review and Meta-Analysis. Nutrients 2019, 11, 2512. [Google Scholar] [CrossRef]
- Palmas, V.; Pisanu, S.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Loviselli, A.; Manzin, A.; et al. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021, 11, 5532. [Google Scholar] [CrossRef] [PubMed]
- Osborne, G.; Wu, F.; Yang, L.; Kelly, D.; Hu, J.; Li, H.; Jasmine, F.; Kibriya, M.G.; Parvez, F.; Shaheen, I.; et al. The association between gut microbiome and anthropometric measurements in Bangladesh. Gut Microbes 2020, 11, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Angelakis, E.; Bibi, F.; Azhar, E.I.; Bachar, D.; Lagier, J.C.; Gaborit, B.; Hassan, A.M.; Jiman-Fatani, A.A.; Alshali, K.Z.; et al. Comparison of the gut microbiota of people in France and Saudi Arabia. Nutr. Diabetes 2015, 5, e153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelakis, E.; Yasir, M.; Bachar, D.; Azhar, E.I.; Lagier, J.-C.; Bibi, F.; Jiman-Fatani, A.A.; Alawi, M.; Bakarman, M.A.; Robert, C.; et al. Gut microbiome and dietary patterns in different Saudi populations and monkeys. Sci. Rep. 2016, 6, 32191. [Google Scholar] [CrossRef] [Green Version]
- Harakeh, S.; Angelakis, E.; Karamitros, T.; Bachar, D.; Bahijri, S.; Ajabnoor, G.; Alfadul, S.M.; Farraj, S.A.; Al Amri, T.; Al-Hejin, A.; et al. Impact of smoking cessation, coffee and bread consumption on the intestinal microbial composition among Saudis: A cross-sectional study. PLoS ONE 2020, 15, e0230895. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [Green Version]
- Black, A.E. Critical evaluation of energy intake using the Goldberg cut-off for energy intake:basal metabolic rate. A practical guide to its calculation, use and limitations. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 1119–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Waist Circumference and Waist-Hip Ratio: Report of a WHO Expert Consultation, Geneva, 8–11 December 2008; World Health Organization: Geneva, Switzerland, 2011.
- So, W.-Y.; Swearingin, B.; Crooms, B.; Lee, R.; Choi, Y.; Dail, T.K.; Melton, D.; Fuller, T.M.; Ha, C.-H. Body composition measurements determined by air displacement plethysmography and eight-polar bioelectrical impedance analysis are equivalent in African American college students. HealthMED 2012, 6, 1902–1905. [Google Scholar]
- Dickey, R.A.; Bartuska, D.; Bray, G.W.; Callaway, C.W.; Davidson, E.T.; Feld, S.; Ferraro, R.T.; Hodgson, S.F.; Jellinger, P.S.; Kennedy, F.P. AACE/ACE Position statement on the prevention, diagnosis, and treatment of obesity (1998 revision). Endocr. Pract. 1998, 4, 297–350. [Google Scholar]
- Alkahtani, S.A. Convergent validity: Agreement between accelerometry and the Global Physical Activity Questionnaire in college-age Saudi men. BMC Res. Notes 2016, 9, 436. [Google Scholar] [CrossRef] [Green Version]
- Suleiman, K.H.; Yates, B.C.; Berger, A.M.; Pozehl, B.; Meza, J. Translating the Pittsburgh Sleep Quality Index into Arabic. West. J. Nurs. Res. 2010, 32, 250–268. [Google Scholar] [CrossRef]
- Alkhalaf, M.M.; Edwards, C.A.; Combet, E. Validation of a food frequency questionnaire specific for salt intake in Saudi Arabian adults using urinary biomarker and repeated multiple pass 24-hour dietary recall. Proc. Nutr. Soc. 2015, 74, E337. [Google Scholar] [CrossRef]
- Roe, M.; Pinchen, H.; Church, S.; Finglas, P. McCance and Widdowson’s The Composition of Foods Seventh Summary Edition and updated Composition of Foods Integrated Dataset. Nutr. Bull. 2015, 40, 36–39. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P. Vegan: Community Ecology Package. 2019. R Package Version 2.5-6. 2019. Available online: https://github.com/vegandevs/vegan (accessed on 13 February 2021).
- Kassambara, A. ggpubr: ‘ggplot2′ Based Publication Ready Plots. R Package Version 0.2.5. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 13 February 2021).
