Analysis of ANRIL Isoforms and Key Genes in Patients with Severe Coronary Artery Disease
Abstract
:1. Introduction
2. Results
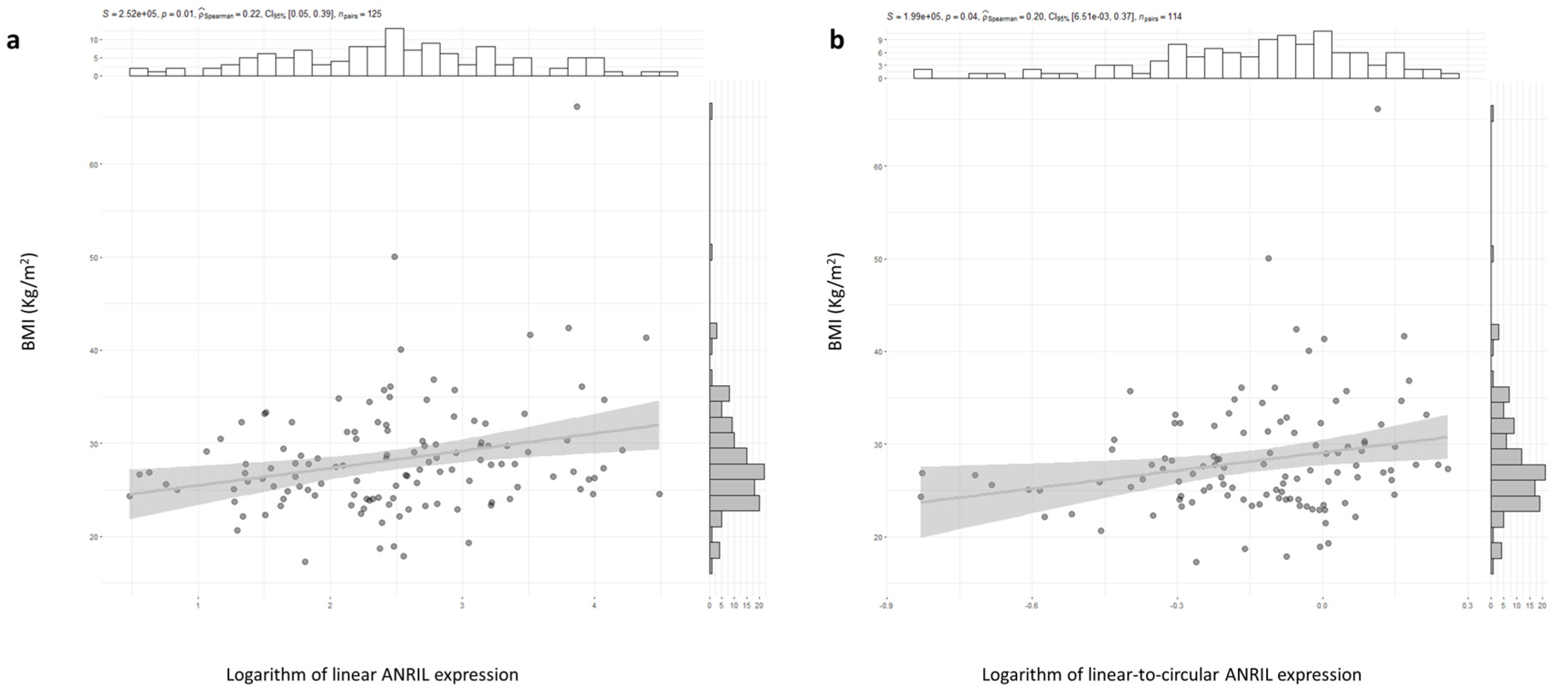
2.1. ANRIL and CDKN2A Gene Expression and CAD Risk Factor
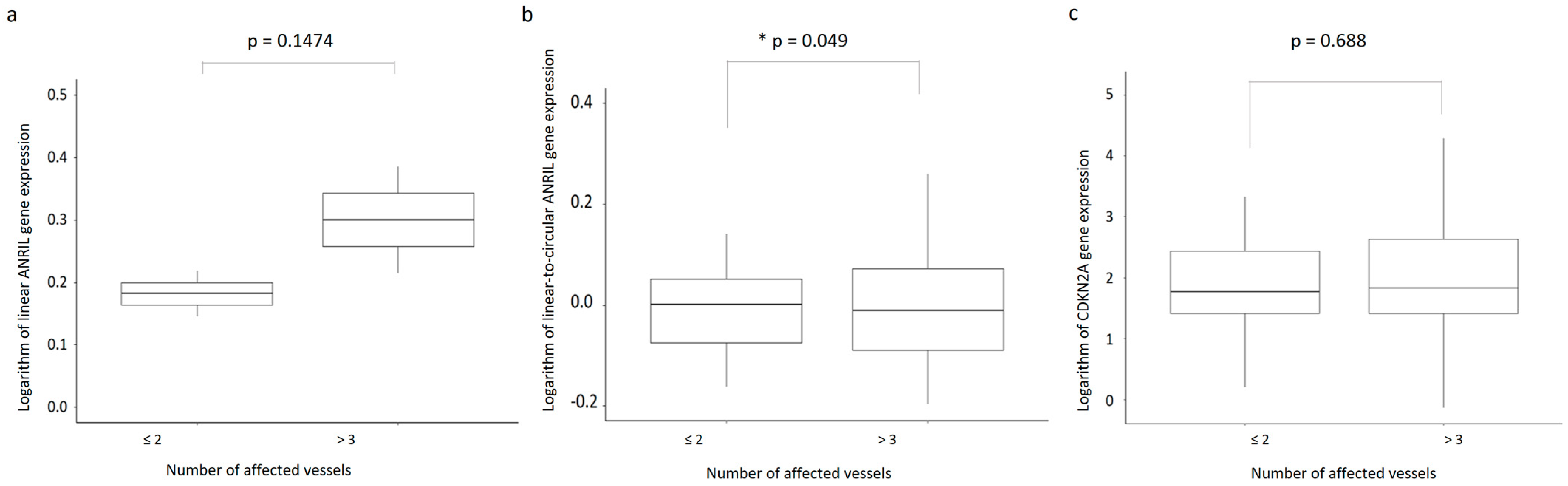
2.2. ANRIL and CDKN2A Gene Expression and CAD Severity
2.3. Plasma VEGF, VEGF, and KDR Gene Expression and CAD Risk Factor
2.4. Plasma VEGF, VEGF, and KDR Gene Expression and CAD Severity
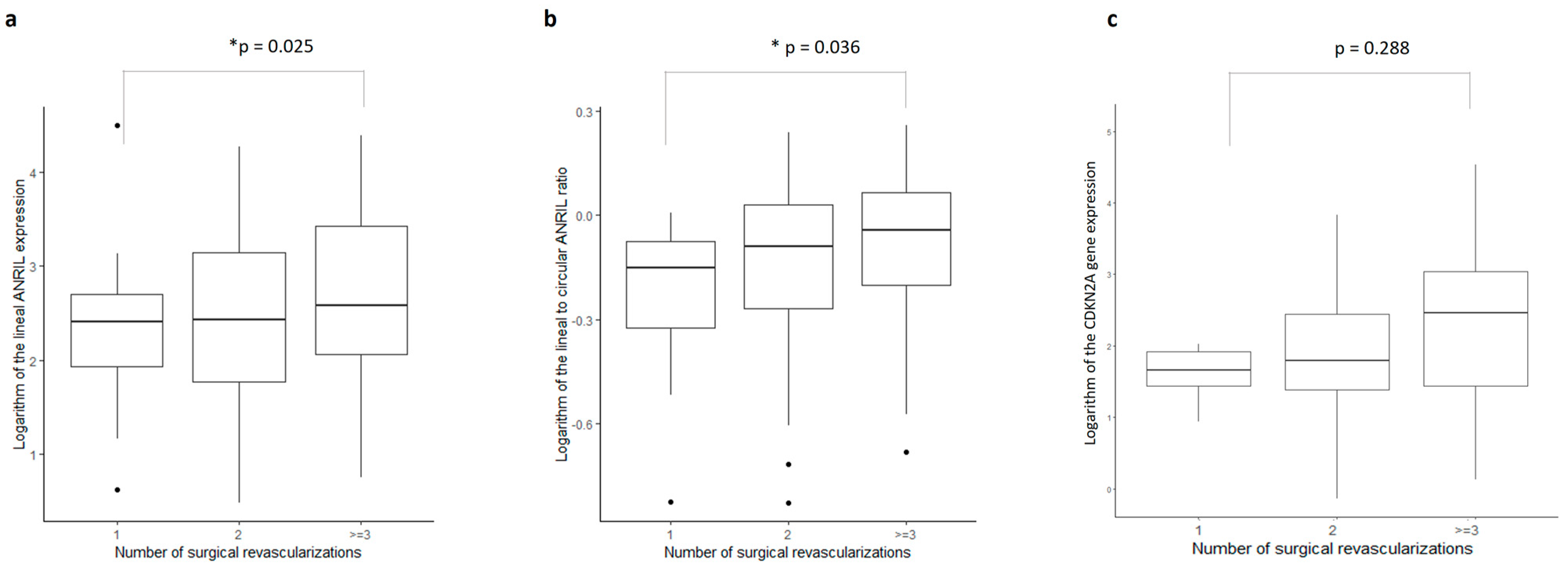
2.5. Gene Expressions as Predictors of Surgical Revascularizations
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Biochemical Determinations
4.3. Gene Expression
4.4. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, K.; Tanaka, T. Molecular Genetics of Coronary Artery Disease. J. Hum. Genet. 2016, 61, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Burton, P.R.; Clayton, D.G.; Cardon, L.R.; Craddock, N.; Deloukas, P.; Duncanson, A.; Kwiatkowski, D.P.; McCarthy, M.I.; Ouwehand, W.H.; Samani, N.J.; et al. Genome-Wide Association Study of 14,000 Cases of Seven Common Diseases and 3000 Shared Controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef]
- McPherson, R.; Pertsemlidis, A.; Kavaslar, N.; Stewart, A.; Roberts, R.; Cox, D.R.; Hinds, D.A.; Pennacchio, L.A.; Tybjaerg-Hansen, A.; Folsom, A.R.; et al. A Common Allele on Chromosome 9 Associated with Coronary Heart Disease. Science 2007, 316, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.-E.; et al. Genomewide Association Analysis of Coronary Artery Disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; et al. A Common Variant on Chromosome 9p21 Affects the Risk of Myocardial Infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef]
- Schunkert, H.; Götz, A.; Braund, P.; McGinnis, R.; Tregouet, D.-A.; Mangino, M.; Linsel-Nitschke, P.; Cambien, F.; Hengstenberg, C.; Stark, K.; et al. Repeated Replication and a Prospective Meta-Analysis of the Association Between Chromosome 9p21.3 and Coronary Artery Disease. Circulation 2008, 117, 1675–1684. [Google Scholar] [CrossRef]
- Aragam, K.G.; Jiang, T.; Goel, A.; Kanoni, S.; Wolford, B.N.; Atri, D.S.; Weeks, E.M.; Wang, M.; Hindy, G.; Zhou, W.; et al. Discovery and Systematic Characterization of Risk Variants and Genes for Coronary Artery Disease in over a Million Participants. Nat. Genet. 2022, 54, 1803–1815. [Google Scholar] [CrossRef]
- Johnson, A.D.; Hwang, S.-J.; Voorman, A.; Morrison, A.; Peloso, G.M.; Hsu, Y.-H.; Thanassoulis, G.; Newton-Cheh, C.; Rogers, I.S.; Hoffmann, U.; et al. Resequencing and Clinical Associations of the 9p21.3 Region. Circulation 2013, 127, 799–810. [Google Scholar] [CrossRef]
