The Patatin–Like Phospholipase Domain Containing Protein 7 Regulates Macrophage Classical Activation through SIRT1/NF-κB and p38 MAPK Pathways
Abstract
:1. Introduction
2. Results
2.1. Pnpla7 Gene Expression Is Downregulated during Macrophage M1 Polarization
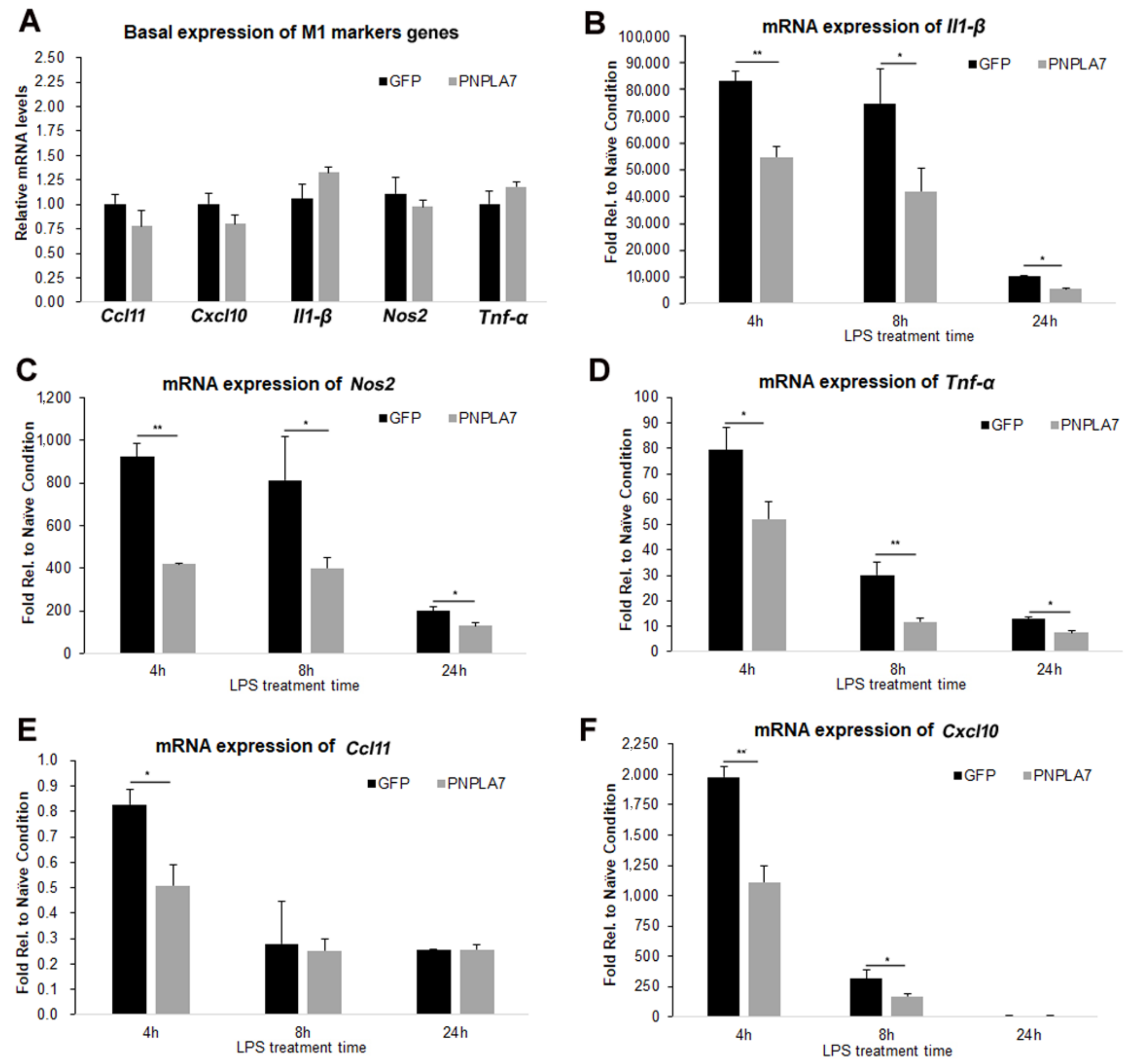
2.2. Overexpression of PNPLA7 Suppresses Proinflammatory Characteristics during M1 Polarization of Macrophages
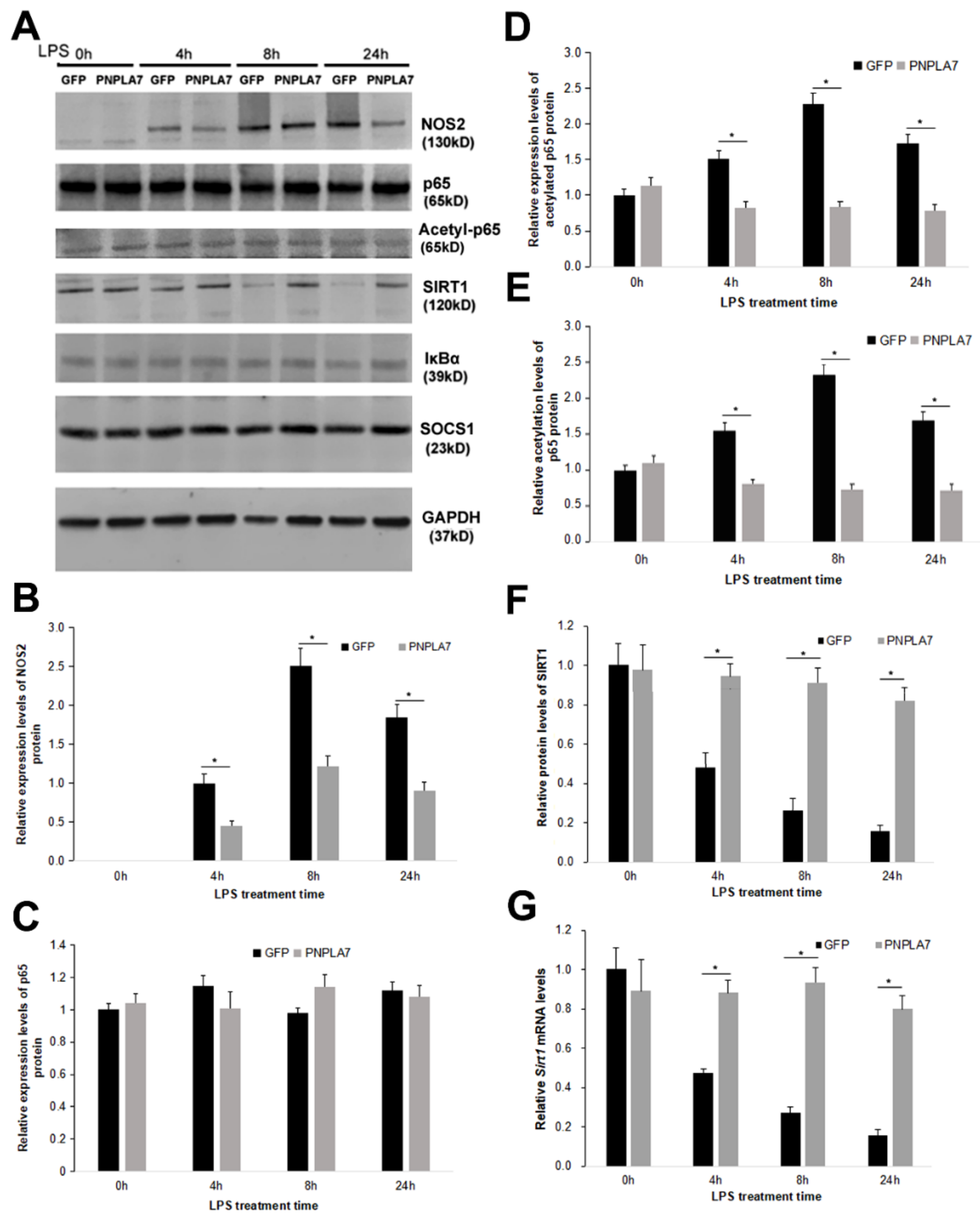
2.3. PNPLA7 Stabilizes Sirt1 Expression to Restrict Acetylation of NF-κB/p65
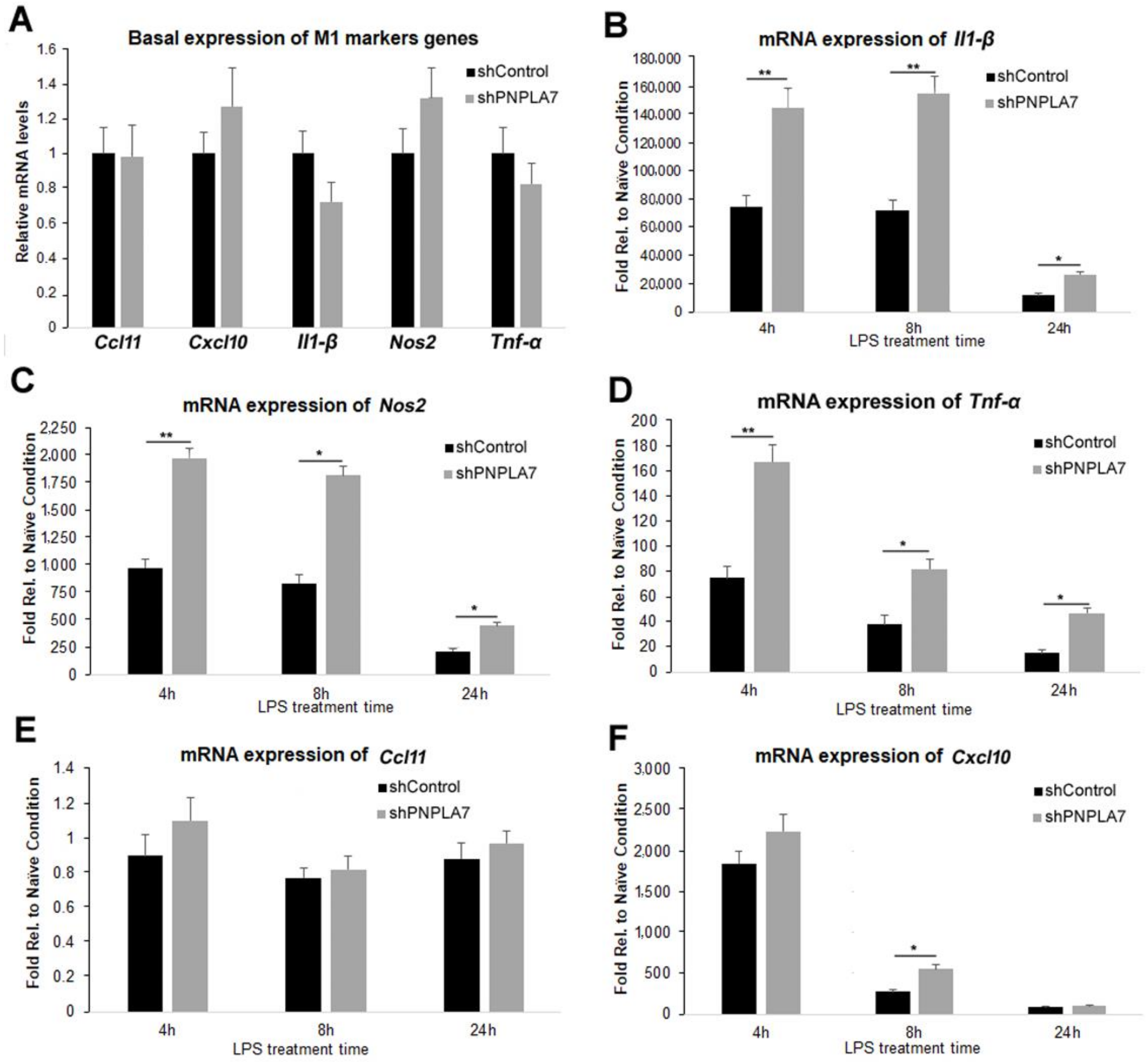
2.4. Knockdown of Pnpla7 Favors Proinflammatory Characteristics during M1 Polarization of Macrophages
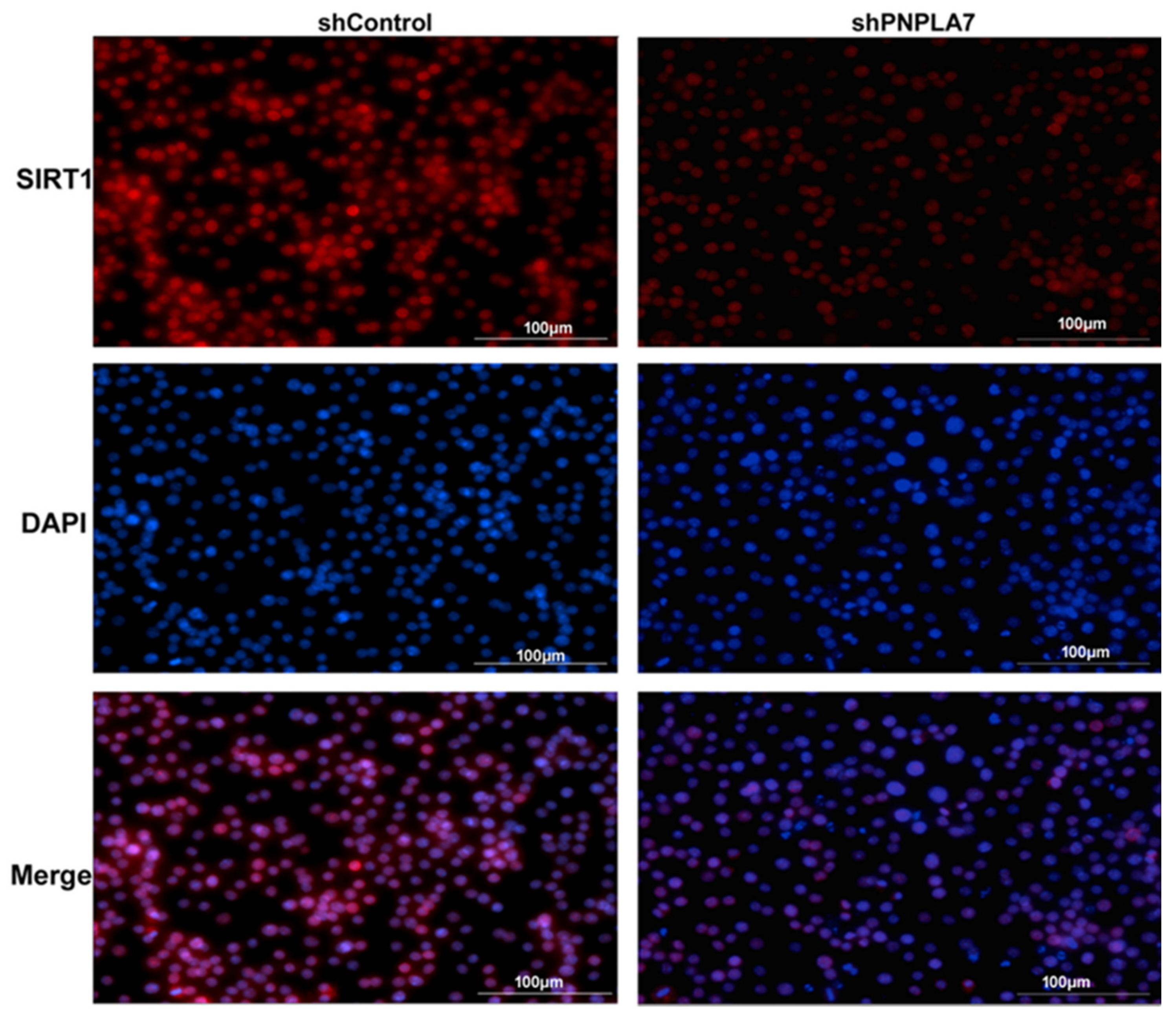
2.5. Reduction of PNPLA7 Downregulates SIRT1 to Promote Acetylation of NF-κB/p65
2.6. PNPLA7 Influences the Phosphorylation of p38 MAPK
3. Discussion
4. Material and Methods
4.1. Materials
4.2. Cell Culture and Treatment
4.3. BMDM Culture and Treatment
4.4. Generation of Lentivirus and Stable Infection of RAW264.7 Cells
4.5. Real-Time Quantitative PCR
4.6. Immunoblotting Analysis
4.7. Immunofluorescence Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [Green Version]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, Y.; Fu, M.; Xin, H.B. Polarizing macrophages in vitro. Methods Mol. Biol. 2018, 1784, 119–126. [Google Scholar]
- Liu, P.; Zhu, W.; Chen, C.; Yan, B.; Zhu, L.; Chen, X.; Peng, C. The mechanisms of lysophosphatidylcholine in the development of diseases. Life Sci. 2020, 247, 117443. [Google Scholar] [CrossRef]
- Knuplez, E.; Marsche, G. An updated review of pro- and anti-inflammatory properties of plasma lysophosphatidylcholines in the vascular system. Int. J. Mol. Sci. 2020, 21, 4501. [Google Scholar] [CrossRef]
