Integrating Common Risk Factors with Polygenic Scores Improves the Prediction of Type 2 Diabetes
Abstract
:1. Introduction
2. Results
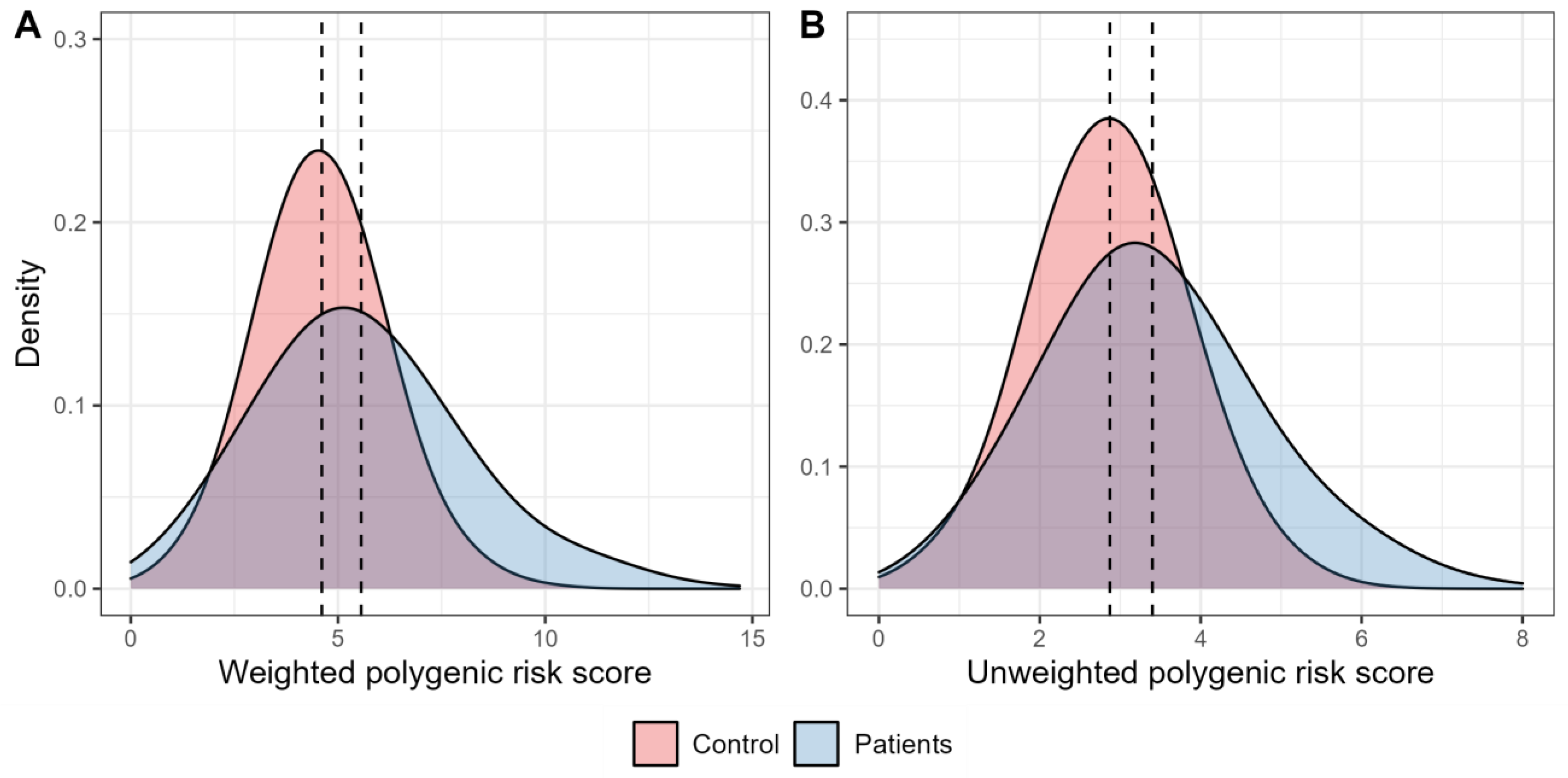
2.1. Association Analysis
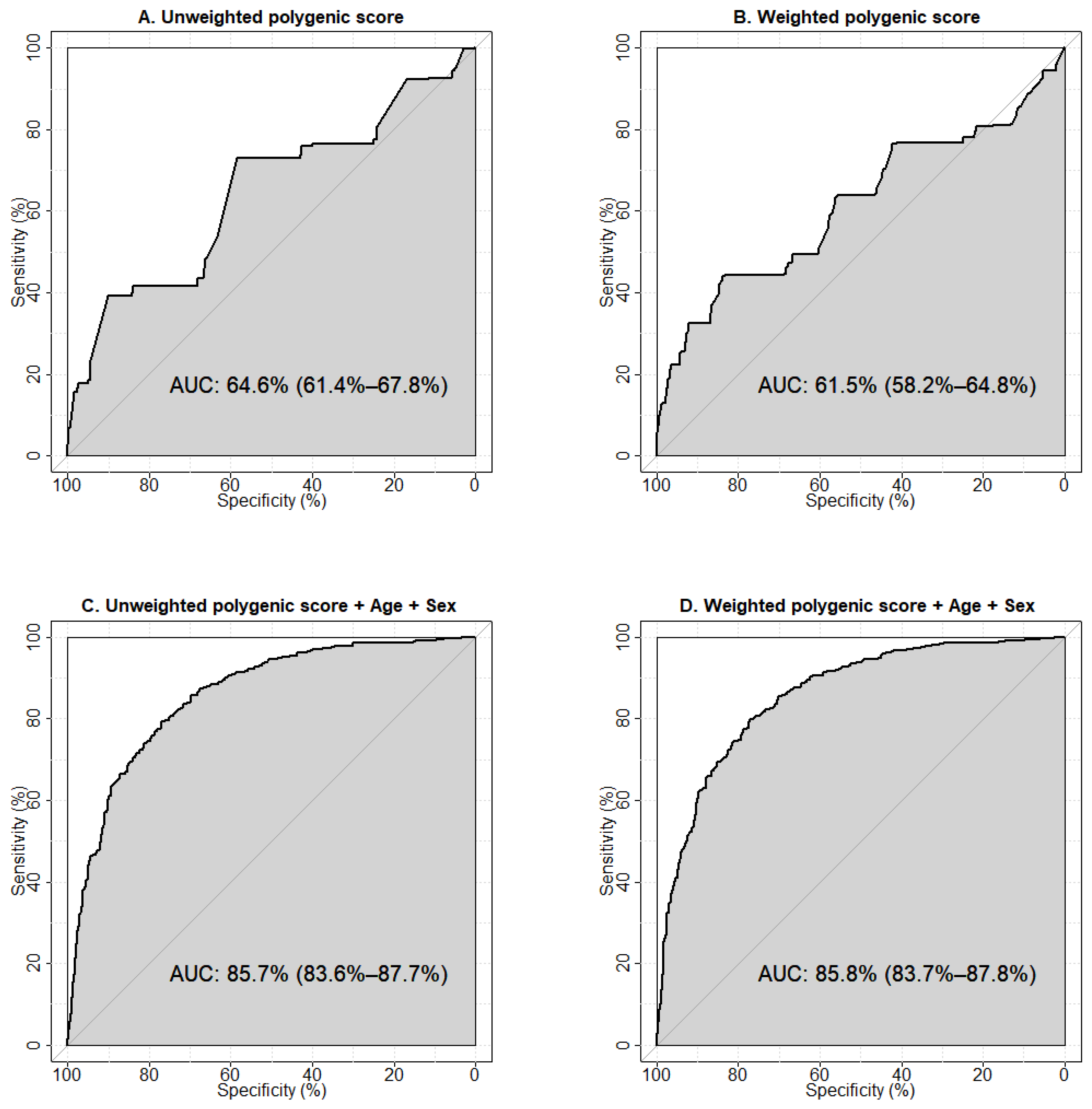
2.2. Receiver Operator Characteristic (ROC) Analysis
2.3. Net Reclassification Improvement Analysis
3. Discussion
4. Materials and Methods
4.1. Study Sample
4.2. Anthropometric Measurements and Biochemical Assays
4.3. Genotyping and Quality Control
4.4. Power Analysis
4.5. Association Analysis
4.6. Polygenic Score Calculation
4.7. Receiver Operator Characteristic Analysis
4.8. Net Reclassification Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Preventing and Managing the Global Epidemic: Report on a WHO Consultation (WHO Technical Report Series 894); World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Chen, H. Cellular inflammatory responses: Novel insights for obesity and insulin resistance. Pharmacol. Res. 2006, 53, 469–477. [Google Scholar] [CrossRef]
- Tourniaire, F.; Romier-Crouzet, B.; Lee, J.H.; Marcotorchino, J.; Gouranton, E.; Salles, J.; Malezet, C.; Astier, J.; Darmon, P.; Blouin, E.; et al. Chemokine Expression in Inflamed Adipose Tissue Is Mainly Mediated by NF-kappaB. PLoS ONE 2013, 8, e66515. [Google Scholar] [CrossRef]
- Flak, J.N.; Myers, M.G., Jr. CNS Mechanisms of Leptin Action. Mol. Endocrinol. 2016, 30, 3–12. [Google Scholar] [CrossRef] [Green Version]
- da Silva, A.A.; do Carmo, J.M.; Hall, J.E. CNS Regulation of Glucose Homeostasis: Role of the Leptin-Melanocortin System. Curr. Diabetes Rep. 2020, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Berger, C.; Kloting, N. Leptin Receptor Compound Heterozygosity in Humans and Animal Models. Int. J. Mol. Sci. 2021, 22, 4475. [Google Scholar] [CrossRef] [PubMed]
- do Carmo, J.M.; da Silva, A.A.; Gava, F.N.; Moak, S.P.; Dai, X.; Hall, J.E. Impact of leptin deficiency compared with neuronal-specific leptin receptor deletion on cardiometabolic regulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019, 317, R552–R562. [Google Scholar] [CrossRef] [PubMed]
- Howlader, M.; Sultana, M.I.; Akter, F.; Hossain, M.M. Adiponectin gene polymorphisms associated with diabetes mellitus: A descriptive review. Heliyon 2021, 7, e07851. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, F.; Meyre, D.; Froguel, P. Adiponectin, type 2 diabetes and the metabolic syndrome: Lessons from human genetic studies. Expert Rev. Mol. Med. 2006, 8, 1–12. [Google Scholar] [CrossRef]
- Vasseur, F.; Helbecque, N.; Dina, C.; Lobbens, S.; Delannoy, V.; Gaget, S.; Boutin, P.; Vaxillaire, M.; Lepretre, F.; Dupont, S.; et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum. Mol. Genet. 2002, 11, 2607–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.M.; Doss, H.M.; Kim, K.S. Multifaceted Physiological Roles of Adiponectin in Inflammation and Diseases. Int. J. Mol. Sci. 2020, 21, 1219. [Google Scholar] [CrossRef]
- Burhans, M.S.; Hagman, D.K.; Kuzma, J.N.; Schmidt, K.A.; Kratz, M. Contribution of Adipose Tissue Inflammation to the Development of Type 2 Diabetes Mellitus. Compr. Physiol. 2018, 9, 1–58. [Google Scholar] [CrossRef]
- Pan, X.; Kaminga, A.C.; Wen, S.W.; Liu, A. Chemokines in Prediabetes and Type 2 Diabetes: A Meta-Analysis. Front. Immunol. 2021, 12, 622438. [Google Scholar] [CrossRef]
- Sterk, M.; Krizancic Bombek, L.; Skelin Klemen, M.; Slak Rupnik, M.; Marhl, M.; Stozer, A.; Gosak, M. NMDA receptor inhibition increases, synchronizes, and stabilizes the collective pancreatic beta cell activity: Insights through multilayer network analysis. PLoS Comput. Biol. 2021, 17, e1009002. [Google Scholar] [CrossRef]
