1. Introduction
In adults, hypertension (HT) is diagnosed when repeated office measurement of systolic blood pressure (SBP) is ≥140 mmHg, or diastolic blood pressure (DBP) is ≥90 mmHg [
1]. HT can be classified into different categories based on the office measurement (
Table 1) [
1]. HT increase the force exerted by by the blood against the inner walls of the arteries, which transport oxygen-rich blood pumped out of the heart to the rest of the body [
2]. As such, chronic HT can inflict damage to various vital organs of the body, such as lung, brain, heart, and kidneys [
2]. The World Health Organization estimates that nearly 1.3 billion people suffered from HT in 2015 globally, and less than 20% received management [
2]. HT is largely asymptomatic, but symptoms can sometimes occur, including headaches, panic attacks and dizziness.
1.1. ECG Signals and Blood Pressure (BP) Measurements
The electrocardiogram (ECG) records the electrical potentials on the body surface that originate from heart, and the signals can provide information on the rhythm as well as structure and function of the heart [
3,
4,
5,
6]. Using advanced analysis, the ECG signals in HT subjects can be correlated to BP measurements and even discriminate for higher clinical risk [
7,
8,
9,
10]. In HT, the heart observes more force and over time becomes hypertrophied, which induces the ECG.
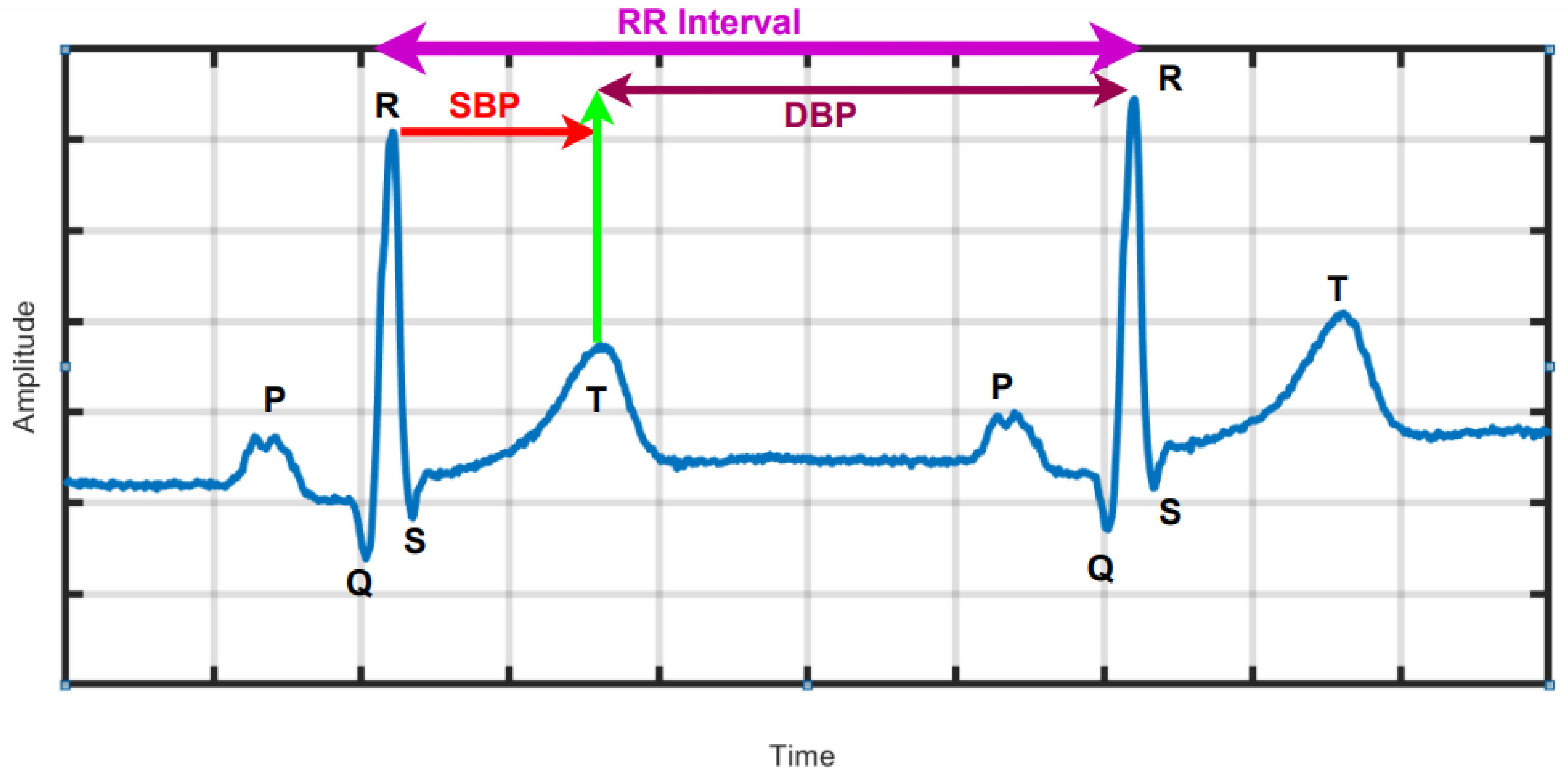
Figure 1 is a graphical depiction of a typical normal ECG waveform, which comprise the P wave, QRS complex and T wave representing atrial depolarization, ventricular depolarization and ventricular re-polarization, as well as standard ECG intervals, including RR interval, PR interval, QT interval and lengths of the PR and ST segments. In [
7], associations were found between SBP and DBP and changes in the ECG at two intervals delineated by the peak of the R wave to the middle of the T wave and the mid of the T-wave to the peak of the R wave as indicated in
Figure 1, respectively, using machine learning (ML) [
11,
12].
