Development of Single-Segment Substitution Lines and Fine-Mapping of qSPP4 for Spikelets Per Panicle and qGW9 for Grain Width Based on Rice Dual-Segment Substitution Line Z783
Abstract
:1. Introduction
2. Results
2.1. Identification of Substitution Segments in Z783
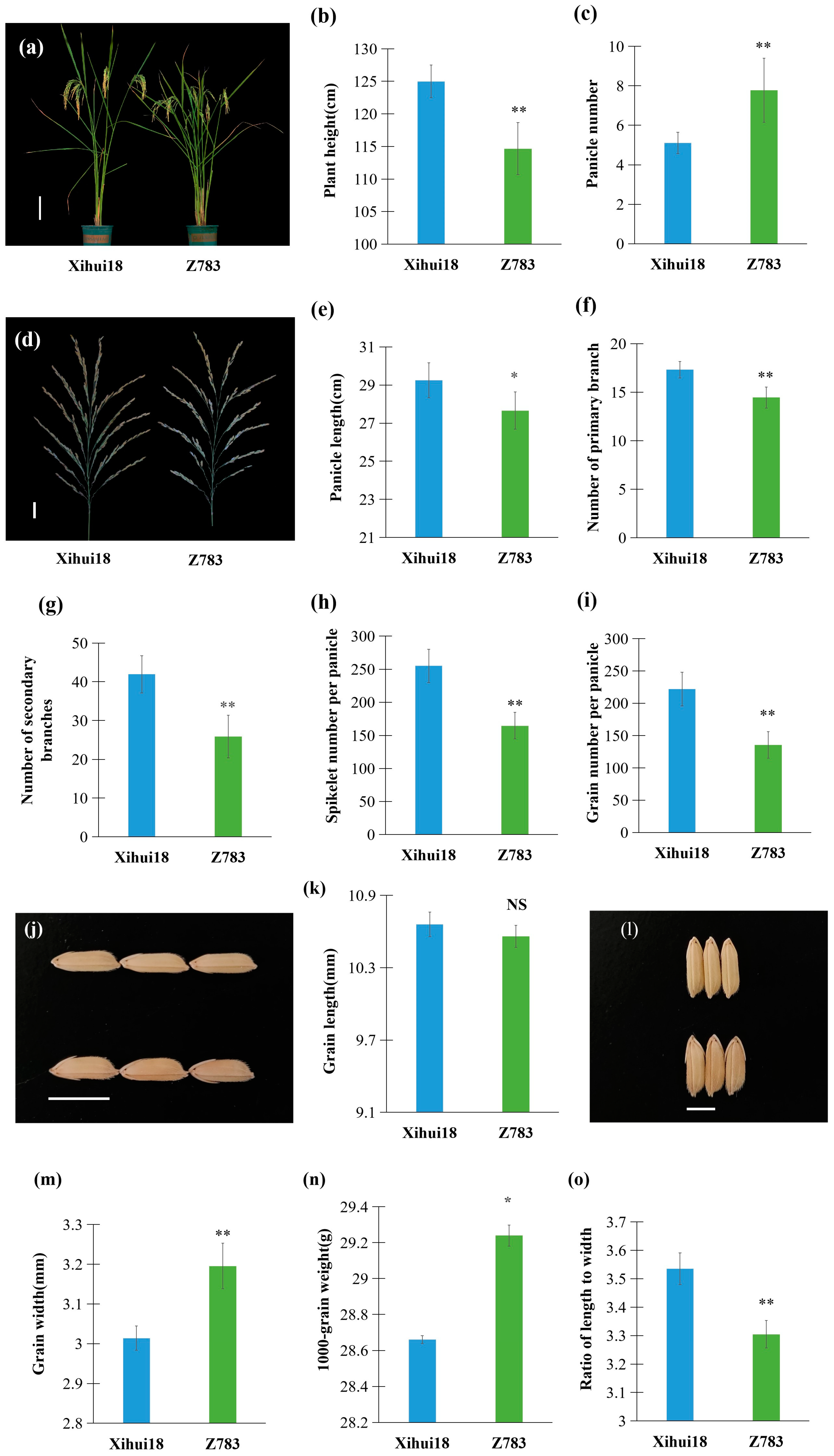
2.2. Phenotype Analysis of Z783
2.3. Identification of QTL by a Secondary F2 Population from Xihui18/Z783
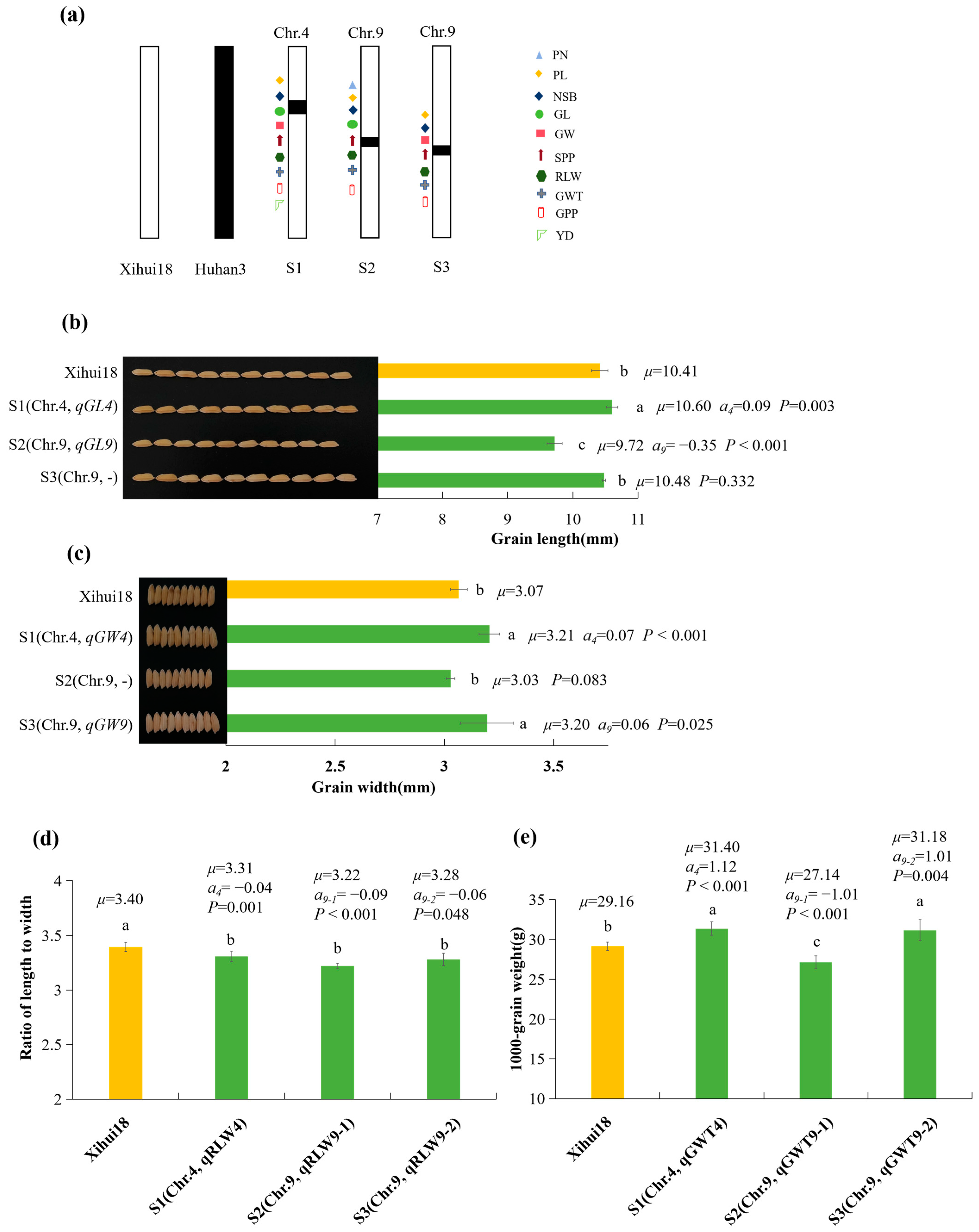
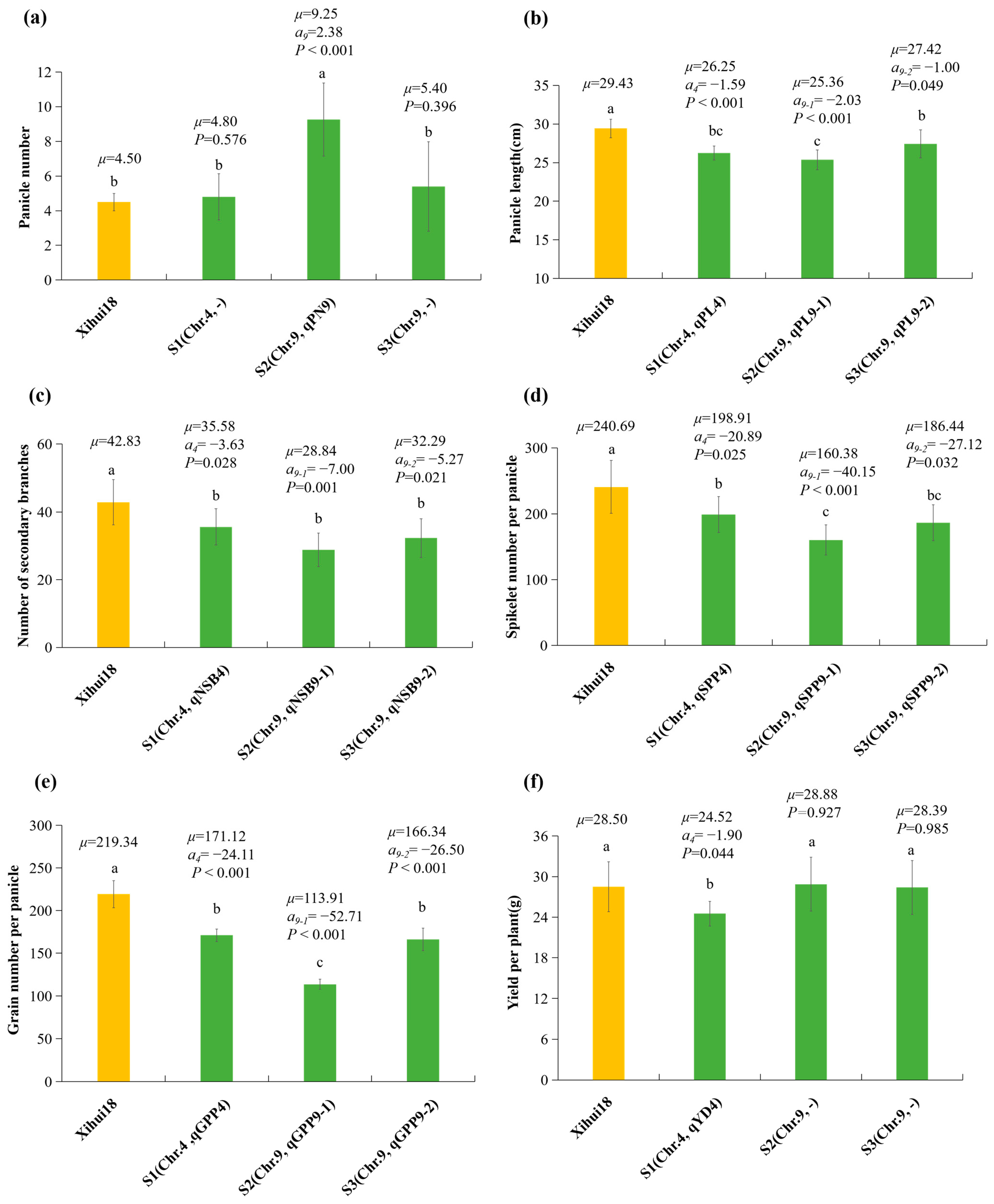
2.4. Construction of SSSLs and Additive Effects Analysis of QTLs Identified by the SSSLs
2.5. Genetic Analysis of qGW9 for Grain Width and qSPP4 for Spikelet Number per Panicle
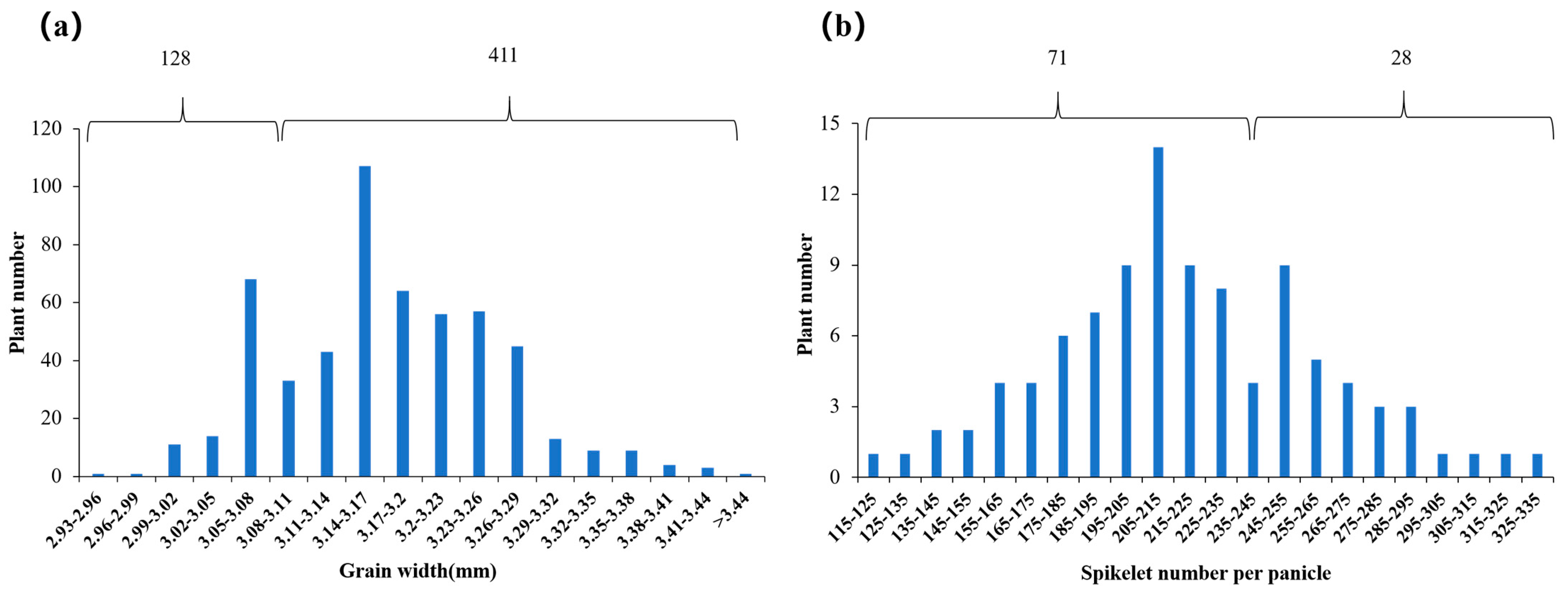
2.6. Fine Mapping of qGW9 for Wide Grain
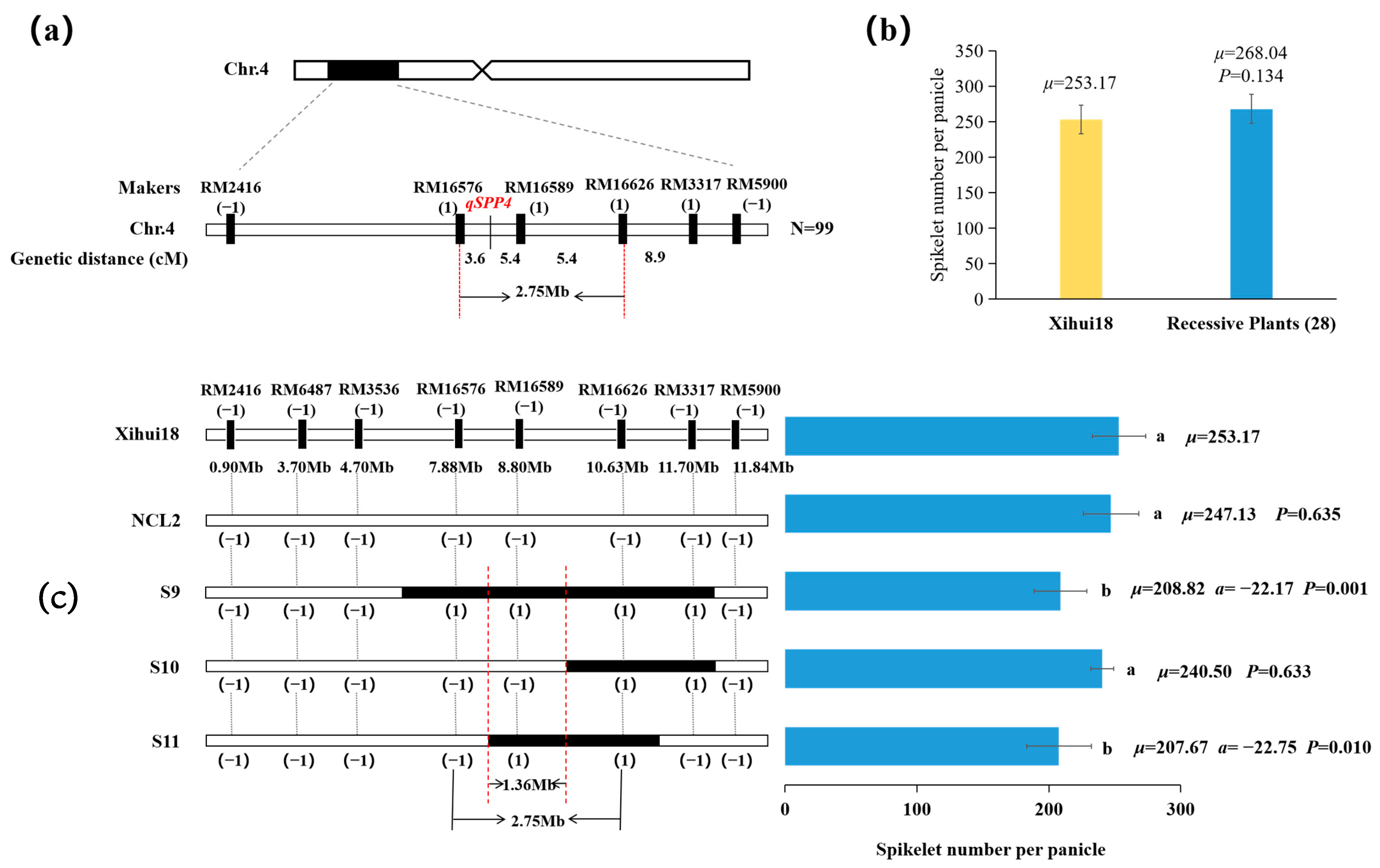
2.7. Fine Mapping of qSPP4 for Spikelets per Panicle
3. Discussion
3.1. The 11 Single Segment Substitution Lines Dissected from CSSL-Z783 Has Important Application Value in Rice Breeding by Design
3.2. Comparison of QTLs Identified in the Study with the Reported Genes
4. Materials and Methods
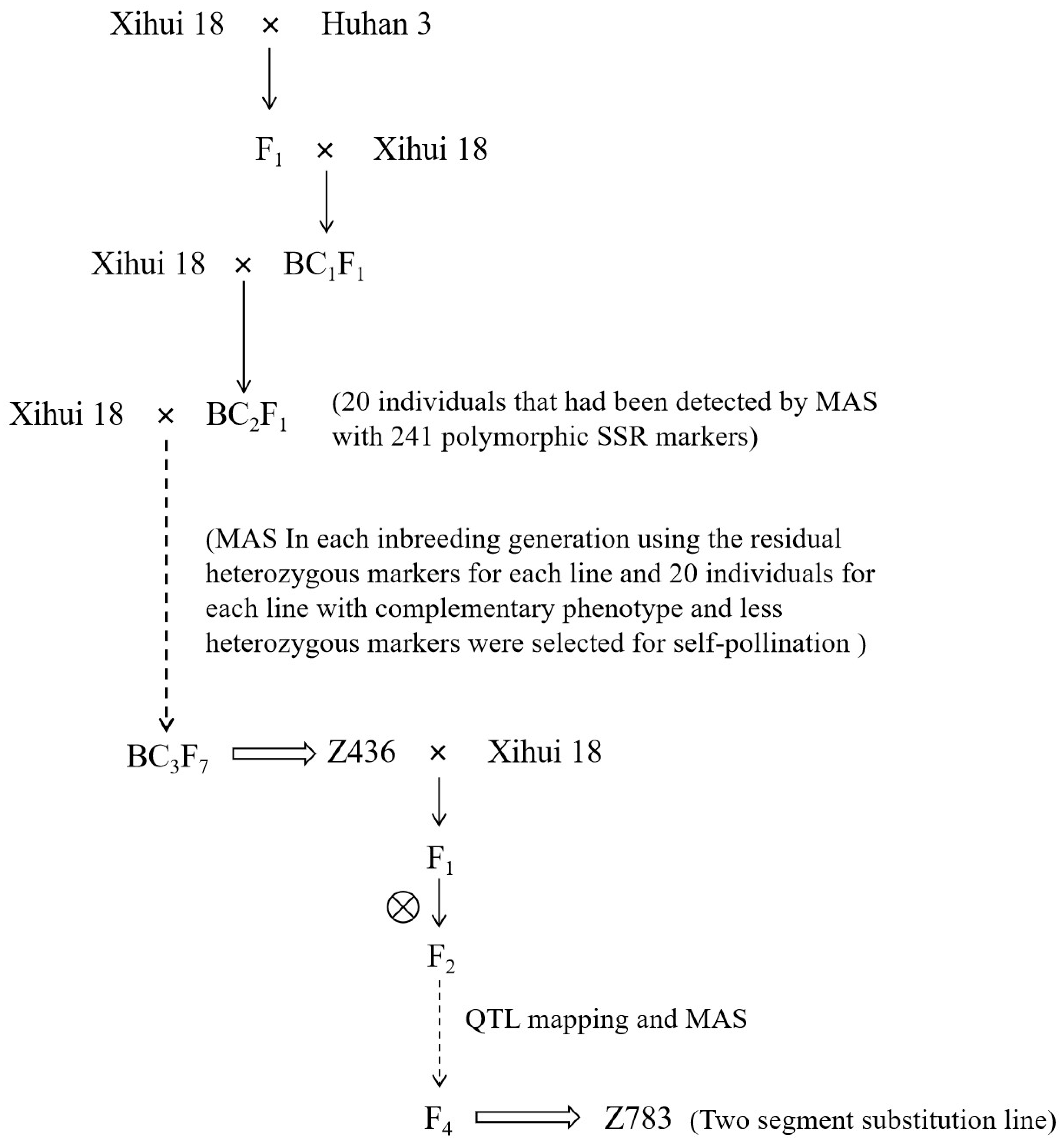
4.1. Experimental Materials
4.2. Material Planting Methods
4.3. Assessment of Agronomic Traits
4.4. QTL Mapping
4.5. Development of SSSLs
4.6. Identification and Additive Effect Analysis of QTL by Three SSSLs
4.7. Fine-Mapping and Overlapping Substitution Mapping of qGW9 and qSPP4