- Lotta, L.A.; Stewart, I.D.; Sharp, S.J.; Day, F.R.; Burgess, S.; Luan, J.; Bowker, N.; Cai, L.; Li, C.; Wittemans, L.B.L.; et al. Association of Genetically Enhanced Lipoprotein Lipase-Mediated Lipolysis and Low-Density Lipoprotein Cholesterol-Lowering Alleles With Risk of Coronary Disease and Type 2 Diabetes. JAMA Cardiol. 2018, 3, 957–966. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Cheng, R.; Huang, C.; Takahashi, Y.; Yang, Y.; Benyajati, S.; Chen, Y.; Zhang, X.A.; Ma, J.X. A novel role of LRP5 in tubulointerstitial fibrosis through activating TGF-β/Smad signaling. Signal Transduct. Target. Ther. 2020, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Leanza, G.; Fontana, F.; Lee, S.Y.; Remedi, M.S.; Schott, C.; Ferron, M.; Hamilton-Hall, M.; Alippe, Y.; Strollo, R.; Napoli, N.; et al. Gain-of-Function Lrp5 Mutation Improves Bone Mass and Strength and Delays Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes. J. Bone Miner. Res. 2021, 36, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.F.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Manolescu, A.; Sainz, J.; Helgason, A.; Stefansson, H.; Emilsson, V.; Helgadottir, A.; et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006, 38, 320–323. [Google Scholar] [CrossRef]
- Di Narzo, A.; Frades, I.; Crane, H.M.; Crane, P.K.; Hulot, J.-S.; Kasarskis, A.; Hart, A.; Argmann, C.; Dubinsky, M.; Peter, I.; et al. Meta-analysis of sample-level dbGaP data reveals novel shared genetic link between body height and Crohn’s disease. Hum. Genet. 2021, 140, 865–877. [Google Scholar] [CrossRef]
- Hodgson, S.; Huang, Q.Q.; Sallah, N.; Genes & Health Research Team; Griffiths, C.J.; Newman, W.G.; Trembath, R.C.; Wright, J.; Lumbers, R.T.; Kuchenbaecker, K.; et al. Integrating polygenic risk scores in the prediction of type 2 diabetes risk and subtypes in British Pakistanis and Bangladeshis: A population-based cohort study. PLoS Med. 2022, 19, e1003981. [Google Scholar] [CrossRef]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef]
- Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013, 9, e1003348. [Google Scholar] [CrossRef]
- Tamlander, M.; Mars, N.; Pirinen, M.; Palotie, A.; Daly, M.; Riley-Gills, B.; Jacob, H.; Paul, D.; Runz, H.; John, S.; et al. Integration of questionnaire-based risk factors improves polygenic risk scores for human coronary heart disease and type 2 diabetes. Commun. Biol. 2022, 5, 158. [Google Scholar] [CrossRef]
- He, Y.; Patel, C.J. Shared exposure liability of type 2 diabetes and other chronic conditions in the UK Biobank. Acta Diabetol. 2022, 59, 851–860. [Google Scholar] [CrossRef]
- Cheng, H.; Sewda, A.; Marquez-Luna, C.; White, S.R.; Whitney, B.M.; Williams-Nguyen, J.; Nance, R.M.; Lee, W.J.; Kitahata, M.M.; Saag, M.S.; et al. Genetic architecture of cardiometabolic risks in people living with HIV. BMC Med. 2020, 18, 288. [Google Scholar] [CrossRef]
- Zhao, J.; Arafat, D.; Brigham, K.L.; Gibson, G. Genetic risk prediction in a small cohort of healthy adults in Atlanta. Genet. Res. 2013, 95, 30–37. [Google Scholar] [CrossRef]
- Morieri, M.L.; Gao, H.; Pigeyre, M.; Shah, H.S.; Sjaarda, J.; Mendonca, C.; Hastings, T.; Buranasupkajorn, P.; Motsinger-Reif, A.A.; Rotroff, D.M.; et al. Genetic Tools for Coronary Risk Assessment in Type 2 Diabetes: A Cohort Study From the ACCORD Clinical Trial. Diabetes Care 2018, 41, 2404–2413. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Pineda, A.; Vernekar, M.; Moreno-Grau, S.; Rojas-Muñoz, A.; Moatamed, B.; Lee, M.T.M.; Nava-Aguilar, M.A.; Gonzalez-Arroyo, G.; Numakura, K.; Matsuda, Y.; et al. Validating and automating learning of cardiometabolic polygenic risk scores from direct-to-consumer genetic and phenotypic data: Implications for scaling precision health research. Hum. Genom. 2022, 16, 37. [Google Scholar] [CrossRef]