- Musunuru, K. Enduring Mystery of the Chromosome 9p21.3 Locus. Circ. Cardiovasc. Genet. 2013, 6, 224–225. [Google Scholar] [CrossRef]
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A LncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef]
- Razeghian-Jahromi, I.; Karimi Akhormeh, A.; Zibaeenezhad, M.J. The Role of ANRIL in Atherosclerosis. Dis. Markers 2022, 2022, 8859677. [Google Scholar] [CrossRef]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gäbel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL Expression Is Associated with Atherosclerosis Risk at Chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef]
- Holdt, L.M.; Teupser, D. Long Noncoding RNA ANRIL: Lnc-Ing Genetic Variation at the Chromosome 9p21 Locus to Molecular Mechanisms of Atherosclerosis. Front. Cardiovasc. Med. 2018, 5, 145. [Google Scholar]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular Non-Coding RNA ANRIL Modulates Ribosomal RNA Maturation and Atherosclerosis in Humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E. Circular RNAs: Unexpected Outputs of Many Protein-Coding Genes. RNA Biol. 2017, 14, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Hu, J. Diagnostic Value of Circulating LncRNA ANRIL and Its Correlation with Coronary Artery Disease Parameters. Brazilian J. Med. Biol. Res. 2019, 52, e8309. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, D. Circulating LncRNA ANRIL Level Positively Correlates with Disease Risk, Severity, Inflammation Level and Poor Prognosis of Coronary Artery Disease. Int. J. Clin. Exp. Med. 2019, 12, 8964–8970. [Google Scholar]
- Liu, Z.F.; Hu, W.W.; Li, R.; Gao, Y.; Yan, L.L.; Su, N. Expression of LncRNA-ANRIL in Patients with Coronary Heart Disease before and after Treatment and Its Short-Term Prognosis Predictive Value. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 376–384. [Google Scholar] [CrossRef]
- Muniz, L.; Lazorthes, S.; Delmas, M.; Ouvrard, J.; Aguirrebengoa, M.; Trouche, D.; Nicolas, E. Circular ANRIL Isoforms Switch from Repressors to Activators of P15/CDKN2B Expression during RAF1 Oncogene-Induced Senescence. RNA Biol. 2021, 18, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.; Zheng, P.; Tu, R.; Huang, J.; Cao, X. Integrated Bioinformatics Analysis for the Identification of Hub Genes and Signaling Pathways Related to CircANRIL. PeerJ 2022, 10, e13135. [Google Scholar] [CrossRef] [PubMed]