- Hung, N.D.; Sok, D.E.; Kim, M.R. Prevention of 1-palmitoyl lysophosphatidylcholine-induced inflammation by polyunsaturated acyl lysophosphatidylcholine. Inflamm. Res. 2012, 61, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Qiu, C.; Zhao, L. Lysophosphatidylcholine perpetuates macrophage polarization toward classically activated phenotype in inflammation. Cell Immunol. 2014, 289, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, K.E.; Andersson, L.; Nilsson, J.; Björkbacka, H. Nanomolar concentrations of lysophosphatidylcholine recruit monocytes and induce proinflammatory cytokine production in macrophages. Biochem. Biophys. Res. Commun. 2008, 370, 348–352. [Google Scholar] [CrossRef]
- Tian, C.; Huang, R.; Tang, F.; Lin, Z.; Cheng, N.; Han, X.; Li, S.; Zhou, P.; Deng, S.; Huang, H.; et al. Transient receptor potential Ankyrin 1 contributes to lysophosphatidylcholine-induced intracellular calcium regulation and THP-1-derived macrophage activation. J. Membr. Biol. 2020, 253, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Assunção, L.S.; Magalhães, K.G.; Carneiro, A.B.; Molinaro, R.; Almeida, P.E.; Atella, G.C.; Castro-Faria-Neto, H.C.; Bozza, P.T. Schistosomal-derived lysophosphatidylcholine triggers M2 polarization of macrophages through PPARγ dependent mechanisms. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Law, S.H.; Chan, M.L.; Marathe, G.K.; Parveen, F.; Chen, C.H.; Ke, L.Y. An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci. 2019, 20, 1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Feng, C.; Li, L.; Xu, G.; Gu, H.; Li, S.; Li, D.; Liu, M.; Han, S.; Zheng, B. Lipid receptor G2A-mediated signal pathway plays a critical role in inflammatory response by promoting classical macrophage activation. J. Immunol. 