- Mahajan, A.; Taliun, D.; Thurner, M.; Robertson, N.R.; Torres, J.M.; Rayner, N.W.; Payne, A.J.; Steinthorsdottir, V.; Scott, R.A.; Grarup, N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50, 1505–1513. [Google Scholar] [CrossRef] [Green Version]
- Sladek, R.; Rocheleau, G.; Rung, J.; Dina, C.; Shen, L.; Serre, D.; Boutin, P.; Vincent, D.; Belisle, A.; Hadjadj, S.; et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007, 445, 881–885. [Google Scholar] [CrossRef]
- Scott, L.J.; Mohlke, K.L.; Bonnycastle, L.L.; Willer, C.J.; Li, Y.; Duren, W.L.; Erdos, M.R.; Stringham, H.M.; Chines, P.S.; Jackson, A.U.; et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007, 316, 1341–1345. [Google Scholar] [CrossRef]
- Steinthorsdottir, V.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Jonsdottir, T.; Walters, G.B.; Styrkarsdottir, U.; Gretarsdottir, S.; Emilsson, V.; Ghosh, S.; et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007, 39, 770–775. [Google Scholar] [CrossRef] [Green Version]
- Rung, J.; Cauchi, S.; Albrechtsen, A.; Shen, L.; Rocheleau, G.; Cavalcanti-Proenca, C.; Bacot, F.; Balkau, B.; Belisle, A.; Borch-Johnsen, K. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat. Genet. 2009, 41, 1110–1115. [Google Scholar] [CrossRef]
- Takeuchi, F.; Serizawa, M.; Yamamoto, K.; Fujisawa, T.; Nakashima, E.; Ohnaka, K.; Ikegami, H.; Sugiyama, T.; Katsuya, T.; Miyagishi, M.; et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 2009, 58, 1690–1699. [Google Scholar] [CrossRef] [Green Version]
- Tabassum, R.; Chauhan, G.; Dwivedi, O.P.; Mahajan, A.; Jaiswal, A.; Kaur, I.; Bandesh, K.; Singh, T.; Mathai, B.J.; Pandey, Y.; et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes 2013, 62, 977–986. [Google Scholar] [CrossRef] [Green Version]
- Saxena, R.; Saleheen, D.; Been, L.F.; Garavito, M.L.; Braun, T.; Bjonnes, A.; Young, R.; Ho, W.K.; Rasheed, A.; Frossard, P.; et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes 2013, 62, 1746–1755. [Google Scholar] [CrossRef] [Green Version]
- Williams, A.L.; Jacobs, S.B.; Moreno-Macías, H.; Huerta-Chagoya, A.; Churchhouse, C.; Márquez-Luna, C.; García-Ortíz, H.; Gómez-Vázquez, M.J.; Burtt, N.P.; Aguilar-Salinas, C.A.; et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 2014, 506, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Qi, Q.; Stilp, A.M.; Sofer, T.; Moon, J.Y.; Hidalgo, B.; Szpiro, A.A.; Wang, T.; Ng, M.C.Y.; Guo, X.; Chen, Y.I.; et al. Genetics of Type 2 Diabetes in U.S. Hispanic/Latino Individuals: Results From the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes 2017, 66, 1419–1425. [Google Scholar] [CrossRef] [Green Version]
- Ng, M.C.; Shriner, D.; Chen, B.H.; Li, J.; Chen, W.M.; Guo, X.; Liu, J.; Bielinski, S.J.; Yanek, L.R.; Nalls, M.A.; et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014, 10, e1004517. [Google Scholar] [CrossRef]
- Chen, J.; Sun, M.; Adeyemo, A.; Pirie, F.; Carstensen, T.; Pomilla, C.; Doumatey, A.P.; Chen, G.; Young, E.H.; Sandhu, M.; et al. Genome-wide association study of type 2 diabetes in Africa. Diabetologia 2019, 62, 1204–1211. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, A.; Go, M.J.; Zhang, W.; Below, J.E.; Gaulton, K.J.; Ferreira, T.; Horikoshi, M.; Johnson, A.D.; Ng, M.C.; Prokopenko, I.; et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014, 46, 234–244. [Google Scholar] [CrossRef]