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruinsma, J. The Resource Outlook to 2050: By How Much Do Land, Water and Crop Yields Need to Increase by 2050? Food and Agriculture Organization (FAO): Rome, Italy, 2009. [Google Scholar]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, J.; Zhu, F.; Paul, P.; Gao, T.; Dhatt, B.K.; Ge, Y.; Staswick, P.; Yu, H.; Walia, H. PI-Plat: A high-resolution image-based 3D reconstruction method to estimate growth dynamics of rice inflorescence traits. Plant Methods 2019, 15, 162. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.Z.; Zhang, Q.F. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Li, G.M.; Tang, J.Y.; Zheng, J.K.; Chu, C.C. Exploration of rice yield potential: Decoding agronomic and physiological traits. Crop J. 2021, 9, 577–589. [Google Scholar] [CrossRef]
- Balakrishnan, D.; Surapaneni, M.; Mesapogu, S.; Neelamraju, S. Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor. Appl. Genet. 2019, 132, 1–25. [Google Scholar] [CrossRef]
- Hirabayashi, H.; Sato, H.; Nonoue, Y.; Kuno-Takemoto, Y.; Takeuchi, Y.; Kato, H.; Nemoto, H.; Ogawa, T.; Yano, M.; Imbe, T. Development of introgression lines derived from Oryza rufipogon and O. glumaepatula in the genetic background of japonica cultivated rice (O. sativa L.) and evaluation of resistance to rice blast. Breed. Sci. 2010, 60, 604–612. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Zeng, R.Z.; Zhang, Z.M.; Ding, X.H.; Lu, Y.G. The construction of a library of single segment substitution lines in rice (Oryza sativa L.). Rice Genet. Newsl. 2004, 21, 85–87. [Google Scholar]
- Monforte, A.J.; Tanksley, S.D. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: A tool for gene mapping and gene discovery. Genome 2000, 43, 803–813. [Google Scholar] [CrossRef]
- Keurentjes, J.J.B.; Bentsink, L.; Alonso-Blanco, C.; Hanhart, C.J.; Blankestijn-De Vries, H.; Effgen, S.; Vreugdenhil, D.; Koornneef, M. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 2000, 175, 891–905. [Google Scholar] [CrossRef]
- Shim, R.A.; Angeles, E.R.; Ashikari, M.; Takashi, T. Development and evaluation of Oryza glaberrima Steud. Chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 2010, 60, 613–619. [Google Scholar] [CrossRef]
- Yoshimura, A.; Nagayama, H.; Kurakazu, T.; Sanchez, P.L.; Doi, K.; Yamagata, Y.; Yasui, H. Introgression lines of rice (Oryza sativa L.) carrying a donor genome from the wild species, O. glumaepatula Steud. and O. meridionalis Ng. Breed. Sci. 2010, 60, 597–603. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Koornneef, M. Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 2000, 5, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Sasaki, T. Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 1997, 35, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, Y. Ubiquitin-mediated control of seed size in plants. Front. Plant Sci. 2014, 5, 332. [Google Scholar] [CrossRef]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Hao, J.Q.; Dang, W.K.; Wu, Y.B.; Huang, K.; Duan, P.; Li, N.; Xu, R.; Zeng, D.; Dong, G.; Zhang, B.; et al. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol Plant 2021, 14, 1266–1280. [Google Scholar] [CrossRef]
- Garg, R.; Jhanwar, S.; Tyagi, A.K.; Jain, M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2012, 17, 353–367. [Google Scholar] [CrossRef]
- Choi, B.S.; Kim, Y.J.; Markkandan, K.; Koo, Y.J.; Song, J.T.; Seo, H.S. GW2 functions as an E3 ubiquitin ligase for rice expansin-like 1. Int. J. Mol. Sci. 2018, 19, 1904. [Google Scholar] [CrossRef]
- Urano, D.; Miura, K.; Wu, Q.; Iwasaki, Y.; Jackson, D.; Jones, A.M. Plant morphology of heterotrimeric G protein mutants. Plant Cell Physiol. 2016, 57, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Kuroha, T.; Ayano, M.; Furuta, T.; Nagai, K.; Komeda, N.; Segami, S.; Miura, K.; Ogawa, D.; Kamura, T.; et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, Y.Y.; Ma, Q.B.; Li, D.; Xu, Z.H.; Chong, K. Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Res. 2006, 16, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Chen, K.; Dong, K.Q.; Shi, C.L.; Ye, W.W.; Gao, J.P.; Shan, J.X.; Lin, H.X. GRAIN SIZE AND NUMBER1 Negatively Regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 2018, 30, 871–888. [Google Scholar] [CrossRef]
