Combined Omics Approaches Reveal Distinct Mechanisms of Resistance and/or Susceptibility in Sugar Beet Double Haploid Genotypes at Early Stages of Beet Curly Top Virus Infection
Abstract
:1. Introduction
2. Results
2.1. Differentially Expressed Sugar Beet Genes at Early Stages of BCTV Infection
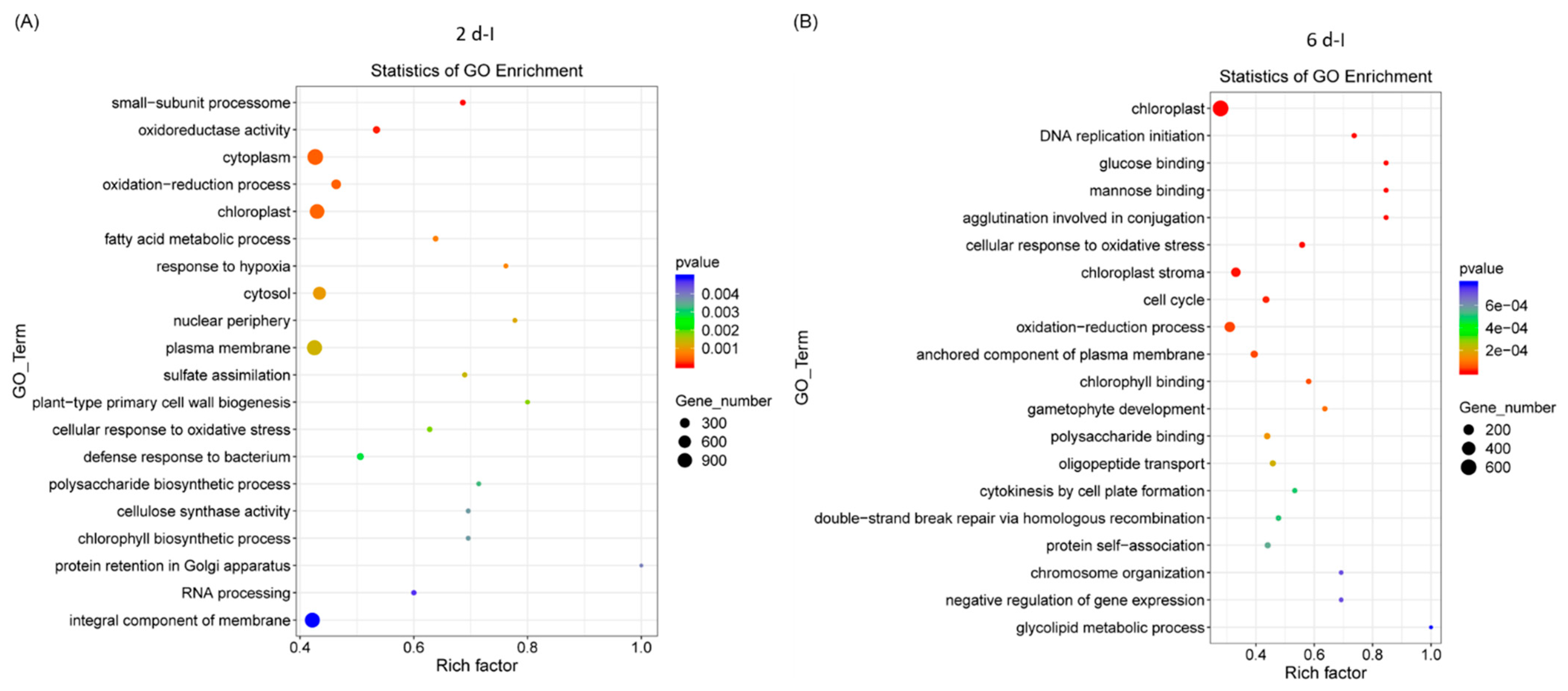
2.2. GO and KEGG Analyses of Differentially Expressed Genes
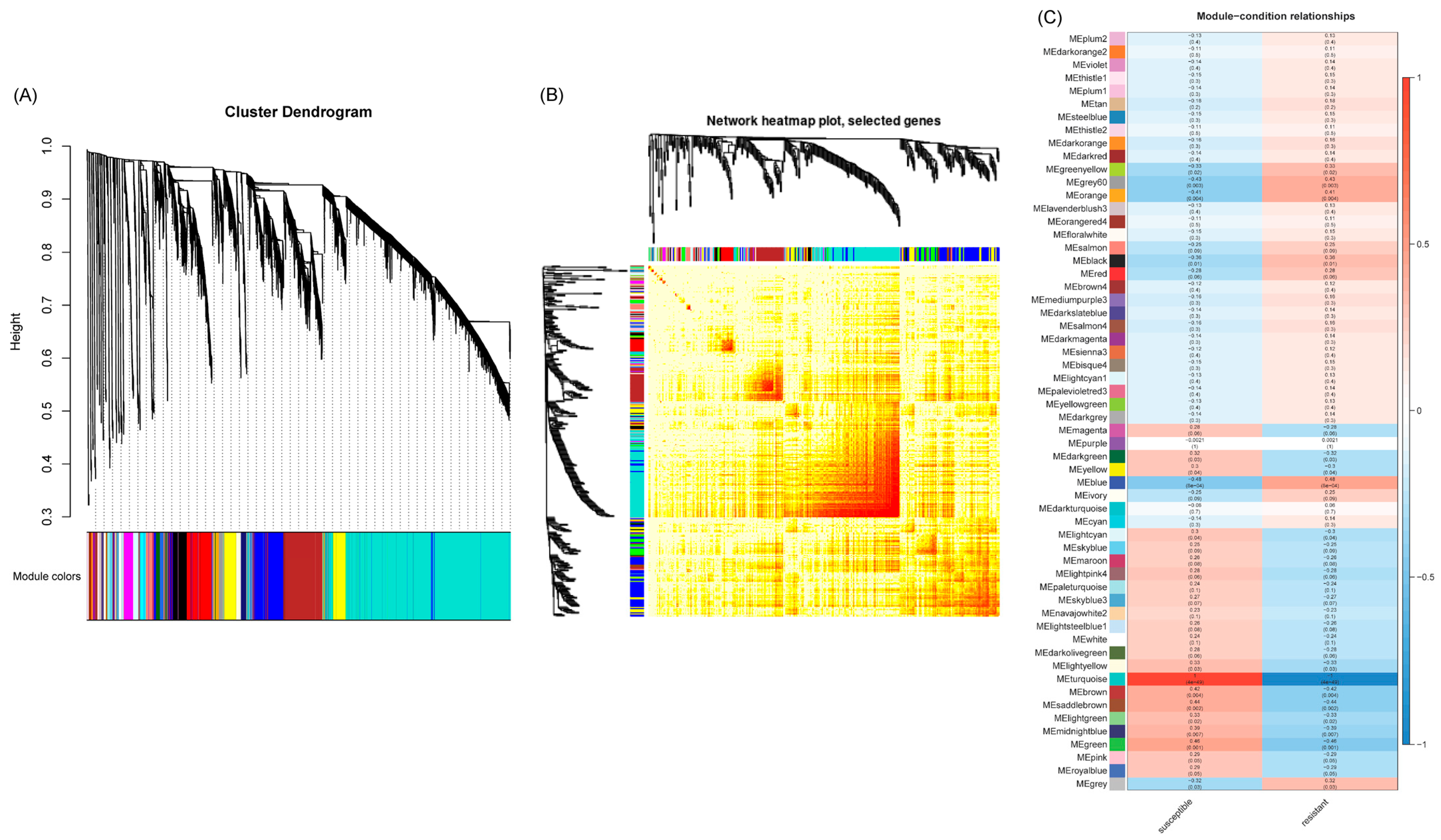
2.3. WGCNA Analyses of Differentially Expressed Genes
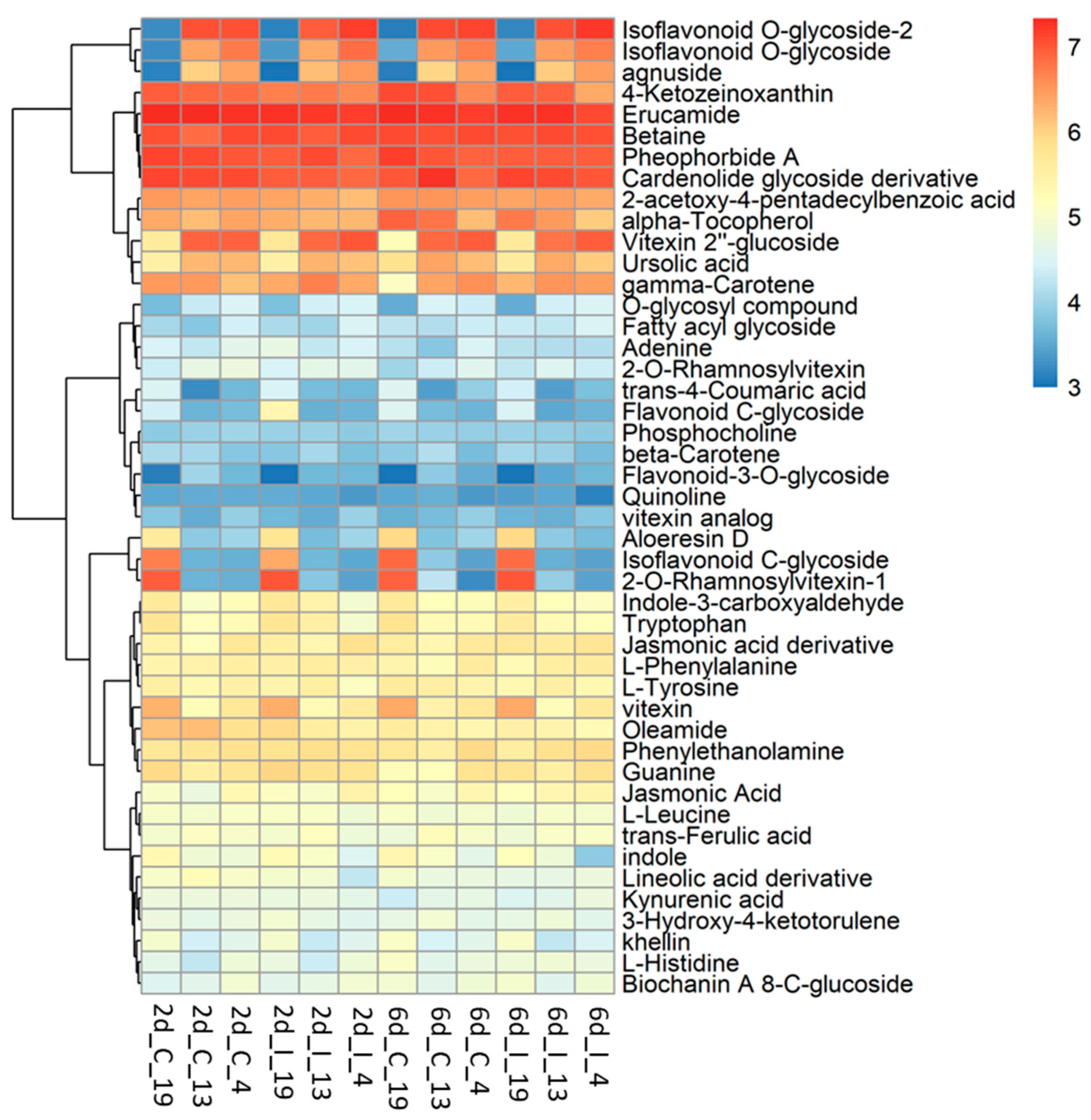
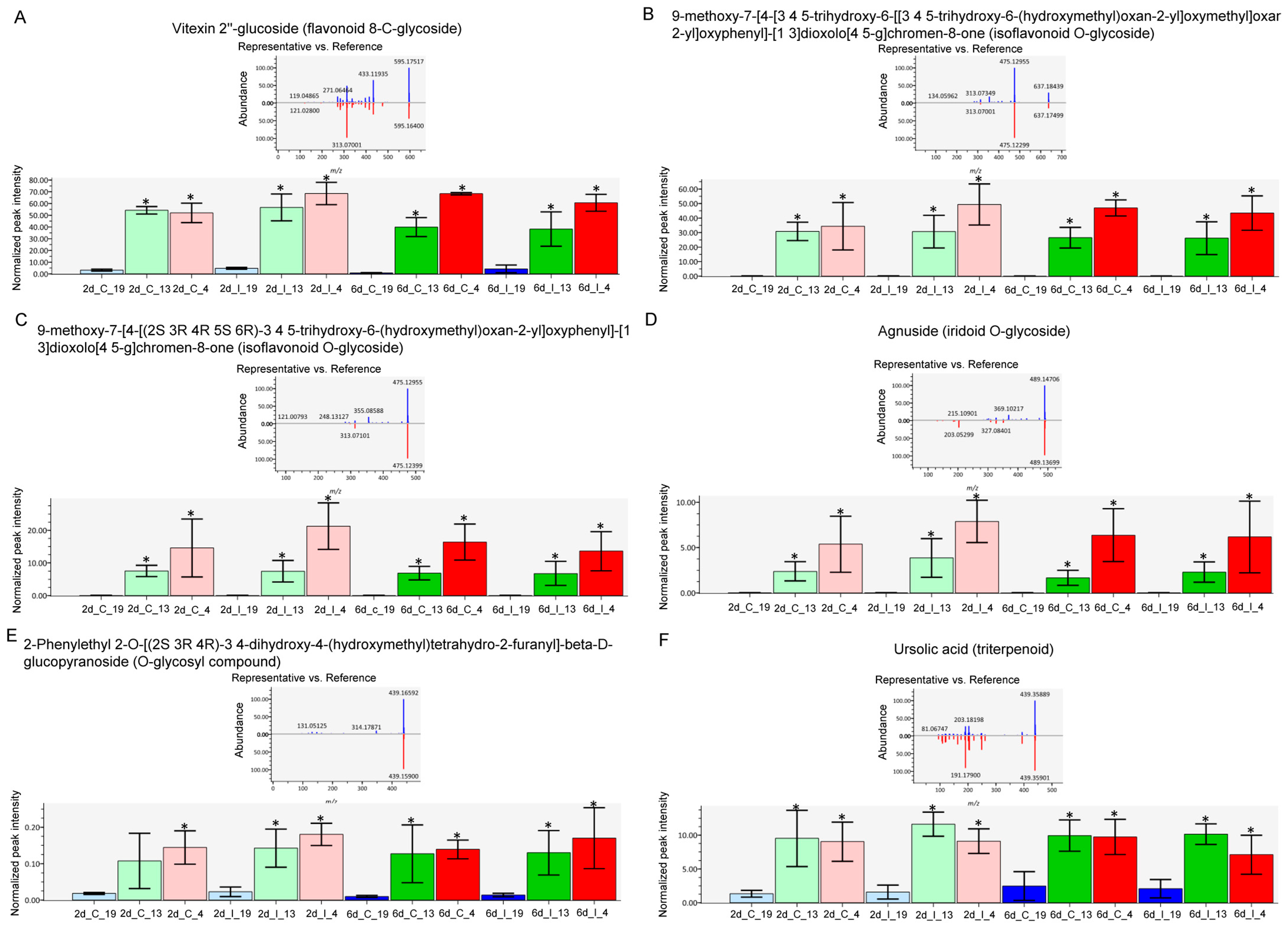
2.4. Metabolome Analysis Reveal Disctinct Differences between Resistant and Susceptible Lines at Early Infection Stages
2.5. BCTV Strain Specific sncRNAs and Their Interaction with Putative Sugar Beet Target Genes
2.6. Validation of BCTV Strain Specific sncRNA Putative Target Sugar Beet Genes through Degradome Sequencing
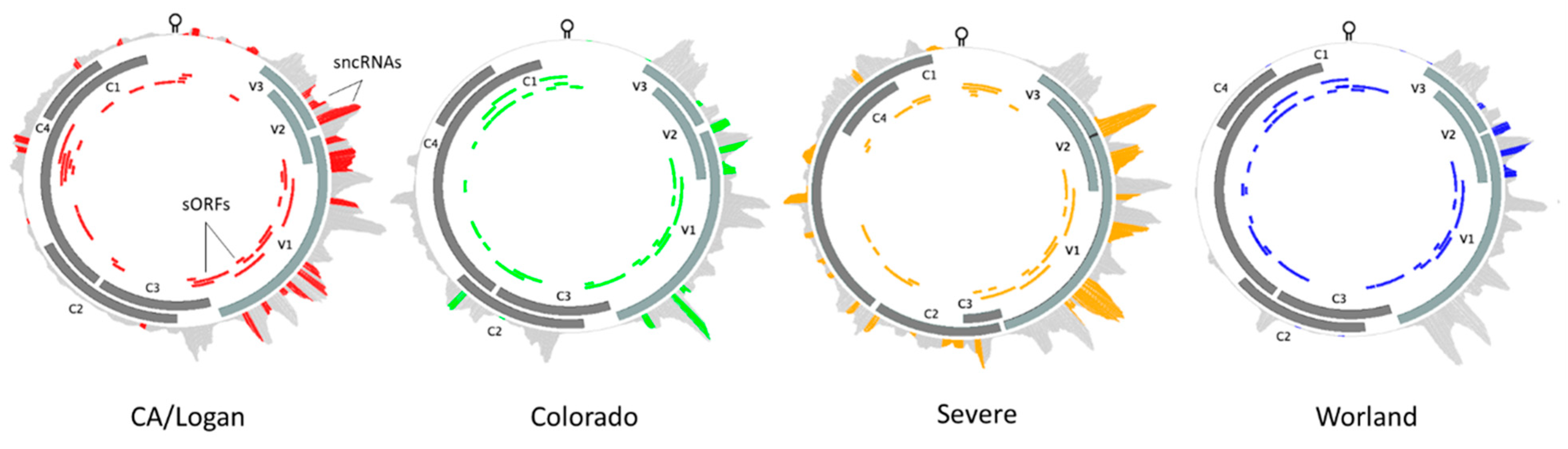
2.7. BCTV Strain Specific Divergence in Relation to Functional Elements
2.8. Differential Regulation of sORF Derived Peptides Originating from the BCTV Strains
3. Discussion
4. Materials and Methods
4.1. Plant Growth Condition, Viral Infection of Sugar Beet Plants, and Sample Collection
4.2. Extraction of Total RNA, sRNA and mRNA Library Preparations, and Sequencing
4.3. Read Mapping and Transcriptome Assembly
4.4. Differential Expression of mRNAs and Bioinformatics Analysis
4.5. Analysis of sncRNAs Derived from the BCTV Genomes
4.6. Visualization of Transcription in the Virus Genomes
4.7. Data and Code Availability
4.8. Population Genomic Analysis
4.9. Degradome Library Construction and Sequencing
4.10. Degradome Data Analysis
4.11. Untargeted Metabolomics: Sample Preparation, Running, and Analysis
4.12. Sample Preparation and Running for Peptidomics Analysis of BCTV Derived Small Peptides
4.13. Bioinformatics Analysis of BCTV sORF Derived Peptides
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beam, K.; Ascencio-Ibanez, J.T. Geminivirus Resistance: A Minireview. Front. Plant Sci. 2020, 11, 1131. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.W. The Curly Top Disease of Sugarbeet and Other Plants, Monograph No. 7; American Phytopathological Society: St. Paul, MN, USA, 1971. [Google Scholar]