- Morris, A.P.; Voight, B.F.; Teslovich, T.M.; Ferreira, T.; Segrè, A.V.; Steinthorsdottir, V.; Strawbridge, R.J.; Khan, H.; Grallert, H.; Mahajan, A.; et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012, 44, 981–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, A.; Wu, Y.; Zhu, Z.; Zhang, F.; Kemper, K.E.; Zheng, Z.; Yengo, L.; Lloyd-Jones, L.R.; Sidorenko, J.; Wu, Y.; et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 2018, 9, 2941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020, 52, 680–691. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Spracklen, C.N.; Zhang, W.; Ng, M.C.Y.; Petty, L.E.; Kitajima, H.; Yu, G.Z.; Rueger, S.; Speidel, L.; Kim, Y.J.; et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 2022, 54, 560–572. [Google Scholar] [CrossRef]
- Timpson, N.J.; Lindgren, C.M.; Weedon, M.N.; Randall, J.; Ouwehand, W.H.; Strachan, D.P.; Rayner, N.W.; Walker, M.; Hitman, G.A.; Doney, A.S.; et al. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes 2009, 58, 505–510. [Google Scholar] [CrossRef] [Green Version]
- Perry, J.R.; Voight, B.F.; Yengo, L.; Amin, N.; Dupuis, J.; Ganser, M.; Grallert, H.; Navarro, P.; Li, M.; Qi, L.; et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012, 8, e1002741. [Google Scholar] [CrossRef] [Green Version]
- Ahlqvist, E.; Prasad, R.B.; Groop, L. Subtypes of Type 2 Diabetes Determined From Clinical Parameters. Diabetes 2020, 69, 2086–2093. [Google Scholar] [CrossRef]
- Mansour Aly, D.; Dwivedi, O.P.; Prasad, R.B.; Karajamaki, A.; Hjort, R.; Thangam, M.; Akerlund, M.; Mahajan, A.; Udler, M.S.; Florez, J.C.; et al. Genome-wide association analyses highlight etiological differences underlying newly defined subtypes of diabetes. Nat. Genet. 2021, 53, 1534–1542. [Google Scholar] [CrossRef]
- Lyssenko, V.; Lupi, R.; Marchetti, P.; Del Guerra, S.; Orho-Melander, M.; Almgren, P.; Sjogren, M.; Ling, C.; Eriksson, K.F.; Lethagen, A.L.; et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Investig. 2007, 117, 2155–2163. [Google Scholar] [CrossRef] [Green Version]
- Gaulton, K.J.; Nammo, T.; Pasquali, L.; Simon, J.M.; Giresi, P.G.; Fogarty, M.P.; Panhuis, T.M.; Mieczkowski, P.; Secchi, A.; Bosco, D.; et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010, 42, 255–259. [Google Scholar] [CrossRef]
- Hivert, M.F.; Manning, A.K.; McAteer, J.B.; Florez, J.C.; Dupuis, J.; Fox, C.S.; O’Donnell, C.J.; Cupples, L.A.; Meigs, J.B. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: The Framingham Offspring Study. Diabetes 2008, 57, 3353–3359. [Google Scholar] [CrossRef] [Green Version]
- Warren, L.L.; Li, L.; Nelson, M.R.; Ehm, M.G.; Shen, J.; Fraser, D.J.; Aponte, J.L.; Nangle, K.L.; Slater, A.J.; Woollard, P.M.; et al. Deep Resequencing Unveils Genetic Architecture of ADIPOQ and Identifies a Novel Low-Frequency Variant Strongly Associated With Adiponectin Variation. Diabetes 2012, 61, 1297–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, D.J.; Plagnol, V.; Walker, N.M.; Cooper, J.D.; Downes, K.; Yang, J.H.; Howson, J.M.; Stevens, H.; McManus, R.; Wijmenga, C.; et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008, 359, 2767–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahsan, M.; Ek, W.E.; Rask-Andersen, M.; Karlsson, T.; Lind-Thomsen, A.; Enroth, S.; Gyllensten, U.; Johansson, Å. The relative contribution of DNA methylation and genetic variants on protein biomarkers for human diseases. PLoS Genet. 2017, 13, e1007005. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.N.; Hawa, M.I.; Roden, M.; Schernthaner, G.; Pozzilli, P.; Buzzetti, R.; Scherbaum, W.A.; Seissler, J.; Hunter, S.; Leslie, R.D.; et al. Increased serum concentrations of adhesion molecules but not of chemokines in patients with Type 2 diabetes compared with patients with Type 1 diabetes and latent autoimmune diabetes in adult age: Action LADA 5. Diabet. Med. 2012, 29, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.T.; Lin, L.Y.; Chen, J.W. A Novel Resolution of Diabetes: C-C Chemokine Motif Ligand 4 Is a Common Target in Different Types of Diabetes by Protecting Pancreatic Islet Cell and Modulating Inflammation. Front. Immunol. 2021, 12, 650626. [Google Scholar] [CrossRef] [PubMed]