2021, 206, 2338–2352. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Hikiji, H.; Okinaga, T.; Hashidate-Yoshida, T.; Shindou, H.; Ariyoshi, W.; Shimizu, T.; Tominaga, K.; Nishihara, T. Essential role of lysophosphatidylcholine acyltransferase 3 in the induction of macrophage polarization in PMA-treated U937 cells. J. Cell. Biochem. 2015, 116, 2840–2848. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Varin, A.; Filomenko, R.; Lopez, T.; Athias, A.; Gambert, P.; Blache, D.; Thomas, C.; Gautier, T.; Lagrost, L.; et al. Liver x receptor regulates arachidonic acid distribution and eicosanoid release in human macrophages: A key role for lysophosphatidylcholine acyltransferase 3. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1171–1179. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Z.; Huan, C.; Jiang, X.C. Macrophage lysophosphatidylcholine acyltransferase 3 deficiency-mediated inflammation is not sufficient to induce atherosclerosis in a mouse model. Front. Cardiovasc. Med. 2019, 5, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heier, C.; Kien, B.; Huang, F.; Eichmann, T.O.; Xie, H.; Zechner, R.; Chang, P.A. The phospholipase PNPLA7 functions as a lysophosphatidylcholine hydrolase and interacts with lipid droplets through its catalytic domain. J. Biol. Chem. 2017, 292, 19087–19098. [Google Scholar] [CrossRef] [Green Version]