- Razeghian-Jahromi, I.; Zibaeenezhad, M.J.; Karimi Akhormeh, A.; Dara, M. Expression Ratio of Circular to Linear ANRIL in Hypertensive Patients with Coronary Artery Disease. Sci. Rep. 2022, 12, 1802. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.-D.; Wagner, N. The Senescence Markers P16INK4A, P14ARF/P19ARF, and P21 in Organ Development and Homeostasis. Cells 2022, 11, 1966. [Google Scholar] [CrossRef]
- Collado, M.; Blasco, M.A.; Serrano, M. Cellular Senescence in Cancer and Aging. Cell 2007, 130, 223–233. [Google Scholar] [CrossRef] [PubMed]
- González-Navarro, H.; Abu Nabah, Y.N.; Vinué, Á.; Andrés-Manzano, M.J.; Collado, M.; Serrano, M.; Andrés, V. P19ARFDeficiency Reduces Macrophage and Vascular Smooth Muscle Cell Apoptosis and Aggravates Atherosclerosis. J. Am. Coll. Cardiol. 2010, 55, 2258–2268. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long Non-Coding RNA ANRIL Is Required for the PRC2 Recruitment to and Silencing of P15INK4B Tumor Suppressor Gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Zhou, X.; Han, X.; Wittfeldt, A.; Sun, J.; Liu, C.; Wang, X.; Gan, L.M.; Cao, H.; Liang, Z. Long Non-Coding RNA ANRIL Regulates Inflammatory Responses as a Novel Component of NF-ΚB Pathway. RNA Biol. 2016, 13, 98–108. [Google Scholar] [CrossRef]
- Xu, B.; Xu, Z.; Chen, Y.; Lu, N.; Shu, Z.; Tan, X.; Franco, N.R.; Massi, M.C.; Ieva, F.; Manzoni, A.; et al. Genetic and Epigenetic Associations of ANRIL with Coronary Artery Disease and Risk Factors. BMC Med. Genom. 2021, 14, 240. [Google Scholar] [CrossRef]
- Han, T.S.; Hur, K.; Cho, H.S.; Ban, H.S. Epigenetic Associations between LncRNA/CircRNA and MiRNA in Hepatocellular Carcinoma. Cancers 2020, 12, 2622. [Google Scholar] [CrossRef]
- Maguire, E.M.; Xiao, Q. Noncoding RNAs in Vascular Smooth Muscle Cell Function and Neointimal Hyperplasia. FEBS J. 2020, 287, 5260–5283. [Google Scholar] [CrossRef]
- Zeng, Z.; Xia, L.; Fan, S.; Zheng, J.; Qin, J.; Fan, X.; Liu, Y.; Tao, J.; Liu, Y.; Li, K.; et al. Circular RNA CircMAP3K5 Acts as a MicroRNA-22-3p Sponge to Promote Resolution of Intimal Hyperplasia Via TET2-Mediated Smooth Muscle Cell Differentiation. Circulation 2021, 143, 354–371. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Condorelli, G. Long Noncoding RNAs and MicroRNAs in Cardiovascular Pathophysiology. Circ. Res. 2015, 116, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, D.; Ji, T.-F.; Shi, L.; Yu, J.-L. Overexpression of LncRNA ANRIL Up-Regulates VEGF Expression and Promotes Angiogenesis of Diabetes Mellitus Combined with Cerebral Infarction by Activating NF-ΚB Signaling Pathway in a Rat Model. Oncotarget 2017, 8, 17347–17359. [Google Scholar] [CrossRef]