- Chiginsky, J.; Langemeier, K.; MacWilliams, J.; Albrecht, T.; Cranshaw, W.; Fulladolsa, A.C.; Kapuscinski, M.; Stenglein, M.; Nachappa, P. First Insights Into the Virus and Viroid Communities in Hemp (Cannabis sativa). Front. Agron. 2021, 3, 778433. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Eujayl, I.A.; Wintermantel, W.M. Beet curly top virus Strains Associated with Sugar Beet in Idaho, Oregon, and a Western U.S. Collection. Plant Dis. 2017, 101, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Dewar, A.M. Integrated Pest Management in Action—Derogation for Use of the Neonicotinoid, Thiamethoxam in Sugar Beet in2022. Outlooks Pest Manag. 2022, 33, 37–40. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Gillen, A.M.; Gallian, J.J.; Camp, S.; Stander, J.R. Influence of Host Resistance and Insecticide Seed Treatments on Curly Top in Sugar Beets. Plant Dis. 2006, 90, 1539–1544. [Google Scholar] [CrossRef]
- Strausbaugh, C.A.; Wenninger, E.J.; Eujayl, I.A. Length of Efficacy for Control of Curly Top in Sugar Beet With Seed and Foliar Insecticides. Plant Dis. 2016, 100, 1364–1370. [Google Scholar] [CrossRef]
- Panella, L.; Kaffka, S.R.; Lewellen, R.T.; Mitchell McGrath, J.; Metzger, M.S.; Strausbaugh, C.A. Sugarbeet Chapter 13. In Yield Gains in Major U.S. Field Crops; CSSA: Madison, WI, USA, 2014; pp. 357–395. [Google Scholar]
- Mishra, J.; Srivastava, R.; Trivedi, P.K.; Verma, P.C. Effect of virus infection on the secondary metabolite production and phytohormone biosynthesis in plants. 3 Biotech 2020, 10, 547. [Google Scholar] [CrossRef]
- Gupta, N.; Reddy, K.; Bhattacharyya, D.; Chakraborty, S. Plant responses to geminivirus infection: Guardians of the plant immunity. Virol. J. 2021, 18, 143. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef]
- Stare, T.; Stare, K.; Weckwerth, W.; Wienkoop, S.; Gruden, K. Comparison between Proteome and Transcriptome Response in Potato (Solanum tuberosum L.) Leaves Following Potato Virus Y (PVY) Infection. Proteomes 2017, 5, 14. [Google Scholar] [CrossRef]
- Lyu, S.; Gao, L.; Zhang, R.; Zhang, C.; Hou, X. Correlation Analysis of Expression Profile and Quantitative iTRAQ-LC-MS/MS Proteomics Reveals Resistance Mechanism Against TuMV in Chinese Cabbage (Brassica rapa ssp. pekinensis). Front. Genet. 2020, 11, 963. [Google Scholar] [CrossRef]
- Akbar, S.; Yao, W.; Yu, K.; Qin, L.; Ruan, M.; Powell, C.A.; Chen, B.; Zhang, M. Photosynthetic characterization and expression profiles of sugarcane infected by Sugarcane mosaic virus (SCMV). Photosynth. Res. 2021, 150, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Saez, C.; Flores-Leon, A.; Montero-Pau, J.; Sifres, A.; Dhillon, N.P.S.; Lopez, C.; Pico, B. RNA-Seq Transcriptome Analysis Provides Candidate Genes for Resistance to Tomato Leaf Curl New Delhi Virus in Melon. Front. Plant Sci. 2021, 12, 798858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, T.; Jia, Z.; Jia, X.; Liu, Y.; Xuan, J.; Wang, G.; Zhang, F. Transcriptome Analysis Reveals a Comprehensive Virus Resistance Response Mechanism in Pecan Infected by a Novel Badnavirus Pecan Virus. Int. J. Mol. Sci. 2022, 23, 13576. [Google Scholar] [CrossRef] [PubMed]