1.2. HRV Signal
The temporal variation of sequential heartbeats (RR intervals) is termed HRV [
13]. From the ECG signal, R-peaks are first extracted and then HRV is deduced using computer programming based on the difference in RR intervals (
Figure 1). HRV reflects the activity of automatic nervous system and provides a window into the cardiac sympathetic and parasympathetic activities, which have significant physiological impact on heart rate rhythm and contractile function [
13]. HRV measurement is non-intrusive easy to perform and results are reproducible. Importantly, it confers both diagnostic and prognostic implications for wellness and cardiac disease [
14,
15]. High HRV is associated with normal subjects and reduced HRV may be pathological. HRV can be analyzed over either long or short durations [
14]. Long duration analysis encompasses activity throughout the day and night (24 h analysis), where as short-duration analysis uses only five-minutes of HRV data. In HT patients, HRV is affected by the presence of cardiovascular risk factors. In depressed patients with HT, HRV is associated with vascular cardiac and renal target organ damage [
13,
16]. The long-term (24-h) HRV is useful in the diagnosis of severe HT conditions. An increased sympathetic activity saturate the ability to modulate heart rate, hence HRV is depressed. To identify severe HRV or high-risk HRV, standard deviation of NN intervals should be less than 50 to 70 msec and HRV triangular index is less than 20 units. Similarly, the NN interval duration is 7.8 msec [
17]. In summary, HRV is a simple non-invasive method which can be used to assess the cardiovascular system [
18].
1.3. Photoplethysmography (PPG Signal)
PPG uses low-intensity infrared (IR) light sensor to detect the amount of light absorbed by or reflected from tissues supplied by the blood vessels. It produces photo electric signal either transmissive or reflective, which reflect the pulsatile blood volume in the area covered by sensors [
19,
20]. The PPG signal contains information about the arterial and venous circulatory system [
19,
20]. The PPG is correlated with and has been applied to the measurement of heart rate, BP, and blood oxygenation there by providing clinically useful information for physiological monitoring.
1.4. Ballistocardiogram (BCG) Signal
BCG measures the whole body motion in terms of displacement, velocity, and acceleration in response to the cyclical ejection of blood from the heart [
21]. It reflects the sum of factors linked to heart and blood vessel function, and used to diagnose various cardio vascular diseases [
21].
In this paper, we reviewed PPG, BCG, ECG and HRV signals related computer-aided diagnosis systems developed for the arterial HT. To the best of our knowledge this is the first review to provide unique ranges for nonlinear features for healthy control (HC), low-risk hypertension (LRHT), and high-risk hypertension (HRHT) ECG classes.