- Teler, J.; Tarnowski, M.; Safranow, K.; Maciejewska, A.; Sawczuk, M.; Dziedziejko, V.; Sluczanowska-Glabowska, S.; Pawlik, A. CCL2, CCL5, IL4 and IL15 Gene Polymorphisms in Women with Gestational Diabetes Mellitus. Horm. Metab. Res. 2017, 49, 10–15. [Google Scholar] [CrossRef]
- Jeon, H.J.; Choi, H.J.; Park, B.H.; Lee, Y.H.; Oh, T. Association of monocyte chemoattractant protein-1 (MCP-1) 2518A/G polymorphism with proliferative diabetic retinopathy in Korean type 2 diabetes. Yonsei Med. J. 2013, 54, 621–625. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Hennein, L.; Xu, Y.; Bao, N.; Coh, P.; Tao, L. Elevated serum monocyte chemoattractant protein-1 levels and its genetic polymorphism is associated with diabetic retinopathy in Chinese patients with Type 2 diabetes. Diabet. Med. 2016, 33, 84–90. [Google Scholar] [CrossRef]
- Raina, P.; Matharoo, K.; Bhanwer, A.J. Monocyte chemoattractant protein-1 (MCP-1) g.-2518A>G polymorphism and susceptibility to type 2 diabetes (T2D) and end stage renal disease (ESRD) in the North-West Indian population of Punjab. Ann. Hum. Biol. 2015, 42, 276–282. [Google Scholar] [CrossRef]
- Gonzalez, E.; Rovin, B.H.; Sen, L.; Cooke, G.; Dhanda, R.; Mummidi, S.; Kulkarni, H.; Bamshad, M.J.; Telles, V.; Anderson, S.A.; et al. HIV-1 infection and AIDS dementia are influenced by a mutant MCP-1 allele linked to increased monocyte infiltration of tissues and MCP-1 levels. Proc. Natl. Acad. Sci. USA 2002, 99, 13795–13800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, D.H.; Yang, Q.; Kathiresan, S.; Cupples, L.A.; Massaro, J.M.; Keaney, J.F., Jr.; Larson, M.G.; Vasan, R.S.; Hirschhorn, J.N.; O’Donnell, C.J.; et al. CCL2 polymorphisms are associated with serum monocyte chemoattractant protein-1 levels and myocardial infarction in the Framingham Heart Study. Circulation 2005, 112, 1113–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letendre, S.; Marquie-Beck, J.; Singh, K.K.; de Almeida, S.; Zimmerman, J.; Spector, S.A.; Grant, I.; Ellis, R. The monocyte chemotactic protein-1 -2578G allele is associated with elevated MCP-1 concentrations in cerebrospinal fluid. J. Neuroimmunol. 2004, 157, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Mühlbauer, M.; Bosserhoff, A.K.; Hartmann, A.; Thasler, W.E.; Weiss, T.S.; Herfarth, H.; Lock, G.; Schölmerich, J.; Hellerbrand, C. A novel MCP-1 gene polymorphism is associated with hepatic MCP-1 expression and severity of HCV-related liver disease. Gastroenterology 2003, 125, 1085–1093. [Google Scholar] [CrossRef]
- Karrer, S.; Bosserhoff, A.K.; Weiderer, P.; Distler, O.; Landthaler, M.; Szeimies, R.M.; Müller-Ladner, U.; Schölmerich, J.; Hellerbrand, C. The -2518 promotor polymorphism in the MCP-1 gene is associated with systemic sclerosis. J. Investig. Dermatol. 2005, 124, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Melzer, D.; Perry, J.R.; Hernandez, D.; Corsi, A.M.; Stevens, K.; Rafferty, I.; Lauretani, F.; Murray, A.; Gibbs, J.R.; Paolisso, G.; et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 2008, 4, e1000072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochetova, O.V.; Avzaletdinova, D.S.; Morugova, T.V.; Mustafina, O.E. Chemokine gene polymorphisms association with increased risk of type 2 diabetes mellitus in Tatar ethnic group, Russia. Mol. Biol. Rep. 2019, 46, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Valverde-Villegas, J.M.; de Medeiros, R.M.; de Andrade, K.P.; Jacovas, V.C.; dos Santos, B.R.; Simon, D.; de Matos Almeida, S.E.; Chies, J.A.B. Novel genetic associations and gene-gene interactions of chemokine receptor and chemokine genetic polymorphisms in HIV/AIDS. AIDS 2017, 31, 1235–1243. [Google Scholar] [CrossRef]