- Anjali; Kumar, S.; Korra, T.; Thakur, R.; Arutselvan, R.; Kashyap, A.S.; Nehela, Y.; Chaplygin, V.; Minkina, T.; Keswani, C. Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant Stress 2023, 8, 100154. [Google Scholar] [CrossRef]
- Tang, J.; Gu, X.; Liu, J.; He, Z. Roles of small RNAs in crop disease resistance. Stress Biol. 2021, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, L.; Yang, Y.; Schmid, M.; Wang, Y. miRNA Mediated Regulation and Interaction between Plants and Pathogens. Int. J. Mol. Sci. 2021, 22, 2913. [Google Scholar] [CrossRef]
- Majumdar, R.; Galewski, P.J.; Eujayl, I.; Minocha, R.; Vincill, E.; Strausbaugh, C.A. Regulatory Roles of Small Non-coding RNAs in Sugar Beet Resistance Against Beet curly top virus. Front. Plant Sci. 2021, 12, 780877. [Google Scholar] [CrossRef]
- Devendran, R.; Namgial, T.; Reddy, K.K.; Kumar, M.; Zarreen, F.; Chakraborty, S. Insights into the multifunctional roles of geminivirus-encoded proteins in pathogenesis. Arch. Virol. 2022, 167, 307–326. [Google Scholar] [CrossRef]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhang, X.; Rong, J.; Fu, X.; Lozano-Duran, R.; et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef]
- Syller, J.; Grupa, A. Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Yogindran, S.; Gnanasekaran, P.; Chakraborty, S.; Winter, S.; Pappu, H.R. Virus and Viroid-Derived Small RNAs as Modulators of Host Gene Expression: Molecular Insights Into Pathogenesis. Front. Microbiol. 2021, 11, 614231. [Google Scholar] [CrossRef] [PubMed]
- Rondon, S.I.; Roster, M.S.; Hamlin, L.L.; Green, K.J.; Karasev, A.V.; Crosslin, J.M. Characterization of Beet curly top virus Strains Circulating in Beet Leafhoppers (Hemiptera: Cicadellidae) in Northeastern Oregon. Plant Dis. 2016, 100, 1586–1590. [Google Scholar] [CrossRef] [PubMed]
- Strausbaugh, C.A.; Wenninger, E.J. Beet leafhopper and BCTV strain survey. Sugarbeet 2021. [Google Scholar]
- Eujayl, I.A.; Strausbaugh, C.A. Beet curly top resistance in USDA-ARS Kimberly germplasm, 2019. PDMR 2020, 14, CF042. [Google Scholar]
- Eujayl, I.A.; Strausbaugh, C.A. Beet curly top resistance in USDA-ARS Kimberly germplasm, 2020. PDMR 2021, 15, V013. [Google Scholar]