2. Methods and Material Used in Article Searching
This review was carried out based on the PRISMA model for the period between 09 October 2008 and 31 March 2021 [
22]. Science Direct, Web of Science, Google Scholar, PubMed, IEEE Explorer, and ResearchGate databases were systematically searched using the following keywords: (“hypertension”, “ECG and hypertension”, “HRV or hypertension”), “photoplethysmography (PPG) and hypertension”, “photoplethysmography (PPG) and ballistocardiogram (BCG)”, “unsupervised machine learning and hypertension”, “supervised machine learning and hypertension”, “detection of hypertension”, “convolution neural network (CNN)”, “Hypertension and Convolution neural network”, “machine learning and hypertension”, “hypertension and deep learning”, “recurrent neural network (RNN)”, “support vector machine (SVM)”, “automated detection of hypertension”, “detection of hypertension using ECG signal”, “hypertension or HRV signal”, “systolic blood pressure (SBP)”, “diastolic blood pressure (DBP)”, “electrocardiogram (ECG)”, “systolic blood pressure and hypertension”, “diastolic blood pressure and hypertension”, and “physiological signal and hypertension”. We identified 250 research articles containing these keywords on initial screening. Science Direct = 50, Web of Science = 40, Google Scholar = 40, PubMed = 50, IEEE Explorer = 50, and ResearchGate = 20. Among these, 103 duplicate articles were omitted.
We further excluded non-English articles and works that were not explicitly designed for diagnosis of HT.
Finally, 23 articles were selected for this review.
Figure 2 shows the flow diagram of article selection, where n is the number of articles.
7. Proposed Work after Understanding Review Studies
An outline of the proposed work is shown in
Figure 4. In this work, we used the public database used by authors in [
23,
24]. A total of 3694 ECG segments were obtained from (SHAREE, and PTB data base) [
23,
24]. The LRHT have 3172 (SHAREE database), HRHT (SHAREE database) have 442, and HC class have 80 (PTB database) ECG signals segmented in to 2 minute duration. A single ECG-lead V5 have been chosen from both databases. To match the sampling frequency of SHAREE and PTB database, we have down-sampled the data obtained from PTB database. The details of the extracted features are discussed below:
7.1. Features Extracted to the Proposed Work
7.1.1. Sample Entropy (SeEn)
It is a measure of uncertainty in the non-linear signal [
2,
23]. SeEn can be computed as;
where a is the
length of vector, and b is the length
l of vector with
[
23].
7.1.2. Approximate Entropy (ApEn)
It is an approximation technique which is useful to measure homogeneity and complexities in time-series data containing noise because of its stability to distinguish closely linked stochastic processes. It is effective in short data intervals to differentiate between chaotic and noisy time-series data [
2,
27,
28,
30]. For finite length N, ApEn is computed as:
is a vector, while
m = 2. The tolerance value
r = 0.20 lies between 0.15 to 0.25 [
28].
7.1.3. Renyi Entropy (ReEn)
It is used to measure the spectral intricacy of time-series signals and generalized as the Shannon entropy [
2,
13]. ReEn is computed as :
where
K = discrete random variable,
is order (
) of ReEn,
= denoted the total spectral-power [
24,
50].
7.1.4. Wavelet Entropy (WlEn)
It is used to calculate the degree of disorder in an ECG signals [
2,
23,
40,
51].
where
i is a resolution level and
is probabilities with respect to
i [
52].
7.1.5. Log Energy (LOGE)
It is the logarithm of an energy of the ECG signal. The mathematical expression of LOGE is given below [
53].
where
is define the log energy of
rth time-series, and the amplitude of
nth sample of
rth time-series is
.
7.1.6. Signal Fractal Dimension (SLFD)
It is used to measure similarity and complexity in physiological signals. It is the ratio of fractal pattern with respect to which it is measured [
53]. It is given by:
7.1.7. Hurst Exponent (HE)
It is a measure of repeat-ability [
54]. The generalize equation of HE is as follows:
Here, Y represents the length of time-series data, while
denoted the re-scaled range value. The difference of maximum and minimum value of mean is considered as A. However, B represents the standard deviation.
7.1.8. Largest Lyapunov Exponent (LLE)
LLE is used to identity chaos in the physiological signal [
55].
7.1.9. HOS Bispectrum (HOSB)
A third-order statistics computation is know as an HOS bispectrum [
56]. The Fourier transform of third order cumulant is the bispectrum
of a signal [
57]. In a two-dimensional frequency plot, the bispectrum shows the cross-correlation between frequency components. Hence, the HOSB is define as:
However, HOSB represents the triple product of its two frequencies. To define principle region or non-redundant region sufficient condition
0,
,
≤
must satisfied [
56]. To analyze the bispectrum plots, different parameters namely moments, centroid, and entropies of the distribution may be extracted [
56]. These features and parameters are as follows:
- (a)
Normalized bispectral entropy (NBE) [
56,
58]:
where
and
represents the principle region.