- Cho, H.; Shen, G.Q.; Wang, X.; Wang, F.; Archacki, S.; Li, Y.; Yu, G.; Chakrabarti, S.; Chen, Q.; Wang, Q.K. Long Noncoding RNA ANRIL Regulates Endothelial Cell Activities Associated with Coronary Artery Disease by Up-Regulating CLIP1, EZR, and LYVE1 Genes. J. Biol. Chem. 2019, 294, 3881–3898. [Google Scholar] [CrossRef]
- Cho, H.; Li, Y.; Archacki, S.; Wang, F.; Yu, G.; Chakrabarti, S.; Guo, Y.; Chen, Q.; Wang, Q.K. Splice Variants of LncRNA RNA ANRIL Exert Opposing Effects on Endothelial Cell Activities Associated with Coronary Artery Disease. RNA Biol. 2020, 17, 1391–1401. [Google Scholar] [CrossRef]
- Hu, D.-J.; Li, Z.-Y.; Zhu, Y.-T.; Li, C.-C. Overexpression of Long Noncoding RNA ANRIL Inhibits Phenotypic Switching of Vascular Smooth Muscle Cells to Prevent Atherosclerotic Plaque Development in Vivo. Aging 2021, 13, 4299–4316. [Google Scholar] [CrossRef]
- Chen, L.; Qu, H.; Guo, M.; Zhang, Y.; Cui, Y.; Yang, Q.; Bai, R.; Shi, D. ANRIL and Atherosclerosis. J. Clin. Pharm. Ther. 2020, 45, 240–248. [Google Scholar] [CrossRef]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long Noncoding RNAs in Patients with Acute Myocardial Infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Khotina, V.A.; Omelchenko, A.V.; Kalmykov, V.A.; Orekhov, A.N. The Role of the VEGF Family in Atherosclerosis Development and Its Potential as Treatment Targets. Int. J. Mol. Sci. 2022, 23, 931. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Wang, L.; Feng, G.; Li, G.; Yu, M.; Li, Y.; Liu, C.; Yuan, X.; Zang, G.; et al. Impaired Lipid Metabolism by Age-Dependent DNA Methylation Alterations Accelerates Aging. Proc. Natl. Acad. Sci. USA 2020, 117, 4328–4336. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shirvani-Farsani, Z.; Hussen, B.M.; Taheri, M.; Samsami, M. The Key Roles of Non-Coding RNAs in the Pathophysiology of Hypertension. Eur. J. Pharmacol. 2022, 931, 175220. [Google Scholar] [CrossRef]
- Wijesinghe, S.N.; Nicholson, T.; Tsintzas, K.; Jones, S.W. Involvements of Long Noncoding RNAs in Obesity-associated Inflammatory Diseases. Obes. Rev. 2021, 22, e13156. [Google Scholar] [CrossRef] [PubMed]
- Yau, M.; Xu, L.; Huang, C.-L.; Wong, C.-M. Long Non-Coding RNAs in Obesity-Induced Cancer. Non-Coding RNA 2018, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, S.; Nakanishi, C.; Mori, M.; Yoshimuta, T.; Yoshida, S.; Shimojima, M.; Yokawa, J.; Kawashiri, M.; Yamagishi, M.; Hayashi, K. Determination of Early and Late Endothelial Progenitor Cells in Peripheral Circulation and Their Clinical Association with Coronary Artery Disease. Int. J. Vasc. Med. 2015, 2015, 674213. [Google Scholar] [CrossRef]