- Burke, S.J.; Karlstad, M.D.; Regal, K.M.; Sparer, T.E.; Lu, D.; Elks, C.M.; Grant, R.W.; Stephens, J.M.; Burk, D.H.; Collier, J.J. CCL20 is elevated during obesity and differentially regulated by NF-κB subunits in pancreatic β-cells. Biochim. Biophys. Acta 2015, 1849, 637–652. [Google Scholar] [CrossRef] [Green Version]
- Kochetova, O.V.; Avzaletdinova, D.S.; Korytina, G.F.; Morugova, T.V.; Mustafina, O.E. The association between eating behavior and polymorphisms in GRIN2B, GRIK3, GRIA1 and GRIN1 genes in people with type 2 diabetes mellitus. Mol. Biol. Rep. 2020, 47, 2035–2046. [Google Scholar] [CrossRef] [PubMed]
- Korytina, G.F.; Aznabaeva, Y.G.; Akhmadishina, L.Z.; Kochetova, O.V.; Nasibullin, T.R.; Zagidullin, N.S.; Zagidullin, S.Z.; Viktorova, T.V. The Relationship Between Chemokine and Chemokine Receptor Genes Polymorphisms and Chronic Obstructive Pulmonary Disease Susceptibility in Tatar Population from Russia: A Case Control Study. Biochem. Genet. 2022, 60, 54–79. [Google Scholar] [CrossRef]
- Avzaletdinova, D.S.; Sharipova, L.F.; Kochetova, O.V.; Morugova, T.V.; Mustafina, O.E. Association of adiponectin gene alleles with type 2 diabetes mellitus in residents of Bashkortostan. Probl. Endokrinol. 2019, 65, 31–38. [Google Scholar] [CrossRef]
- Krylov, M.Y.e.; Benevolenskaya, L.; Myakotkin, V.; Krylov, M.Y. Leptin A19G polymorphism and leptin receptor Gln223Arg and Lys109Arg polymorphismsin postmenopausal osteoporosis. Nauchno-Prakt. Revmatol. 2010, 48, 27–31. [Google Scholar] [CrossRef]
- Khan, I.A.; Jahan, P.; Hasan, Q.; Rao, P. Validation of the association of TCF7L2 and SLC30A8 gene polymorphisms with post-transplant diabetes mellitus in Asian Indian population. Intractable Rare Dis. Res. 2015, 4, 87–92. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yan, G.; Zhang, J.; Gao, K.; Zhang, M.; Li, L.; Wang, Y.; Wang, Q.; Zhai, Y.; You, H.; et al. Association of LRP5, TCF7L2, and GCG variants and type 2 diabetes mellitus as well as fasting plasma glucose and lipid metabolism indexes. Hum. Immunol. 2015, 76, 339–343. [Google Scholar] [CrossRef]
- Munoz-Barrios, S.; Guzman-Guzman, I.P.; Munoz-Valle, J.F.; Salgado-Bernabe, A.B.; Salgado-Goytia, L.; Parra-Rojas, I. Association of the HindIII and S447X polymorphisms in LPL gene with hypertension and type 2 diabetes in Mexican families. Dis. Markers 2012, 33, 313–320. [Google Scholar] [CrossRef]
- Aoki, M.N.; da Silva do Amaral Herrera, A.C.; Amarante, M.K.; do Val Carneiro, J.L.; Fungaro, M.H.P.; Watanabe, M.A.E. CCR5 and p53 codon 72 gene polymorphisms: Implications in breast cancer development. Int. J. Mol. Med. 2009, 23, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Bikbov, M.; Gilmanshin, Т.; Zainullin, R.; Kudoyarova, K. On the Epidemiology of Diabetic Retinopathy in the Republic of Bashkortostan. Acta Biomed. Sci. 2019, 4, 66–69. [Google Scholar]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Carstensen, B.; Plummer, M.; Laara, E.; Hills, M. Epi: Statistical Analysis in Epidemiology; R package version 2.47; 2020. Available online: https://CRAN.R-project.org/package=Epi (accessed on 21 September 2022).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Pencina, M.J.; D’Agostino, R.B., Sr.; D’Agostino, R.B., Jr.; Vasan, R.S. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat. Med. 2008, 27, 157–172. [Google Scholar] [CrossRef]
- Inoue, E. Nricens: Nri for Risk Prediction Models with Time to Event and Binary Response Data; R package version 1.6. 2018. Available online: https://CRAN.R-project.org/package=nricens (accessed on 21 September 2022).