- (b)
Normalized bispectral square entropy (NBSE) [
58]:
where
. Both entropies are computed for parameters which have values between 0 and 1.
- (c)
A weighted center feature of bispectrum (WCOB) is described as [
58]:
Here,
k and
l represents the frequency bin index in the principle region of bispectrum plot [
58]. Similarly, some moments related features are given below:
- (d)
Bispectrum logarithmic amplitude feature [
58]:
- (e)
Bispectrum sum of logarithmic amplitude of diagonal elements feature [
58]:
- (f)
Bispectrum first-order spectral moments of amplitude of diagonal elements feature [
58]:
- (g)
Bispectrum mean magnitude feature [
58]:
- (h)
Bispectrum phase entropy feature [
58] :
Here, the number of points is represented by L of the principle region,
is the phase angle, and
refer the space of the region [
58].
7.1.10. Higher Order Spectral Cumulant (HOSC)
Obtaining the nonlinear dynamical characteristics of ECG signal using lower-order (first) of statistics is complex [
57]. Therefore, higher order statistics, such as second, third and fourth, are widely used to analyze the ECG signal. Hence, HOS cumulant higher order statistics features are used in analysis of non-stationary ECG signals in various applications [
57].
Let
is representing a zero mean random process for
q dimensional multivariate. While
,
,
, and
are the order of moments from first to fourth [
57]. As well as
represents the lag-parameters. Hence, using the non-linear combinations of the moments, cumulant can be computed.
Here
,
,
, and
are the order of cumulants from first to fourth. In this work, we have computed second, third, and fourth order cumulants.
7.1.11. Recurrence Plot (RP)
For the physiological signal in the time domain, RP can find hidden patterns which are not clearly identifiable [
57]. Recurrence can be defined as the value of
k and
l dropped below the threshold value
is known as recurrence. Assuming
in an
L dimensional space be the
point. However, a dot is considered at
as the distance between the
and
is closer. When
, the recurrence plots are symmetric along the diagonal, as
is close to
, then
is close to
. Hence, a RP is
square of an array dots [
57]. This may also be presented in time-related space as a
matrix. The yellow dot implies there has been a recurrence.
7.1.12. Recurrence Quantification Analysis (RQA)
RQA is better choice for dynamical system used to measure the number and duration of recurrence. It helps to measure and analyze the recurrence plot of non-stationary physiological signal [
57]. In the time domain, RQA evaluates the non-stationary and hidden periodicity of signals. The following RQA features are used in this work:
(a) Recurrence rate (RR), (b) Determinism (DET), (c) Entropy (ENT), (d) Laminarity (LMR) [
57].
7.2. Results
The proposed work is performed on LRHT, HRHT and HC subjects. A total of 3694 ECG segments were obtained from SHAREE and PTB databases. The LRHT have 3172, HRHT have 442, and HC class have 80 ECG signal segments of 2 min duration.
An experiment is performed on MATLAB 2016b version 9.1.10 (licensed) and work station (personal computer) with Intel i7 processor, 16GB RAM, 1TB HD, and 4 GB graphics card. We have tried several non-linear features to obtain the optimum results. However, higher order spectral cumulant, bispectrum and recurrence quantitative analysis (RQA) yielded optimum results.
In addition to this, the optimum performance were obtained using the combination of HOS bispectrum, cumulant and RQA feature. A total of 9 bispectrum-based features are extracted and shown in
Table 5. The detailed of RQA and HOS cumulant features are presented in
Table 6 and
Table 7 respectively.
The highest classification accuracy, sensitivity and specificity of 98.05%, 95.66%, and 96.58%, respectively are obtained using support vector machine classifier with ten-fold cross-validation strategy.
Table 8 represents the confusion matrix obtained for SVM classifier using all bispectrum, cumulant and RQA features.
Table 9 shows the performance measures obtained for each class using HOS bispectrum, cumulants and RQA features with SVM classifier. The highest AUC of 1.00 is obtained using SVM classifier (
Figure 5).
Figure 6 shows the plot of accuracy % versus the number of features. HOS bispectrum magnitude and contour plots for HC, LRHT and HRHT class are shown in the
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11 and
Figure 12.
Figure 13 shows the recurrence plots for HC, LRHT, and HRHT classes. Summary of classification performance obtained using various combination of features is shown in
Table 10.