- Eujayl, I.A.; Strausbaugh, C.A.; Galewski, P.J. Beet curly top resistance in USDA-ARS Kimberly germplasm, 2022. PDMR 2023, 17, V043. [Google Scholar]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Deng, Z.; Ma, L.; Zhang, P.; Zhu, H. Small RNAs Participate in Plant-Virus Interaction and Their Application in Plant Viral Defense. Int. J. Mol. Sci. 2022, 23, 696. [Google Scholar] [CrossRef]
- Hellinger, R.; Gruber, C.W. Peptide-based protease inhibitors from plants. Drug Discov. Today. 2019, 9, 1877–1889. [Google Scholar] [CrossRef]
- Gatehouse, J.A. Plant resistance towards insect herbivores: A dynamic interaction. New Phytol. 2002, 2, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Haggag, W.M.; Mahmoud, Y.S.; Farag, E.M. Signaling Necessities and Function of Polyamines/Jasmonate-Dependent Induced Resistance in Sugar Beet Against Beet Mosaic Virus (BtMV) Infection. N. Y. Sci. J. 2010, 3, 95–103. [Google Scholar]
- Chen, D.; Mubeen, B.; Hasnain, A.; Rizwan, M.; Adrees, M.; Naqvi, S.A.H.; Iqbal, S.; Kamran, M.; El-Sabrout, A.M.; Elansary, H.O.; et al. Role of Promising Secondary Metabolites to Confer Resistance Against Environmental Stresses in Crop Plants: Current Scenario and Future Perspectives. Front. Plant Sci. 2022, 13, 881032. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, L.; Wang, H.; Xiong, Y. Recent Advances in Antiviral Activities of Triterpenoids. Pharmaceuticals 2022, 15, 1169. [Google Scholar] [CrossRef]
- Jesus, J.A.; Lago, J.H.; Laurenti, M.D.; Yamamoto, E.S.; Passero, L.F. Antimicrobial activity of oleanolic and ursolic acids: An update. Evid. Based Complement. Altern. Med. 2015, 2015, 620472. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, W.; Jia, H.; Feng, H.; Wei, X.; Chen, H.; Wang, D.; Xue, Y.; Sun, X. Plant-derived compounds: A potential source of drugs against Tobacco mosaic virus. Pestic. Biochem. Physiol. 2020, 169, 104589. [Google Scholar] [CrossRef]
- de Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, P.; Sun, J.; Zhao, Y.; Zhang, Y.; Yang, Q.; Wang, W.; Chen, Z.; Mai, T.; Zou, Y.; et al. Research Progress of PPR Proteins in RNA Editing, Stress Response, Plant Growth and Development. Front. Genet. 2021, 12, 765580. [Google Scholar] [CrossRef]
- Gobert, A.; Quan, Y.; Arrive, M.; Waltz, F.; Da Silva, N.; Jomat, L.; Cohen, M.; Jupin, I.; Giege, P. Towards plant resistance to viruses using protein-only RNase P. Nat. Commun. 2021, 12, 1007. [Google Scholar] [CrossRef]