- Yuan, R.Z.; Zhao, N.; Usman, B.; Luo, L.; Liao, S.; Qin, Y.; Nawaz, G.; Li, R. Development of chromosome segment substitution lines (CSSLs) derived from guangxi wild rice (Oryza rufipogon Griff.) under Rice (Oryza sativa L.) background and the identification of QTLs for plant architecture, agronomic traits and cold tolerance. Genes 2020, 11, 980. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.K.; Wu, K.; Yuan, Q.B.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Taguchi-Shiobara, F.; Kojima, Y.; Ebitani, T.; Yano, M.; Ebana, K. Variation in domesticated rice inflorescence architecture revealed by principal component analysis and quantitative trait locus analysis. Breed. Sci. 2011, 61, 52–60. [Google Scholar] [CrossRef]
- Deveshwar, P.; Prusty, A.; Sharma, S.; Tyagi, A.K. Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 2020, 11, 586462. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.Y.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mi, X.F.; Shan, J.X.; Li, X.M.; Xu, J.L.; Lin, H.X. The QTL GNP1 Encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016, 120, e1006386. [Google Scholar] [CrossRef]
- Nadir, S.; Khan, S.; Zhu, Q.; Henry, D.; Wei, L.; Lee, D.S.; Chen, L. An overview on reproductive isolation in Oryza sativa complex. AoB Plants 2018, 10, ply060. [Google Scholar] [CrossRef]
- Furuta, T.; Uehara, K.; Angeles-Shim, R.B.; Shim, J.; Ashikari, M.; Takashi, T. Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 2014, 63, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Akagi, H.; Nakamura, A.; Yokozeki-Misono, Y.; Inagaki, A.; Takahashi, H.; Mori, K.; Fujimura, T. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 2004, 108, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Etsuko, I.; Natsuko, I.; Sota, F.; Kazama, T.; Toriyama, K. The fertility restorer gene, Rf2, for lead rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J. 2001, 65, 359–367. [Google Scholar] [CrossRef]
- Cai, J.; Liao, Q.P.; Dai, Z.J.; Zhu, H.T.; Zeng, R.Z.; Zhang, Z.M.; Zhang, G.Q. Allelic differentiations and effects of the Rf3 and Rf4 genes on fertility restoration in rice with wild abortive cytoplasmic male sterility. Biol. Plant. 2013, 57, 274–280. [Google Scholar] [CrossRef]
- Farkhanda, N.; Muhammad, A.; Sun, S.F.; Zhang, J.Y.; Wang, D.C.; Zhou, K.; Zhao, F.M. Identification and pyramiding of QTLs for rice grain size based on short-Wide grain CSSL-Z436 seven SSSLs & eight DSSLs. Int. Trans. J. Eng. Manag. Appl. Sci. Technol. 2022, 13, 35. [Google Scholar]