8. Discussion
Table 2,
Table 3 and
Table 4 summarizes all studies using ECG, BCG, PPG, and HRV signals. It is also evident from
Table 2 that the ECG-based computed aided diagnosis system obtained the highest area under receiver operating characteristics (AUC=1.00) performance compared to rest of methods. Moreover,
Table 2 represents the highest classification accuracy of 99.99% using ECG signals.
Table 11 lists the summary of artificial intelligence (AI) techniques used to classify HT based on ECG and HRV signals. On the other hand,
Table 2,
Table 3 and
Table 4 summarize the methods, features, subjects, results and type of databases that have been used to diagnose HT using HRV, ECG, BCG, and PPG signals. In both ECG and HRV signal-based studies, authors have used transformational approaches, converted time-domain signals into the frequency domain, extracted non-linear features and classified using SVM and KNN classifiers. The summary of automated systems developed for HT are as follows:
Rajput et al. [
8] developed an HDI accurately using ECG signals to stratify low-risk versus high-risk HT with a single numeric value.
Poddar et al. [
28] used HRV signals to classify HT and normal subjects using SVM classifier with 100% accuracy using 20 features. They have used a balanced data set of 56 normal and 57 HT subjects in their study.
Rajput et al. [
23] classified ECG signals into three classes (LRHT, HRHT, and HC) using features extracted from the five-level wavelet decomposition of ECG signals. They have obtained 99.95% classification accuracy using SeEn and WeEn features with unbalanced data set. Testing error is found to be only 3.26% with hold-out validation method.
Soh et al. [
24] developed a CNN architecture for the classification of normal and HT ECG classes and achieved an accuracy of 99.99%, sensitivity of 100% and specificity of 99.97%.
In this work, bispectrum based features obtained the highest classification accuracy of 96.5% among the nonlinear features. The summary of classification performance obtained using various combination of features is shown in
Table 10.
It can be noted from
Table 5 that bispectrum features are clinically significant and show clear difference between three classes. On the other hand, Hos cumulant order-three mean and standard values yielded the large difference among three class as mentioned in
Table 7, hence it is useful for 1-D signal features extraction. However, RQA RR feature also comprising distinct difference in all three class.
It is well recognized that the bispectrum conserves the phase information [
57]. Because of this property, it is used to examine quadratic nonlinear differences between various frequency components of ECG signals. Such interactions have been observed between different frequencies of three classes of ECG signals. This analysis may be useful for detecting changes in ECG signals. For a normal, LRHT, and HRHT ECG signal, the magnitude and its contour representation are shown in the
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11 and
Figure 12. It can be noted from
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11 and
Figure 12 that, these plots are unique and can be used to discriminate the class of the ECG signal (HC, LRHT, or HRHT). Similarly,
Figure 13 shows the recurrence plots for HC, LRHT and HRHT ECG signals. These plots are unique and can also be used to differentiate the three classes. We have obtained the highest classification performance only using HOS bispectrum features without transforming the ECG signals (
Table 10). The advantages of proposed study are: (i) many nonlinear features are employed which can be used for the classification. (ii) Proposed unique HOS bispectrum and recurrence plots for three classes. (iii) HOS-based features are more robust to noise.
In general, works conducted using ML and DL coupled with ECG signals have yielded the highest and optimum performance. The works done in
Table 2,
Table 3 and
Table 4,
2,
8,
13,
16,
21,
23,
24,
25,
26,
34,
35,
26] have used public (open source) databases, while the rest of the studies have used private databases. This underscores the importance of public databases for computer aided diagnosis systems development.
Liang et al. [
20,
34,
35] detected HT using PPG signal in three separate studies using public and private databases. They achieved a best classification F-score of 94.84%.
Liu et al. [
21] diagnosed HT from BCG-derived HRV signal, and achieved highest classification accuracy of 84.4% using ML.
Such results demonstrate the effectiveness of transformation methods that combine nonlinear and entropy-based features. ML methods work well with balanced and smaller databases. The performance of ML models also depends on the features extracted and classifiers used.
In the future, we intend to use deep learning architectures to detect the HT ECG signals using large database [
59]. The biggest challenge for this study is the availability of the large public database.
Figure 14 illustrate the cloud based proposed model. Initially, the ECG, PPG, BCG, and HRV signal recorded from patients and stored in hospital database. The stored signals were sent to the cloud based model, where it is installed. The cloud based model analyze the provided data and detect the hypertension accurately. To the same, the results were revert from cloud to hospital. Hence, the Doctors can compare the results obtained by cloud based model as well as manually finding.
Table 12 have all the list of abbreviation used in the paper.