- Sun, M.S.; Zhang, J.; Jiang, L.Q.; Pan, Y.X.; Tan, J.Y.; Yu, F.; Guo, L.; Yin, L.; Shen, C.; Shu, H.B.; et al. TMED2 Potentiates Cellular IFN Responses to DNA Viruses by Reinforcing MITA Dimerization and Facilitating Its Trafficking. Cell Rep. 2018, 25, 3086–3098. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EBMnet 2011, 17, 3. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zhang, H.; Meltzer, P.; Davis, S. RCircos: An R package for Circos 2D track plots. BMC Bioinform. 2013, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Addoquaye, C.; Miller, W.; Axtell, M.J. Cleaveland: A pipeline for using degradome data to find cleaved small rna targets. Bioinformatics 2009, 25, 130. [Google Scholar]
- Hemu, X.; Serra, A.; Darwis, D.A.; Cornvik, T.; Sze, S.K.; Tam, J.P. Peptidomic identification of cysteine-rich peptides from plants. Methods Mol. Biol. 2018, 1719, 379–393. [Google Scholar] [PubMed]



| BCTV sncRNA | Sequence | Sugar Beet Gene ID | Gene Name | CA/Logan | Colorado | Severe | Worland |
|---|---|---|---|---|---|---|---|
| sncRNA_1 | CTGGAGGAGGAAGAAAA | EL10Ac1g00033 | Nitrate reductase [NADH] | x | x | ||
| sncRNA_2 | GTGGCCGAAGAAGAGGA | EL10Ac1g00783 | Homeobox-leucine zipper protein HAT3 | x | x | x | x |
| sncRNA_3 | GCTTCATTTTCTGAGTTA | EL10Ac1g01113 | Protein TRANSPARENT TESTA 12 | x | |||
| sncRNA_4 | GTTCAAAAGATTGTGATGTTGAAGG | EL10Ac1g01206 | Leucine-rich repeat-containing protein 46 | x | |||
| sncRNA_5 | TATCAACCCCAAAATAT | EL10Ac1g02347 | AP2-like ethylene-responsive transcription factor ANT | x | |||
| sncRNA_6 | GGGCTCTCTTCAAATCCCC | EL10Ac2g02425 | Pentatricopeptide repeat-containing protein | x | |||
| sncRNA_7 | TTTCGGAGGAGGAAGAAAAA | EL10Ac2g02734 | Cytosolic sulfotransferase 15 | x | |||
| sncRNA_8 | GAAGAAGCTAGTGAGGT | EL10Ac2g04408 | Kanadaptin | x | x | ||
| sncRNA_9 | CTTCAATATTTGAAGTA | EL10Ac2g04434 | Auxin transport protein BIG | x | |||
| sncRNA_10 | ATCACTTTAAGTTTTTA | EL10Ac2g04915 | Staphylococcal-like nuclease CAN1 | x | x | ||
| sncRNA_11 | AAAGAAGAAAGAGGAAA | EL10Ac3g06583 | Zinc finger CCCH domain-containing protein 32 | x | x | ||
| sncRNA_12 | TTTTTCAAGAAATTGTT | EL10Ac3g06769 | Pentatricopeptide repeat-containing protein | x | |||
| sncRNA_13 | CCCAAAATATGCATCAT | EL10Ac3g07263 | Putative SWI/SNF-related matrix-associated actin-dependent chromatin regulator | x | |||
| sncRNA_14 | GTTGTGGTTGAATCTTT | EL10Ac3g07325 | Putative disease resistance protein RGA3 | x | x | ||
| sncRNA_15 | TGTAGCTCTCTGGCATT | EL10Ac4g08785 | Heat shock 70 kDa protein 16 | x | |||
| sncRNA_16 | TGCAGTGGAATTGTTTG | EL10Ac4g08848 | Pentatricopeptide repeat-containing protein At1g11290 | x | |||
| sncRNA_17 | TAATGATGAATTGTGAAA | EL10Ac4g09996 | Aspartic proteinase-like protein 2 | x | x | ||
| sncRNA_18 | AAGGAAGTGAAGAAGCT | EL10Ac4g10022 | Domain of unknown function (DUF3411) | x | x | x | x |