- Li, J.; Yang, H.X.; Xu, G.Y.; Deng, K.L.; Yu, J.J.; Xiang, S.Q.; Zhou, K.; Zhang, Q.L.; Li, R.X.; Li, M.M.; et al. QTL analysis of Z414, a chromosome segment substitution line with short/wide grains, and substitution mapping of qGL11 in rice. Rice 2022, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Wang, H.; Zhang, Q.; Zhou, K.; Zhao, F.M. Identification and pyramiding of QTLs for rice grain size based on short-eide grain CSSL-Z563 and fine-mapping of qGL3–2. Rice 2021, 14, 35. [Google Scholar] [CrossRef]
- Sun, S.F.; Wang, Z.B.; Xiang, S.Q.; Lv, M.; Zhou, K.; Li, J.; Liang, P.X.; Li, M.M.; Li, R.X.; Ling, Y.H.; et al. Identification, pyramid, and candidate gene of QTL for yield-related traits based on rice CSSLs in indica Xihui18 background. Mol. Breed. 2022, 42, 19. [Google Scholar] [CrossRef]
- Xu, G.Y.; Deng, K.L.; Yu, J.J.; Li, Q.L.; Li, L.; Xiang, A.N.; Ling, Y.H.; Zhao, F.M. Genetic effects analysis of QTL for rice grain size based on CSSL-Z403 and its dissected single and dual-segment substitution lines. Int. J. Mol. Sci. 2023, 24, 12013. [Google Scholar] [CrossRef]
- Zhang, G.Q. Target chromosome-segment substitution: A way to breeding by design in rice. Crop J. 2021, 9, 658–668. [Google Scholar] [CrossRef]