- Roszkowska-Gancarz, M.; Kurylowicz, A.; Polosak, J.; Mossakowska, M.; Franek, E.; Puzianowska-Kuźnicka, M. Functional polymorphisms of the leptin and leptin receptor genes are associated with longevity and with the risk of myocardial infarction and of type 2 diabetes mellitus. Endokrynologia Polska 2014, 65, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Shah, R.; Hinkle, C.C.; Ferguson, J.F.; Mehta, N.N.; Li, M.; Qu, L.; Lu, Y.; Putt, M.E.; Ahima, R.S.; Reilly, M.P. Fractalkine Is a Novel Human Adipochemokine Associated With Type 2 Diabetes. Diabetes 2011, 60, 1512–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Słomiński, B.; Ławrynowicz, U.; Myśliwska, J.; Ryba-Stanisławowska, M.; Skrzypkowska, M.; Myśliwiec, M.; Brandt, A. CCR5-Δ32 gene polymorphism is related to celiac disease and autoimmune thyroiditis coincidence in patients with type 1 diabetes. J. Diabetes Its Complicat. 2017, 31, 615–618. [Google Scholar] [CrossRef]
- Tetik Vardarlı, A.; Harman, E.; Bozok Çetintaş, V.; Kayıkçıoğlu, M.; Vardarlı, E.; Zengi, A.; Küçükaslan, A.; Eroğlu, Z. Polymorphisms of lipid metabolism enzyme-coding genes in patients with diabetic dyslipidemia. Anatol. J. Cardiol. 2017, 17, 313–321. [Google Scholar] [CrossRef]
- Souza, K.S.C.d.; Ururahy, M.A.G.; Oliveira, Y.M.d.C.; Loureiro, M.B.; Silva, H.P.V.d.; Bortolin, R.H.; Luchessi, A.D.; Arrais, R.F.; Hirata, R.D.C.; Almeida, M.d.G. The low-density lipoprotein receptor-related protein 5 (LRP5) 4037C> T polymorphism: Candidate for susceptibility to type 1 diabetes mellitus. Archives Endocrinol. Metab. 2018, 62, 480–484. [Google Scholar] [CrossRef]
- Zhernakova, A.; Alizadeh, B.Z.; Eerligh, P.; Hanifi-Moghaddam, P.; Schloot, N.C.; Diosdado, B.; Wijmenga, C.; Roep, B.O.; Koeleman, B.P.C. Genetic variants of RANTES are associated with serum RANTES level and protection for type 1 diabetes. Genes Immun. 2006, 7, 544–549. [Google Scholar] [CrossRef] [Green Version]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuckovic, D.; Bao, E.L.; Akbari, P.; Lareau, C.A.; Mousas, A.; Jiang, T.; Chen, M.H.; Raffield, L.M.; Tardaguila, M.; Huffman, J.E.; et al. The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell 2020, 182, 1214–1231.e1211. [Google Scholar] [CrossRef]
- Harshfield, E.L.; Fauman, E.B.; Stacey, D.; Paul, D.S.; Ziemek, D.; Ong, R.M.Y.; Danesh, J.; Butterworth, A.S.; Rasheed, A.; Sattar, T.; et al. Genome-wide analysis of blood lipid metabolites in over 5000 South Asians reveals biological insights at cardiometabolic disease loci. BMC Med. 2021, 19, 232. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.B.; Rivadeneira, F.; Inouye, M.; Pastinen, T.M.; Soranzo, N.; Wilson, S.G.; Andrew, T.; Falchi, M.; Gwilliam, R.; Ahmadi, K.R.; et al. Bone mineral density, osteoporosis, and osteoporotic fractures: A genome-wide association study. Lancet 2008, 371, 1505–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emilsson, V.; Ilkov, M.; Lamb, J.R.; Finkel, N.; Gudmundsson, E.F.; Pitts, R.; Hoover, H.; Gudmundsdottir, V.; Horman, S.R.; Aspelund, T.; et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018, 361, 769–773. [Google Scholar] [CrossRef] [PubMed]
| Chr 1 | Position GRCh37 2 | Gene | SNP 3 | EA 4 | MA 5 | MAF 6 | PHWE 7 | OR 8 (95% CIOR) 9 | P 10 | PFDR 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | T2D | ||||||||||
| 1 | 66,036,441 | LEPR | rs1137100 | G | G | 0.28 | 0.31 | 0.429 | 1.24 (0.89–1.72) | 0.202 | 0.348 |
| 2 | 228,677,842 | CCL20 | rs6749704 * | C | C | 0.28 | 0.39 | 0.094 | 1.68 (1.35–2.09) | 2.61 × 10−6 | 3.40 × 10−5 |
| 3 | 39,307,162 | CX3CR1 | rs3732378 | A | A | 0.21 | 0.21 | 0.092 | 1.01 (0.77–1.31) | 0.954 | 0.954 |
| 3 | 46,414,944 | CCR5 | rs333 * | D | D | 0.06 | 0.10 | 1.000 | 1.99 (1.16–3.42) | 0.013 | 0.033 |
| 3 | 186,572,089 | ADIPOQ | rs17366743 * | C | C | 0.03 | 0.09 | 1.000 | 3.17 (1.64–6.12) | 6.10 × 10−4 | 2.64 × 10−3 |
| 8 | 19,819,077 | LPL | rs320 | G | G | 0.24 | 0.26 | 0.294 | 1.21 (0.90–1.62) | 0.214 | 0.348 |
| 10 | 114,758,349 | TCF7L2 | rs7903146 * | T | T | 0.25 | 0.39 | 0.616 | 1.77 (1.37–2.29) | 1.44 × 10−5 | 9.37 × 10−5 |
| 11 | 68,201,295 | LRP5 | rs3736228 | T | T | 0.10 | 0.12 | 0.490 | 0.97 (0.64–1.46) | 0.874 | 0.947 |