- Wilson, P.A.; Gardner, S.D.; Lambie, N.M.; Commans, S.A.; Crowther, D.J. Characterization of the human patatin-like phospholipase family. J. Lipid Res. 2006, 47, 1940–1949. [Google Scholar] [CrossRef] [Green Version]
- Kienesberger, P.C.; Oberer, M.; Lass, A.; Zechner, R. Mammalian patatin domain containing proteins: A family with diverse lipolytic activities involved in multiple biological functions. J. Lipid Res. 2009, 50, S63–S68. [Google Scholar] [CrossRef] [Green Version]
- Kienesberger, P.C.; Lass, A.; Preiss-Landl, K.; Wolinski, H.; Kohlwein, S.D.; Zimmermann, R.; Zechner, R. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J. Biol. Chem. 2008, 283, 5908–5917. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.A.; Wang, Z.X.; Long, D.X.; Qin, W.Z.; Wei, C.Y.; Wu, Y.J. Identification of two novel splicing variants of murine NTE-related esterase. Gene 2012, 497, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Sun, T.; Heie, C.; Gao, H.; Xu, H.; Huang, F. Interaction of the lysophospholipase PNPLA7 with lipid droplets through the catalytic region. Mol. Cells 2020, 43, 286–297. [Google Scholar]
- Wang, X.; Guo, M.; Wang, Q.; Wang, Q.; Zuo, S.; Zhang, X.; Tong, H.; Chen, J.; Wang, H.; Chen, X.; et al. The patatin-like phospholipase domain containing protein 7 facilitates VLDL secretion by modulating ApoE stability. Hepatology 2020, 72, 1569–1585. [Google Scholar] [CrossRef]
- Mazgaeen, L.; Gurung, P. Recent advances in lipopolysaccharide recognition systems. Int. J. Mol. Sci. 2020, 21, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciesielska, A.; Matyjek, M.; Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 2021, 78, 1233–1261. [Google Scholar] [CrossRef]