| sncRNA_19 | AAGTGGGCCCCACAGGAA | EL10Ac5g10458 | Hexose carrier protein HEX6 | x | x | ||
| sncRNA_20 | GCTTCTTCTTTTGAAAG | EL10Ac5g12605 | 7-deoxyloganetic acid glucosyltransferase | x | |||
| sncRNA_21 | GAGATATGAACAAGAGG | EL10Ac6g14074 | Transmembrane emp24 domain-containing protein p24delta3 | x | |||
| sncRNA_22 | CATTTGAAGTTTGATAT | EL10Ac6g14625 | DNA-directed RNA polymerase subunit beta | x | x | ||
| sncRNA_23 | CATTTGAAGTTTGATATA | EL10Ac6g14832 | Myosin heavy chain kinase B | x | x | x | x |
| sncRNA_24 | GATGTTGAAGGAAGTAA | EL10Ac6g15173 | (R,S)-reticuline 7-O-methyltransferase | x | |||
| sncRNA_25 | AATATTGAGGAAGTCTT | EL10Ac6g15406 | Putative pentatricopeptide repeat-containing protein | x | |||
| sncRNA_26 | AGGTTTATTGTGAAGAA | EL10Ac7g16816 | UPF0554 protein C2orf43 homolog | x | x | x | x |
| sncRNA_27 | TGTCTGTTTACCTCCTC | EL10Ac7g16868 | Casparian strip membrane protein 2 | x | |||
| sncRNA_28 | ATTATACTATTATATCT | EL10Ac7g17297 | Hypothetical | x | x | x | x |
| sncRNA_29 | AAGGATATGGAGGGAAGGAGA | EL10Ac7g17983 | Ras-related protein RABA5e | x | x | ||
| sncRNA_30 | AGAGGACTTGTGAGAGC | EL10Ac7g18186 | Exocyst complex component EXO70A1 | x | x | ||
| sncRNA_31 | ATATTAACATATCTATT | EL10Ac8g18763 | Heparanase-like protein 3 | x | |||
| sncRNA_32 | TTTTTCAAGACTTTCAAAAA | EL10Ac8g19534 | Domain of unknown function (DUF4216) | x | x | ||
| sncRNA_33 | TTGAGGAAATACCAATT | EL10Ac9g21413 | MADS-box protein AGL24 | x | |||
| sncRNA_34 | AACTTTACTTTATTTAA | EL10Ac9g21740 | Protein RAFTIN 1A | x | |||
| sncRNA_35 | ATGATGATATGTTGGGT | EL10Ac9g22691 | Plant transposase | x | x | ||
| sncRNA_36 | AATGAAAGAAAAGAAAG | EL10Ac9g22982 | Transmembrane protein 53 | x | x | x | x |
| sncRNA_37 | CATTACCACCTTTAATGA | EL10As5g23617 | Putative pectinesterase inhibitor 45 | x | x | x | x |
| BCTV Strain | BCTV sncRNA | Sequence | Sugar Beet Gene ID | Gene Name | sncRNA Abundance per Sample | Target Gene Expression (FPKM) | Correlation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KDH13 (R) | KDH19-17 (S) | KDH4-9 (R) | KDH13 (R) | KDH19-17 (S) | KDH4-9 (R) | ||||||
| CA/Logan | sncRNA_4 | GTTCAAAAGATTGTGATGTTGAAGG | EL10Ac1g01206 | Leucine-rich repeat-containing protein 46 | 5 | 162 | 3 | 30.37 | 24.22 | 34.26 | −0.93 |
| CA/Logan | sncRNA_20 | GCTTCTTCTTTTGAAAG | EL10Ac5g12605 | 7-deoxyloganetic acid glucosyltransferase | 0 | 23 | 0 | 2.46 | 1.13 | 3.33 | −0.92 |
| CA/Logan | sncRNA_21 | GAGATATGAACAAGAGG | EL10Ac6g14074 | Transmembrane emp24 domain-containing protein p24delta3 | 1 | 35 | 1 | 14.65 | 12.75 | 16.20 | −0.89 |
| CA/Logan | sncRNA_36 | AATGAAAGAAAAGAAAG | EL10Ac9g22982 | Transmembrane protein 53 | 0 | 1 | 0 | 1.46 | 1.41 | 1.71 | −0.63 |