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khubrani, Y.M.; Wetton, J.H.; Jobling, M.A. Extensive geographical and social structure in the paternal lineages of Saudi Arabia revealed by analysis of 27 Y-STRs. Forensic Sci. Int. Genet. 2018, 33, 98–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrocino, I.; Di Cagno, R.; De Angelis, M.; Turroni, S.; Vannini, L.; Bancalari, E.; Rantsiou, K.; Cardinali, G.; Neviani, E.; Cocolin, L. Fecal microbiota in healthy subjects following omnivore, vegetarian and vegan diets: Culturable populations and rRNA DGGE profiling. PLoS ONE 2015, 10, e0128669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salazar, N.; Arboleya, S.; Fernández-Navarro, T.; de Los Reyes-Gavilán, C.G.; Gonzalez, S.; Gueimonde, M. Age-associated changes in gut microbiota and dietary components related with the immune system in adulthood and old age: A cross-sectional study. Nutrients 2019, 11, 1765. [Google Scholar] [CrossRef] [Green Version]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, D.; Heymsfield, S.B.; Heo, M.; Jebb, S.A.; Murgatroyd, P.R.; Sakamoto, Y. Healthy percentage body fat ranges: An approach for developing guidelines based on body mass index. Am. J. Clin. Nutr. 2000, 72, 694–701. [Google Scholar] [CrossRef] [Green Version]
- Jian, C.; Silvestre, M.P.; Middleton, D.; Korpela, K.; Jalo, E.; Broderick, D.; de Vos, W.M.; Fogelholm, M.; Taylor, M.W.; Raben, A.; et al. Gut microbiota predicts body fat change following a low-energy diet: A PREVIEW intervention study. Genome Med. 2022, 14, 54. [Google Scholar] [CrossRef]
- Pinart, M.; Dötsch, A.; Schlicht, K.; Laudes, M.; Bouwman, J.; Forslund, S.K.; Pischon, T.; Nimptsch, K. Gut Microbiome Composition in Obese and Non-Obese Persons: A Systematic Review and Meta-Analysis. Nutrients 2021, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Yassour, M.; Lim, M.Y.; Yun, H.S.; Tickle, T.L.; Sung, J.; Song, Y.M.; Lee, K.; Franzosa, E.A.; Morgan, X.C.; Gevers, D.; et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wei, H.; Zhou, Y.; Szeto, C.-H.; Li, C.; Lin, Y.; Coker, O.O.; Lau, H.C.H.; Chan, A.W.H.; Sung, J.J.Y.; et al. High-Fat Diet Promotes Colorectal Tumorigenesis through Modulating Gut Microbiota and Metabolites. Gastroenterology 2022, 162, 135–149.e2. [Google Scholar] [CrossRef] [PubMed]
- Conlon, M.A.; Bird, A.R. The Impact of Diet and Lifestyle on Gut Microbiota and Human Health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fang, X.; Zhou, Y.; Dou, L.; Dou, T. Machine learning-based investigation of the relationship between gut microbiome and obesity status. Microbes Infect. 2022, 24, 104892. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.W.; Shao, L.; Sun, S.H.; Wu, J.; Song, Q.H.; Zou, H.S.; Ling, Z.X. Gut microbiota dysbiosis in Chinese children with type 1 diabetes mellitus: An observational study. World J. Gastroenterol. 2021, 27, 2394–2414. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.-N.; Yu, Q.-F.; Fu, N.; Liu, X.-W.; Lu, F.-G. Effects of four Bifidobacteria on obesity in high-fat diet induced rats. World J. Gastroenterol. 2010, 16, 3394–3401. [Google Scholar] [CrossRef] [PubMed]
- Sabico, S.; Al-Mashharawi, A.; Al-Daghri, N.M.; Wani, K.; Amer, O.E.; Hussain, D.S.; Ahmed Ansari, M.G.; Masoud, M.S.; Alokail, M.S.; McTernan, P.G. Effects of a 6-month multi-strain probiotics supplementation in endotoxemic, inflammatory and cardiometabolic status of T2DM patients: A randomized, double-blind, placebo-controlled trial. Clin. Nutr. 2019, 38, 1561–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabico, S.; Al-Mashharawi, A.; Al-Daghri, N.M.; Yakout, S.; Alnaami, A.M.; Alokail, M.S.; McTernan, P.G. Effects of a multi-strain probiotic supplement for 12 weeks in circulating endotoxin levels and cardiometabolic profiles of medication naïve T2DM patients: A randomized clinical trial. J. Transl. Med. 2017, 15, 249. [Google Scholar] [CrossRef] [Green Version]