- Kauppinen, A.; Suuronen, T.; Ojala, J.; Kaarniranta, K.; Salminen, A. Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell. Signal. 2013, 25, 1939–1948. [Google Scholar] [CrossRef]
- Alam, S.; Liu, Q.; Liu, S.; Liu, Y.; Zhang, Y.; Yang, X.; Liu, G.; Fan, K.; Ma, J. Up-regulated cathepsin C induces macrophage M1 polarization through FAK-triggered p38 MAPK/NF-κB pathway. Exp. Cell Res. 2019, 382, 111472. [Google Scholar] [CrossRef]
- Aflaki, E.; Balenga, N.A.; Luschnig-Schratl, P.; Wolinski, H.; Povoden, S.; Chandak, P.G.; Bogner-Strauss, J.G.; Eder, S.; Konya, V.; Kohlwein, S.D.; et al. Impaired Rho GTPase activation abrogates cell polarization and migration in macrophages with defective lipolysis. Cell. Mol. Life Sci. 2011, 68, 3933–3947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashley, J.W.; Hancock, W.D.; Nelson, A.J.; Bone, R.N.; Tse, H.M.; Wohltmann, M.; Turk, J.; Ramanadham, S. Polarization of macrophages toward M2 phenotype is favored by reduction in iPLA2β (group VIA phospholipase A2). J. Biol. Chem. 2016, 291, 23268–23281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabral, D.; van den Bogaart, G. The roles of phospholipase A2 in phagocytes. Front. Cell Dev. Biol. 2021, 9, 673502. [Google Scholar]
- Abate, W.; Alrammah, H.; Abate, W.; Kiernan, M.; Tonks, A.J.; Jackson, S.K. Lysophosphatidylcholine acyltransferase 2 (LPCAT2) co-localises with TLR4 and regulates macrophage inflammatory gene expression in response to LPS. Sci. Rep. 2020, 10, 10355. [Google Scholar]
- Ishihara, K.; Kuroda, A.; Sugihara, K.; Kanai, S.; Nabe, T.; Akiba, S. Regulation of macrophage differentiation and polarization by group IVC phospholipase A2. Biochem. Biophys. Res. Commun. 2011, 416, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef]