| 12 | 14,018,777 | GRIN2B | rs7301328 | G | C | 0.43 | 0.40 | 0.103 | 1.16 (0.95–1.41) | 0.150 | 0.325 |
| 16 | 57,447,414 | CCL17 | rs223828 | C | T | 0.14 | 0.12 | 1.000 | 1.20 (0.86–1.65) | 0.280 | 0.405 |
| 17 | 32,579,788 | CCL2 | rs1024611 * | A | G | 0.31 | 0.24 | 0.627 | 1.38 (1.08–1.76) | 0.011 | 0.033 |
| 17 | 32,612,402 | CCL11 | rs16969415 | T | T | 0.06 | 0.06 | 1.000 | 1.13 (0.74–1.74) | 0.565 | 0.734 |
| 17 | 34,207,780 | CCL5 | rs2107538 | C | T | 0.25 | 0.25 | 0.261 | 0.95 (0.73–1.22) | 0.662 | 0.783 |
| Baseline (Age + Sex) | ||||||||
| 5SNP 1 Polygenic Score + Age + Sex | 13SNP Polygenic Score + Age + Sex | |||||||
| NRI 2 | SE 3 | 95% CI 4 | p-Value 5 | NRI | SE | 95% CI | p-Value | |
| Total | 37.62 | 7.80 | 19.29–49.00 | 1.39 × 10−6 | 36.73 | 7.14 | 22.87–50.97 | 2.73 × 10−7 |
| Cases | 5.51 | 5.88 | −4.07–18.18 | 0.349 | 13.98 | 4.10 | 6.19–22.84 | 6.44 × 10−4 |
| Controls | 32.11 | 4.80 | 16.88–36.21 | 2.17 × 10−11 | 22.74 | 4.24 | 13.65–30.23 | 8.30 × 10−8 |
| Baseline (Age + Sex + BMI 6) | ||||||||
| 5SNP Polygenic Score + Age + Sex + BMI | 13SNP Polygenic Score + Age + Sex + BMI | |||||||
| NRI | SE | 95% CI | p-Value | NRI | SE | 95% CI | p-Value | |
| Total | 41.72 | 8.70 | 24.67–55.62 | 8.44 × 10−7 | 41.50 | 8.95 | 21.21–58.79 | 3.53 × 10−6 |
| Cases | 8.70 | 4.65 | 0.26–18.37 | 0.061 | 16.85 | 5.10 | 7.61–27.15 | 9.58 × 10−4 |
| Controls | 33.02 | 6.03 | 20.76–44.44 | 4.27 × 10−8 | 24.65 | 6.28 | 10.44–34.96 | 8.72 × 10−5 |
| Baseline (5SNP Polygenic Score + Age + Sex) | Baseline (5SNP Polygenic Score + Age + Sex + BMI) | |||||||
| 13SNP Polygenic Score + Age + Sex | 13SNP Polygenic Score + Age + Sex + BMI | |||||||
| NRI | SE | 95% CI | p-Value | NRI | SE | 95% CI | p-Value | |
| Total | −17.86 | 10.63 | −37.29–3.61 | 0.093 | 4.80 | 16.72 | −29.60–34.48 | 0.774 |
| Cases | −9.74 | 4.95 | −18.43–1.17 | 0.049 | −2.17 | 7.45 | −16.09–13.49 | 0.770 |
| Controls | −8.11 | 6.81 | −21.65–5.19 | 0.234 | 6.98 | 10.53 | −16.93–24.30 | 0.508 |
| Characteristic | T2D 7, N = 496 Mean ± SD 8 | Control, N = 875 Mean ± SD 8 | p 9 |
|---|---|---|---|
| Age (years) | 55.21 ± 9.77 | 49.65 ± 10.9 | >0.001 * |
| Sex: female (N, %) | 370 (74.5) | 300 (34.3) | >0.001 * |
| Duration of T2D (years) | 7.23 ± 5.66 | — | — |
| Age at onset (years) | 54.53 ± 9.27 | — | — |
| BMI 1 (kg/m2) | 30.23 ± 5.36 | 28.93 ± 5.13 | 0.003 * |
| Fasting blood glucose (mmol/L) | 7.35 ± 2.34 | 4.88 ± 0.71 | >0.001 * |
| PPG 2 (mmol/L) | 9.38 ± 2.62 | — | — |
| HbA1c 3 (%) | 7.41 ± 1.01 | 4.89 ± 0.60 | >0.001 * |
| Total cholesterol | 5.52 ± 1.14 | 5.09 ± 0.64 | >0.001 * |
| Triglycerides | 1.67 ± 1.16 | 1.48 ± 0.60 | 0.036 |
| HDL 4 | 1.19 ± 0.50 | 1.09 ± 0.37 | 0.016 |
| LDL 5 | 3.41 ± 4.30 | 2.96 ± 1.08 | 0.148 |
| AC 6 | 3.76 ± 1.30 | 3.59 ± 0.85 | 0.115 |
| C-peptide | 2.16 ± 1.35 | 2.31 ± 0.94 | 0.166 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timasheva, Y.; Balkhiyarova, Z.; Avzaletdinova, D.; Rassoleeva, I.; Morugova, T.V.; Korytina, G.; Prokopenko, I.; Kochetova, O. Integrating Common Risk Factors with Polygenic Scores Improves the Prediction of Type 2 Diabetes. Int. J. Mol. Sci. 2023, 24, 984. https://doi.org/10.3390/ijms24020984
Timasheva Y, Balkhiyarova Z, Avzaletdinova D, Rassoleeva I, Morugova TV, Korytina G, Prokopenko I, Kochetova O. Integrating Common Risk Factors with Polygenic Scores Improves the Prediction of Type 2 Diabetes. International Journal of Molecular Sciences. 2023; 24(2):984. https://doi.org/10.3390/ijms24020984
Chicago/Turabian StyleTimasheva, Yanina, Zhanna Balkhiyarova, Diana Avzaletdinova, Irina Rassoleeva, Tatiana V. Morugova, Gulnaz Korytina, Inga Prokopenko, and Olga Kochetova. 2023. "Integrating Common Risk Factors with Polygenic Scores Improves the Prediction of Type 2 Diabetes" International Journal of Molecular Sciences 24, no. 2: 984. https://doi.org/10.3390/ijms24020984
APA StyleTimasheva, Y., Balkhiyarova, Z., Avzaletdinova, D., Rassoleeva, I., Morugova, T. V., Korytina, G., Prokopenko, I., & Kochetova, O. (2023). Integrating Common Risk Factors with Polygenic Scores Improves the Prediction of Type 2 Diabetes. International Journal of Molecular Sciences, 24(2), 984. https://doi.org/10.3390/ijms24020984