- Wehling, H.; Lusher, J. People with a body mass index ≥30 under-report their dietary intake: A systematic review. J. Health Psychol. 2017, 24, 2042–2059. [Google Scholar] [CrossRef]
- Zaheer, R.; Noyes, N.; Ortega Polo, R.; Cook, S.R.; Marinier, E.; Van Domselaar, G.; Belk, K.E.; Morley, P.S.; McAllister, T.A. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci. Rep. 2018, 8, 5890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillmann, B.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Zhu, Q.; Gohl, D.M.; Beckman, K.B.; Knight, R.; Knights, D. Evaluating the Information Content of Shallow Shotgun Metagenomics. mSystems 2018, 3, e00069–18. [Google Scholar] [CrossRef] [PubMed]
- Rajan, S.K.; Lindqvist, M.; Brummer, R.J.; Schoultz, I.; Repsilber, D. Phylogenetic microbiota profiling in fecal samples depends on combination of sequencing depth and choice of NGS analysis method. PLoS ONE 2019, 14, e0222171. [Google Scholar] [CrossRef] [PubMed]






| Characteristics | Total (n = 92) | Control Group BMI = 18.50–24.99 (n = 48) | Case Group BMI ≥ 30.00 (n = 44) | p-Value |
|---|---|---|---|---|
| Age (Years) | 21.1 ± 1.5 | 20.6 ± 1.1 | 21.6 ± 1.7 | <0.01 |
| Age of Menarche | 12.4 ± 1.0 | 12.5 ±1.0 | 12.3 ± 1.1 | 0.31 |
| Anthropometric measurements | ||||
| Height (cm) | 157.7 ± 5.2 | 158.0 ± 5.7 | 157.3 ± 4.5 | 0.48 |
| Weight (kg) | 70.7 ± 19.7 | 54.3 ± 6.0 | 89.1 ± 11.8 | <0.001 |
| BMI (kg/m2) | 28.6 ± 8.0 | 21.7 ± 1.9 | 36.0 ± 4.7 | <0.001 |
| Waist (cm) | 80.4 ± 17.4 | 67.7 ± 4.3 | 94.5 ± 15.4 | <0.001 |
| Hip (cm) | 109.2 ± 16.4 | 96.4 ± 7.6 | 123.5 ± 10.8 | <0.001 |
| WHR (ratio) | 0.7 ± 0.1 | 0.7 ± 0.1 | 0.8 ± 0.1 | 0.01 |
| Fat (%) | 42.5 ± 9.4 | 34.8 ± 5.5 | 51.1 ± 3.3 | <0.001 |
| Total body protein (kg) | 7.6 ± 1.1 | 6.9 ± 0.7 | 8.5 ± 0.9 | <0.001 |
| Skeletal muscle mass (kg) | 21.0 ± 3.5 | 18.6 ± 2.2 | 23.6 ± 2.6 | <0.001 |
| Muscle mass (%) | 28.2 ± 7.0 | 29.6 ± 9.3 | 26.6 ± 1.9 | 0.04 |
| Total Body Water (L) | 28.7 ± 4.3 | 25.8 ± 2.7 | 31.9 ± 3.3 | <0.001 |
| Fluid (%) | 42.1 ± 6.8 | 47.7 ± 4.0 | 35.9 ± 2.3 | <0.001 |
| Biochemical data | ||||
| Total cholesterol (mmol/L) | 4.1 ± 1.5 | 3.6 ± 1.7 | 4.5 ± 1.0 | 0.01 |
| FBG (mmol/L) | 4.6 ± 0.7 | 4.5 ± 0.8 | 4.8 ± 0.6 | 0.04 |
| HDL-cholesterol (mmol/L) | 1.0 ± 0.3 | 0.9 ± 0.4 | 1.0 ± 0.3 | 0.24 |
| LDL-cholesterol (mmol/L) | 2.9 ± 1.3 | 2.6 ± 1.5 | 3.3 ± 1.0 | 0.01 |
| Total cholesterol/HDL ratio | 4.3 ± 1.7 | 3.9 ± 1.7 | 4.7 ± 1.7 | 0.05 |
| Triglyceride (mmol/L) # | 0.7 (0.5–1.0) | 0.5 (0.4–0.7) | 1.0 (0.8–1.1) | <0.001 |
| Insulin (µIU/mL) # | 9.9 (6.0–15.8) | 6.4 (5.0–9.3) | 15.2 (12.2–20.3) | <0.001 |
| HOMA-IR # | 2.0 (1.1–3.6) | 1.2 (0.9–1.6) | 3.5 (2.3–4.4) | <0.001 |
| HOMA-β # | 165.7 (112.0–288.1) | 114.5 (71.8–148.3) | 224.5 (182.7–316.1) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aljazairy, E.A.; Al-Musharaf, S.; Abudawood, M.; Almaarik, B.; Hussain, S.D.; Alnaami, A.M.; Sabico, S.; Al-Daghri, N.M.; Clerici, M.; Aljuraiban, G.S. Influence of Adiposity on the Gut Microbiota Composition of Arab Women: A Case-Control Study. Biology 2022, 11, 1586. https://doi.org/10.3390/biology11111586
Aljazairy EA, Al-Musharaf S, Abudawood M, Almaarik B, Hussain SD, Alnaami AM, Sabico S, Al-Daghri NM, Clerici M, Aljuraiban GS. Influence of Adiposity on the Gut Microbiota Composition of Arab Women: A Case-Control Study. Biology. 2022; 11(11):1586. https://doi.org/10.3390/biology11111586
Chicago/Turabian StyleAljazairy, Esra’a A., Sara Al-Musharaf, Manal Abudawood, Basmah Almaarik, Syed D. Hussain, Abdullah M. Alnaami, Shaun Sabico, Nasser M. Al-Daghri, Mario Clerici, and Ghadeer S. Aljuraiban. 2022. "Influence of Adiposity on the Gut Microbiota Composition of Arab Women: A Case-Control Study" Biology 11, no. 11: 1586. https://doi.org/10.3390/biology11111586