- De Gregorio, E.; Colell, A.; Morales, A.; Marí, M. Relevance of SIRT1-NF-κB axis as therapeutic target to ameliorate inflammation in liver disease. Int. J. Mol. Sci. 2020, 21, 3858. [Google Scholar] [CrossRef]
- Shen, Z.; Ajmo, J.M.; Rogers, C.Q.; Liang, X.; Le, L.; Murr, M.M.; Peng, Y.; You, M. Role of SIRT1 in regulation of LPS- or two ethanol metabolites-induced TNF-alpha production in cultured macrophage cell lines. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G1047–G1053. [Google Scholar] [CrossRef]
- Han, S.; Li, Z.; Han, F.; Jia, Y.; Qi, L.; Wu, G.; Cai, W.; Xu, Y.; Li, C.; Zhang, W.; et al. ROR alpha protects against LPS-induced inflammation by down-regulating SIRT1/NF-kappa B pathway. Arch. Biochem. Biophys. 2019, 668, 1–8. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Schenk, S.; Imamura, T.; Babendure, J.L.; Sonoda, N.; Bae, E.J.; Oh, D.Y.; Lu, M.; Milne, J.C.; Westphal, C.; et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E419–E428. [Google Scholar] [CrossRef] [Green Version]
- Xue, B.; Yang, Z.; Wang, X.; Shi, H. Omega-3 polyunsaturated fatty acids antagonize macrophage inflammation via activation of AMPK/SIRT1 pathway. PLoS ONE 2012, 7, e45990. [Google Scholar] [CrossRef]
- Inoue, T.; Tanaka, M.; Masuda, S.; Ohue-Kitano, R.; Yamakage, H.; Muranaka, K.; Wada, H.; Kusakabe, T.; Shimatsu, A.; Hasegawa, K.; et al. Omega-3 polyunsaturated fatty acids suppress the inflammatory responses of lipopolysaccharide-stimulated mouse microglia by activating SIRT1 pathways. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, C.; Fan, S.; Wu, S.; Yang, F.; Fang, Z.; Fu, H.; Li, Y. Omega-3 polyunsaturated fatty acid attenuates the inflammatory response by modulating microglia polarization through SIRT1-mediated deacetylation of the HMGB1/NF-κB pathway following experimental traumatic brain injury. J. Neuroinflamm. 2018, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- Najt, C.P.; Khan, S.A.; Heden, T.D.; Witthuhn, B.A.; Perez, M.; Heier, J.L.; Mead, L.E.; Franklin, M.P.; Karanja, K.K.; Graham, M.J.; et al. Lipid droplet-derived monounsaturated fatty acids traffic via PLIN5 to allosterically activate SIRT1. Mol. Cell 2020, 77, 810–824. [Google Scholar] [CrossRef] [PubMed]
- Bode, J.G.; Ehlting, C.; Häussinger, D. The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell. Signal. 2012, 24, 1185–1194. [Google Scholar] [CrossRef]
- Carneiro, A.B.; Iaciura, B.M.; Nohara, L.L.; Lopes, C.D.; Veas, E.M.; Mariano, V.S.; Bozza, P.T.; Lopes, U.G.; Atella, G.C.; Almeida, I.C.; et al. Lysophosphatidylcholine triggers TLR2- and TLR4-mediated signaling pathways but counteracts LPS-induced NO synthesis in peritoneal macrophages by inhibiting NF-κB translocation and MAPK/ERK phosphorylation. PLoS ONE 2013, 8, e76233. [Google Scholar]
- Quan, H.; Hur, Y.H.; Xin, C.; Kim, J.M.; Choi, J.I.; Kim, M.Y.; Bae, H.B. Stearoyl lysophosphatidylcholine enhances the phagocytic ability of macrophages through the AMP-activated protein kinase/p38 mitogen activated protein kinase pathway. Int. Immunopharmacol. 2016, 39, 328–334. [Google Scholar] [CrossRef]