- Varela-Eirín, M.; Carpintero-Fernández, P.; Sánchez-Temprano, A.; Varela-Vázquez, A.; Paíno, C.L.; Casado-Díaz, A.; Continente, A.C.; Mato, V.; Fonseca, E.; Kandouz, M.; et al. Senolytic Activity of Small Molecular Polyphenols from Olive Restores Chondrocyte Redifferentiation and Promotes a Pro-Regenerative Environment in Osteoarthritis. Aging 2020, 12, 15882–15905. [Google Scholar] [CrossRef] [PubMed]
| N. | Mean | St. Dev. | Min. | Max. | |
|---|---|---|---|---|---|
| Age (years) | 163 | 70.963 | 9.438 | 35 | 101 |
| BMI (Kg/m2) | 135 | 28.206 | 6.115 | 17.290 | 66.120 |
| SBP (mmHg) | 161 | 129.012 | 21.429 | 85 | 200 |
| DBP (mmHg) | 161 | 72.453 | 14.312 | 48 | 169 |
| Cholesterol (mg/dL) | 141 | 139.553 | 41.881 | 59 | 355 |
| HDL cholesterol (mg/dL) | 118 | 40.869 | 13.914 | 17.000 | 138.000 |
| LDL cholesterol (mg/dL) | 104 | 77.618 | 28.203 | 22.900 | 151.000 |
| Triglycerides (mg/dL) | 141 | 145.352 | 71.788 | 55.000 | 585.000 |
| TBARS (µM) | 153 | 2.285 | 1.182 | 1.118 | 7.302 |
| Plasma VEGF (pg/mL) | 40 | 40.951 | 45.445 | 4.432 | 228.102 |
| Glycemia (mg/dL) | 162 | 143.296 | 57.386 | 67 | 442 |
| Urea (mg/dL) | 148 | 54.209 | 36.086 | 13 | 258 |
| Urate (mg/dL) | 54 | 5.244 | 1.857 | 1.180 | 11.140 |
| Creatinine (mg/dL) | 161 | 1.232 | 0.931 | 0.500 | 8.400 |
| GFR (CKD-EPI) (ml/min−1) | 161 | 70.177 | 26.402 | 5.480 | 118.660 |
| GFR (MDRD-IMDS) (ml/min−1) | 160 | 72.657 | 30.549 | 6.200 | 166.880 |
| Folic acid (ng/mL) | 23 | 6.996 | 4.381 | 1.800 | 18.800 |
| Vitamin B12 (pg/mL) | 29 | 357.828 | 197.455 | 159 | 1021 |
| Leukocytes (×103/µL) | 162 | 10.074 | 4.789 | 1.990 | 31.710 |
| Neutrophils (×103/µL) | 162 | 7.214 | 4.732 | 1.260 | 30.310 |
| Lymphocytes (×103/µL) | 162 | 1.832 | 0.884 | 0.240 | 5.080 |
| Monocytes (×103/µL) | 162 | 0.777 | 0.394 | 0.030 | 3.200 |
| Eosinophils (×103/µL) | 162 | 0.205 | 0.192 | 0.000 | 0.950 |
| Basophils (×103/µL) | 162 | 0.043 | 0.031 | 0.000 | 0.180 |
| Red blood cells (×103/µL) | 162 | 4.000 | 0.816 | 2.440 | 6.060 |
| Condition/Habit | Total (%) | Male (%) | Female (%) | p Value |
|---|---|---|---|---|
| Number of Patients | 163 | 131 | 32 | - |
| Hypertensive patients | 82.8 | 81.6 | 87.5 | 0.434 |
| Diabetic patients | 60.7 | 57.2 | 75 | 0.065 |
| Patients with dyslipidemia | 79.7 | 77.8 | 87.5 | 0.224 |
| Smokers | 47.2 | 54.2 | 18.8 | <0.001 |
| Patients with overweight | 15.9 | 15.2 | 18.7 | 0.630 |
| Patients with LVD * | 58.3 | 55.7 | 68.7 | 0.180 |
| Patients with aortic trunk lesion | 33 | 34.6 | 28.1 | 0.485 |
| Patients with ≥2 affected vessels | 92 | 93.9 | 84.4 | 0.075 |
| Patients with ≥2 revascularizations | 82.8 | 87 | 65.6 | 0.004 |
| All causes of death | 26.4 | 26.7 | 25 | 0.843 |