| CA/Logan | sncRNA_26 | AGGTTTATTGTGAAGAA | EL10Ac7g16816 | UPF0554 protein C2orf43 homolog | 14 | 449 | 4 | 7.44 | 4.80 | 4.64 | −0.44 |
| Colorado | sncRNA_3 | GCTTCATTTTCTGAGTTA | EL10Ac1g01113 | Protein TRANSPARENT TESTA 12 | 0 | 1 | 0 | 5.98 | 3.72 | 6.58 | −0.98 |
| Colorado | sncRNA_10 | ATCACTTTAAGTTTTTA | EL10Ac2g04915 | Staphylococcal-like nuclease CAN1 | 0 | 1 | 0 | 8.54 | 6.10 | 7.76 | −0.95 |
| Colorado | sncRNA_26 | AGGTTTATTGTGAAGAA | EL10Ac7g16816 | UPF0554 protein C2orf43 homolog | 0 | 36 | 0 | 7.44 | 4.80 | 4.64 | −0.46 |
| Colorado | sncRNA_30 | AGAGGACTTGTGAGAGC | EL10Ac7g18186 | Exocyst complex component EXO70A1 | 0 | 1 | 0 | 0.14 | 0.11 | 0.19 | −0.78 |
| Colorado | sncRNA_36 | AATGAAAGAAAAGAAAG | EL10Ac9g22982 | Transmembrane protein 53 | 0 | 1 | 0 | 1.46 | 1.41 | 1.71 | −0.63 |
| Severe | sncRNA_25 | AATATTGAGGAAGTCTT | EL10Ac6g15406 | Putative pentatricopeptide repeat-containing protein | 0 | 1 | 0 | 3.41 | 3.03 | 3.20 | −0.83 |
| Severe | sncRNA_26 | AGGTTTATTGTGAAGAA | EL10Ac7g16816 | UPF0554 protein C2orf43 homolog | 14 | 449 | 4 | 7.44 | 4.80 | 4.64 | −0.44 |
| Severe | sncRNA_30 | AGAGGACTTGTGAGAGC | EL10Ac7g18186 | Exocyst complex component EXO70A1 | 0 | 1 | 0 | 0.14 | 0.11 | 0.19 | −0.78 |
| Severe | sncRNA_36 | AATGAAAGAAAAGAAAG | EL10Ac9g22982 | Transmembrane protein 53 | 0 | 1 | 0 | 1.46 | 1.41 | 1.71 | −0.64 |
| Severe | sncRNA_16 | TGCAGTGGAATTGTTTG | EL10Ac4g08848 | Pentatricopeptide repeat-containing protein | 0 | 4 | 0 | 10.97 | 8.59 | 8.73 | −0.54 |
| Worland | sncRNA_26 | AGGTTTATTGTGAAGAA | EL10Ac7g16816 | UPF0554 protein C2orf43 homolog | 0 | 53 | 0 | 7.44 | 4.80 | 4.64 | −0.46 |
| Worland | sncRNA_36 | AATGAAAGAAAAGAAAG | EL10Ac9g22982 | Transmembrane protein 53 | 0 | 1 | 0 | 1.46 | 1.41 | 1.71 | −0.63 |
| BCTV sncRNA | BCTV Strain | Target Sugar Beet Gene ID | Description | Target Transcript Sequence (5′-3′) | Degradome Reads | ||
|---|---|---|---|---|---|---|---|
| I (Mean FPKM) | C (Mean FPKM) | p-Value | |||||
| sncRNA_1 | Colorado, Severe | EL10Ac5g11423 | aspartic protease in guard cell 1 | CCUACUUCCUCUUCCAC & CUGGAGGAGGAAGAAAA | 1044.7 | 0 | 0.10 |
| sncRNA_2 | CA/Logan, Colorado, Severe, Worland | EL10Ac8g19233 | PRA1 family protein F3-like | UCUCUCUUCUUUGGCACC & GUGGCCGAAGAAGAG-GA | 1411.5 | 0 | 0.00 |
| sncRNA_9 | CA/Logan | EL10Ac9g21455 | snakin-2 | ACUUCUCUCUCUUCUUG & AAAGAAGAAAGAGGAAA | 547.0 | 0 | 0.10 |
| sncRNA_10 | Colorado, Severe | Bevul.2G165300 | uncharacterized protein LOC104890896 | UAAGAGCUUAAGG-GAC & AUCACUUUAAGUUUUUA | 362.3 | 0 | 0.02 |