- Baig, M.S.; Zaichick, S.V.; Mao, M.; de Abreu, A.L.; Bakhshi, F.R.; Hart, P.C.; Saqib, U.; Deng, J.; Chatterjee, S.; Block, M.L.; et al. NOS1-derived nitric oxide promotes NF-κB transcriptional activity through inhibition of suppressor of cytokine signaling-1. J. Exp. Med. 2015, 212, 1725–1738. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]




| Gene Name | Primer Sequence (5′–3′) |
|---|---|
| Ccl11 Cxcl10 Il1β Nos2 Pnpla7 Pnpla6 Sirt1 Tnfα 36B4 | GAATCACCAACAACAGATGCAC ATCCTGGACCCACTTCTTCTT GCCGTCATTTTCTGCCTCAT GCTTCCCTATGGCCCTCATT CCATGGCACATTCTGTTCAAA GCCCATCAGAGGCAAGGA CAGCTGGGCTGTACAAACCTT CATTGGAAGTGAAGCGTTTCG CGTGTT TTCCAACGACCACC TCTGCTAGTGCCCTGAGGAT CGGGTGCAGAAAACTCCAG CGCATAATCTTCCGGCCATAGA GTCACACGCCAGCTCTAGT GACAGAAACCCCAGCTCCA TTCGGCTACCCCAAGTTCAT CGCACGTAGTTCCGCTTTC GCTTCATTGTGGGAGCAGACA CATGGTGTTCTTGCCCATCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Heier, C.; Pang, H.; Wang, Y.; Huang, F.; Chang, P. The Patatin–Like Phospholipase Domain Containing Protein 7 Regulates Macrophage Classical Activation through SIRT1/NF-κB and p38 MAPK Pathways. Int. J. Mol. Sci. 2022, 23, 14983. https://doi.org/10.3390/ijms232314983
Zhao Z, Heier C, Pang H, Wang Y, Huang F, Chang P. The Patatin–Like Phospholipase Domain Containing Protein 7 Regulates Macrophage Classical Activation through SIRT1/NF-κB and p38 MAPK Pathways. International Journal of Molecular Sciences. 2022; 23(23):14983. https://doi.org/10.3390/ijms232314983
Chicago/Turabian StyleZhao, Zheng, Christoph Heier, Huimin Pang, Yu Wang, Feifei Huang, and Pingan Chang. 2022. "The Patatin–Like Phospholipase Domain Containing Protein 7 Regulates Macrophage Classical Activation through SIRT1/NF-κB and p38 MAPK Pathways" International Journal of Molecular Sciences 23, no. 23: 14983. https://doi.org/10.3390/ijms232314983