| Variables | Univariate | Multivariate | Optimal Multivariate Model | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Beta | SE | OR | CI 95 | p-Value | Beta | SE | OR | CI 95 | p-Value | Beta | SE | OR | CI 95 | p-Value | |
| Intercept | 2.96 | 2.49 | 19.318 | 0.157–3574.719 | 0.235 | 2.83 | 1.1 | 16.926 | 2.91–328.83 | 0.01 | ||||||
| Female sex | 77 | −0.58 | 0.59 | 0.56 | 0.18–1.86 | 0.325 | −0.9 | 0.8 | 0.405 | 0.08–1.958 | 0.26 | - | - | - | - | - |
| Hypertension | 77 | −1.51 | 1.08 | 0.22 | 0.01–1.25 | 0.161 | −1.49 | 1.19 | 0.226 | 0.011–1.651 | 0.211 | −1.57 | 1.09 | 0.209 | 0.011–1.21 | 0.15 |
| Diabetes | 77 | 0.46 | 0.52 | 1.59 | 0.57–4.49 | 0.375 | 0.72 | 0.64 | 2.063 | 0.599–7.657 | 0.258 | - | - | - | - | - |
| Dyslipidemia | 77 | −0.17 | 0.64 | 0.85 | 0.21–2.81 | 0.795 | 0.21 | 0.81 | 1.239 | 0.234–5.99 | 0.791 | - | - | - | - | - |
| Smoker | 77 | 0.17 | 0.52 | 1.18 | 0.42–3.35 | 0.751 | 0.2 | 0.75 | 1.226 | 0.28–5.506 | 0.785 | - | - | - | - | - |
| LVD | 77 | −0.45 | 0.52 | 0.64 | 0.22–1.78 | 0.392 | −0.53 | 0.61 | 0.59 | 0.174–1.932 | 0.385 | - | - | - | - | - |
| Aortic trunk | 77 | −0.82 | 0.53 | 0.44 | 0.15–1.24 | 0.122 | −1.3 | 0.66 | 0.274 | 0.069–0.965 | 0.051 | −0.85 | 0.54 | 0.426 | 0.14–1.21 | 0.113 |
| BMI | 77 | 0.01 | 0.04 | 1.01 | 0.93–1.1 | 0.857 | −0.03 | 0.05 | 0.972 | 0.884–1.083 | 0.57 | - | - | - | - | - |
| Lineal ANRIL | 77 | 0.32 | 0.35 | 1.38 | 0.7–2.81 | 0.36 | 0.68 | 0.58 | 1.978 | 0.66–6.681 | 0.239 | - | - | - | - | - |
| Circular ANRIL | 77 | −0.14 | 0.31 | 0.87 | 0.48–1.66 | 0.646 | −0.17 | 0.38 | 0.841 | 0.401–1.798 | 0.645 | - | - | - | - | - |
| Ratio ANRIL | 77 | 1.26 | 1.06 | 3.53 | 0.15–86.8 | 0.43 | - | - | - | - | - | - | - | - | - | - |
| CDKN2A | 77 | 0.01 | 0.32 | 1.01 | 0.53–1.93 | 0.983 | −0.56 | 0.58 | 0.571 | 0.169–1.752 | 0.335 | - | - | - | - | - |
| VEGF | 77 | 0.04 | 0.29 | 1.04 | 0.59–1.9 | 0.89 | 0.11 | 0.41 | 1.112 | 0.499–2.563 | 0.794 | - | - | - | - | - |
| KDR | 77 | 0.18 | 0.19 | 1.19 | 0.81–1.74 | 0.357 | 0.11 | 0.22 | 1.115 | 0.719–1.718 | 0.618 | - | - | - | - | - |
| AUC ROC | 0.7509 | 0.643 | ||||||||||||||
| Variables | Univariate | Multivariate | Optimal Multivariate Model | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Beta | SE | OR | CI 95 | p-Value | Beta | SE | OR | CI 95 | p-Value | Beta | SE | OR | CI 95 | p-Value | |
| Intercept | - | - | - | - | - | - | −2.38 | 3.16 | 0.092 | 0–36.039 | 0.451 | −1.41 | 2.37 | 0.245 | 0.001–19.76 | 0.554 |
| Femalesex | 77 | −1 | 0.65 | 0.37 | 0.1–1.4 | 0.128 | −1.59 | 1.28 | 0.204 | 0.013–2.241 | 0.212 | −1.85 | 0.99 | 0.157 | 0.018–0.988 | 0.062 |