| sncRNA_10 | Colorado, Severe | EL10Ac2g04382 | transport protein SEC31 homolog B isoform X2 | AGGGAACUUAAAGAGAA & AUCACUUUAAGUUUUUA | 137.7 | 0 | 0.07 |
| sncRNA_10 | Colorado, Severe | EL10Ac8g20333 | splicing factor 3B subunit 2 | AAAGAAUUUGAGGUGAA & AUCACUUUAAGUUUUUA | 167.1 | 0 | 0.04 |
| sncRNA_10 | Colorado, Severe | Bevul.9G034200 | uncharacterized protein LOC104890896 | UAAGAGCUUAAGG-GAC & AUCACUUUAAGUUUUUA | 341.9 | 0 | 0.03 |
| sncRNA_11 | Colorado, Severe | EL10Ac9g21455 | snakin-2 | ACUUCUCUCUCUUCUUG & AAAGAAGAAAGAGGAAA | 523.2 | 0 | 0.10 |
| sncRNA_16 | Severe | EL10Ac6g13544 | photosynthetic NDH subunit of lumenal location 5, chloroplastic | UAGUAAAUUCCGCUGCU & UGCAGUGGAAUUGUUUG | 491.8 | 0 | 0.10 |
| sncRNA_20 | CA/Logan | EL10Ac4g08680 | ACT domain-containing protein ACR12 | CUUGCAAGGGGAGAAGC & GCUUCUUCUUUUGAAAG | 142.3 | 0 | 0.07 |
| sncRNA_20 | CA/Logan | EL10Ac5g11755 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | UGGUCAAAAGGAUGAGGC & GCUUC-UUCUUUUGAAAG | 212.4 | 0 | 0.10 |
| sncRNA_24 | CA/Logan | EL10Ac1g02173 | E3 ubiquitin-protein ligase CIP8 | CGACUUCC-UCCGCAUC & GAUGUUGAAGGAAGUAA | 372.3 | 0 | 0.07 |
| sncRNA_28 | CA/Logan, Colorado, Severe, Worland | EL10Ac3g06671 | 28 kDa ribonucleoprotein, chloroplastic | CAAUAUGGUAGUGUGGU & AUUAUACUAUUAUAUCU | 544.5 | 0 | 0.07 |
| sncRNA_34 | CA/Logan | EL10Ac1g01692 | pre-mRNA-processing protein 40A isoform X1 | UUGAAGAAAGUGAAGAA & AACUUUACUUUAUUUAA | 249.1 | 0 | 0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galewski, P.J.; Majumdar, R.; Lebar, M.D.; Strausbaugh, C.A.; Eujayl, I.A. Combined Omics Approaches Reveal Distinct Mechanisms of Resistance and/or Susceptibility in Sugar Beet Double Haploid Genotypes at Early Stages of Beet Curly Top Virus Infection. Int. J. Mol. Sci. 2023, 24, 15013. https://doi.org/10.3390/ijms241915013
Galewski PJ, Majumdar R, Lebar MD, Strausbaugh CA, Eujayl IA. Combined Omics Approaches Reveal Distinct Mechanisms of Resistance and/or Susceptibility in Sugar Beet Double Haploid Genotypes at Early Stages of Beet Curly Top Virus Infection. International Journal of Molecular Sciences. 2023; 24(19):15013. https://doi.org/10.3390/ijms241915013
Chicago/Turabian StyleGalewski, Paul J., Rajtilak Majumdar, Matthew D. Lebar, Carl A. Strausbaugh, and Imad A. Eujayl. 2023. "Combined Omics Approaches Reveal Distinct Mechanisms of Resistance and/or Susceptibility in Sugar Beet Double Haploid Genotypes at Early Stages of Beet Curly Top Virus Infection" International Journal of Molecular Sciences 24, no. 19: 15013. https://doi.org/10.3390/ijms241915013