| Hypertension | 77 | −0.91 | 1.09 | 0.4 | 0.02–2.38 | 0.403 | 0.03 | 1.61 | 1.026 | 0.025–22.30 | 0.987 | - | - | - | - | - |
| Diabetes | 77 | 0.6 | 0.61 | 1.82 | 0.54–6.26 | 0.328 | 2.07 | 1.12 | 7.953 | 1.17–109.03 | 0.063 | 1.77 | 0.87 | 5.851 | 1.185–39.824 | 0.043 |
| Dyslipidemia | 77 | 0.07 | 0.72 | 1.07 | 0.22–4.09 | 0.924 | 1.9 | 1.21 | 6.662 | 0.689–93.45 | 0.116 | 1.93 | 1.12 | 6.876 | 0.809–75.708 | 0.085 |
| Smoker | 77 | 0.09 | 0.61 | 1.1 | 0.33–3.75 | 0.881 | 0.1 | 1.14 | 1.11 | 0.122–12.43 | 0.927 | - | - | - | - | - |
| LVD | 77 | 0.41 | 0.62 | 1.5 | 0.45–5.44 | 0.513 | −0.19 | 0.94 | 0.826 | 0.115–5.23 | 0.838 | - | - | - | - | - |
| Aortictrunk | 77 | 0.95 | 0.7 | 2.59 | 0.71–12.38 | 0.176 | 0.99 | 0.98 | 2.699 | 0.433–22.51 | 0.31 | - | - | - | - | - |
| BMI | 77 | −0.01 | 0.04 | 0.99 | 0.91–1.09 | 0.768 | −0.14 | 0.08 | 0.87 | 0.724–1.02 | 0.092 | −0.14 | 0.07 | 0.872 | 0.75–1.009 | 0.06 |
| LinearANRIL | 77 | 0.82 | 0.45 | 2.27 | 0.99–5.86 | 0.067 | 1.69 | 0.88 | 5.419 | 1.184–42.45 | 0.055 | 1.57 | 0.72 | 4.8 | 1.314–23.896 | 0.029 |
| CircularANRIL | 77 | −0.29 | 0.34 | 0.75 | 0.39–1.54 | 0.396 | −0.99 | 0.63 | 0.371 | 0.087–1.1 | 0.118 | −0.85 | 0.5 | 0.429 | 0.14–1.07 | 0.09 |
| RatioANRIL | 77 | 3.28 | 1.93 | 26.54 | 0.66–1436.8 | 0.09 | - | - | - | - | - | - | - | - | - | - |
| CDKN2A | 77 | 0.28 | 0.39 | 1.32 | 0.62–2.96 | 0.481 | −1.26 | 0.8 | 0.284 | 0.046–1.182 | 0.118 | −0.92 | 0.63 | 0.399 | 0.105–1.318 | 0.143 |
| VEGF | 77 | 0.85 | 0.43 | 2.33 | 1.07–5.85 | 0.048 | 2.73 | 1.06 | 15.369 | 2.65–183.23 | 0.01 | 2.35 | 0.84 | 10.516 | 2.378–67.635 | 0.005 |
| KDR | 77 | 0.3 | 0.22 | 1.34 | 0.87–2.06 | 0.172 | 0.38 | 0.32 | 1.459 | 0.783–2.85 | 0.237 | - | - | - | - | - |
| AUCROC | - | 0.887 | 0.863 | |||||||||||||
| Variables | Univariate | Multivariate | Optimal Multivariate Model | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Beta | SE | OR | CI 95 | p-Value | Beta | SE | OR | CI 95 | p-Value | Beta | SE | OR | CI 95 | p-Value | |
| Intercept | - | - | - | - | - | - | 2.77 | 2.94 | 16.019 | 0.064–7938 | 0.346 | 4.1 | 2.04 | 60.087 | 1.399–4712 | 0.045 |
| Femalesex | 77 | 1 | 0.61 | 2.73 | 0.79–9.08 | 0.103 | 1.39 | 0.94 | 4.017 | 0.654–28.5 | 0.14 | 1.16 | 0.72 | 3.18 | 0.772–13.6 | 0.108 |
| Hypertension | 77 | −0.29 | 0.74 | 0.75 | 0.19–3.73 | 0.696 | −0.04 | 0.97 | 0.958 | 0.151–7.521 | 0.965 | - | - | - | - | - |
| Diabetes | 77 | −0.11 | 0.57 | 0.89 | 0.29–2.8 | 0.842 | 0.3 | 0.72 | 1.349 | 0.339–5.932 | 0.676 | - | - | - | - | - |
| Dyslipidemia | 77 | −1.38 | 0.61 | 0.25 | 0.07–0.85 | 0.024 | −1.35 | 0.87 | 0.261 | 0.044–1.437 | 0.122 | −1.49 | 0.72 | 0.225 | 0.052–0.91 | 0.038 |
| Smoker | 77 | −0.22 | 0.57 | 0.8 | 0.26–2.43 | 0.699 | 0.76 | 0.85 | 2.129 | 0.414–12.501 | 0.375 | - | - | - | - | - |
| LVD | 77 | 0.48 | 0.57 | 1.62 | 0.54–5.07 | 0.395 | 0.52 | 0.71 | 1.68 | 0.418–7.226 | 0.467 | - | - | - | - | - |
| Aortictrunk | 77 | −0.87 | 0.63 | 0.42 | 0.11–1.36 | 0.17 | −0.75 | 0.75 | 0.474 | 0.097–1.99 | 0.321 | - | - | - | - | - |
| IMC | 77 | −0.21 | 0.08 | 0.81 | 0.69–0.93 | 0.007 | −0.18 | 0.08 | 0.835 | 0.692–0.968 | 0.031 | −0.18 | 0.08 | 0.838 | 0.708–0.96 | 0.022 |
| LinealANRIL | 77 | −0.35 | 0.38 | 0.71 | 0.33–1.47 | 0.363 | −0.46 | 0.73 | 0.63 | 0.141–2.513 | 0.527 | - | - | - | - | - |
| CircularANRIL | 77 | 0.09 | 0.34 | 1.09 | 0.53–2.05 | 0.797 | 0.1 | 0.43 | 1.105 | 0.451–2.569 | 0.818 | - | - | - | - | - |
| RatioANRIL | 77 | −1.93 | 1.74 | 0.15 | 0–4.31 | 0.267 | - | - | - | - | - | - | - | - | - | - |
| CDKN2A | 77 | 0.08 | 0.35 | 1.09 | 0.54–2.16 | 0.813 | 0.72 | 0.78 | 2.047 | 0.46–9.931 | 0.356 | - | - | - | - | - |
| VEGF | 77 | 0.35 | 0.31 | 1.42 | 0.77–2.63 | 0.261 | 0.19 | 0.52 | 1.211 | 0.424–3.453 | 0.714 | - | - | - | - | - |
| KDR | 77 | −0.13 | 0.21 | 0.88 | 0.59–1.34 | 0.537 | −0.13 | 0.26 | 0.875 | 0.53–1.496 | 0.606 | - | - | - | - | - |
| AUCROC | - | 0.8135 | 0.7684 | |||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Esparragón, F.; Torres-Mata, L.B.; Cazorla-Rivero, S.E.; Serna Gómez, J.A.; González Martín, J.M.; Cánovas-Molina, Á.; Medina-Suárez, J.A.; González-Hernández, A.N.; Estupiñán-Quintana, L.; Bartolomé-Durán, M.C.; et al. Analysis of ANRIL Isoforms and Key Genes in Patients with Severe Coronary Artery Disease. Int. J. Mol. Sci. 2023, 24, 16127. https://doi.org/10.3390/ijms242216127
Rodríguez-Esparragón F, Torres-Mata LB, Cazorla-Rivero SE, Serna Gómez JA, González Martín JM, Cánovas-Molina Á, Medina-Suárez JA, González-Hernández AN, Estupiñán-Quintana L, Bartolomé-Durán MC, et al. Analysis of ANRIL Isoforms and Key Genes in Patients with Severe Coronary Artery Disease. International Journal of Molecular Sciences. 2023; 24(22):16127. https://doi.org/10.3390/ijms242216127
Chicago/Turabian StyleRodríguez-Esparragón, Francisco, Laura B. Torres-Mata, Sara E. Cazorla-Rivero, Jaime A. Serna Gómez, Jesús M. González Martín, Ángeles Cánovas-Molina, José A. Medina-Suárez, Ayose N. González-Hernández, Lidia Estupiñán-Quintana, María C. Bartolomé-Durán, and et al. 2023. "Analysis of ANRIL Isoforms and Key Genes in Patients with Severe Coronary Artery Disease" International Journal of Molecular Sciences 24, no. 22: 16127. https://doi.org/10.3390/ijms242216127