- Sun, W.; Xu, X.H.; Li, Y.P.; Xie, L.X.; He, Y.A.; Li, W.; Lu, X.B.; Sun, H.W.; Xie, X.Z. OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice. New Phytol. 2020, 226, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Qin, P.; Hu, L.; Zhan, S.J.; Wang, S.F.; Gao, P.; Li, J.; Jin, M.Y.; Xu, Z.Y.; Gao, Q.; et al. OsSPL18 controls grain weight and grain number in rice. J. Genet. Genomics. 2019, 46, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, Y.; Ren, X.Y.; Wei, K.; Fan, X.L.; Huang, L.C.; Zhao, D.S.; Zhang, L.; Zhang, C.Q.; Liu, Q.Q.; et al. Co-Overexpression of two key source genes, OsBMY4 and OsISA3, improves multiple key traits of rice seeds. J. Agric. Food Chem. 2023, 71, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Damon, S.; Hewitt, J.D.; Zamir, D.; Rabinowitch, H.D.; Lincoln, S.E.; Lander, E.S.; Tanksley, S.D. Mendelian factors underlying quantitative traits in tomato: Comparison across species, generations, and environments. Genetics 1991, 127, 181–197. [Google Scholar] [CrossRef]
- Zhao, F.M.; Tan, Y.; Zheng, L.Y.; Zhou, K.; He, G.H.; Ling, Y.H.; Zhang, L.H.; Xu, S.Z. Identification of rice chromosome segment substitution line Z322-1-10 and mapping QTL for agronomic traits from the F3 population. Cereal Res. Commun. 2016, 44, 370–380. [Google Scholar] [CrossRef]
- Yang, W.F.; Liang, J.Y.; Hao, Q.W.; Luan, X.; Tan, Q.Y.; Lin, S.W.; Zhu, H.T.; Liu, G.F.; Liu, Z.P.; Bu, S.H.; et al. Fine mapping of two grain chalkiness QTL sensitive to high temperature in rice. Rice 2021, 14, 33. [Google Scholar] [CrossRef]
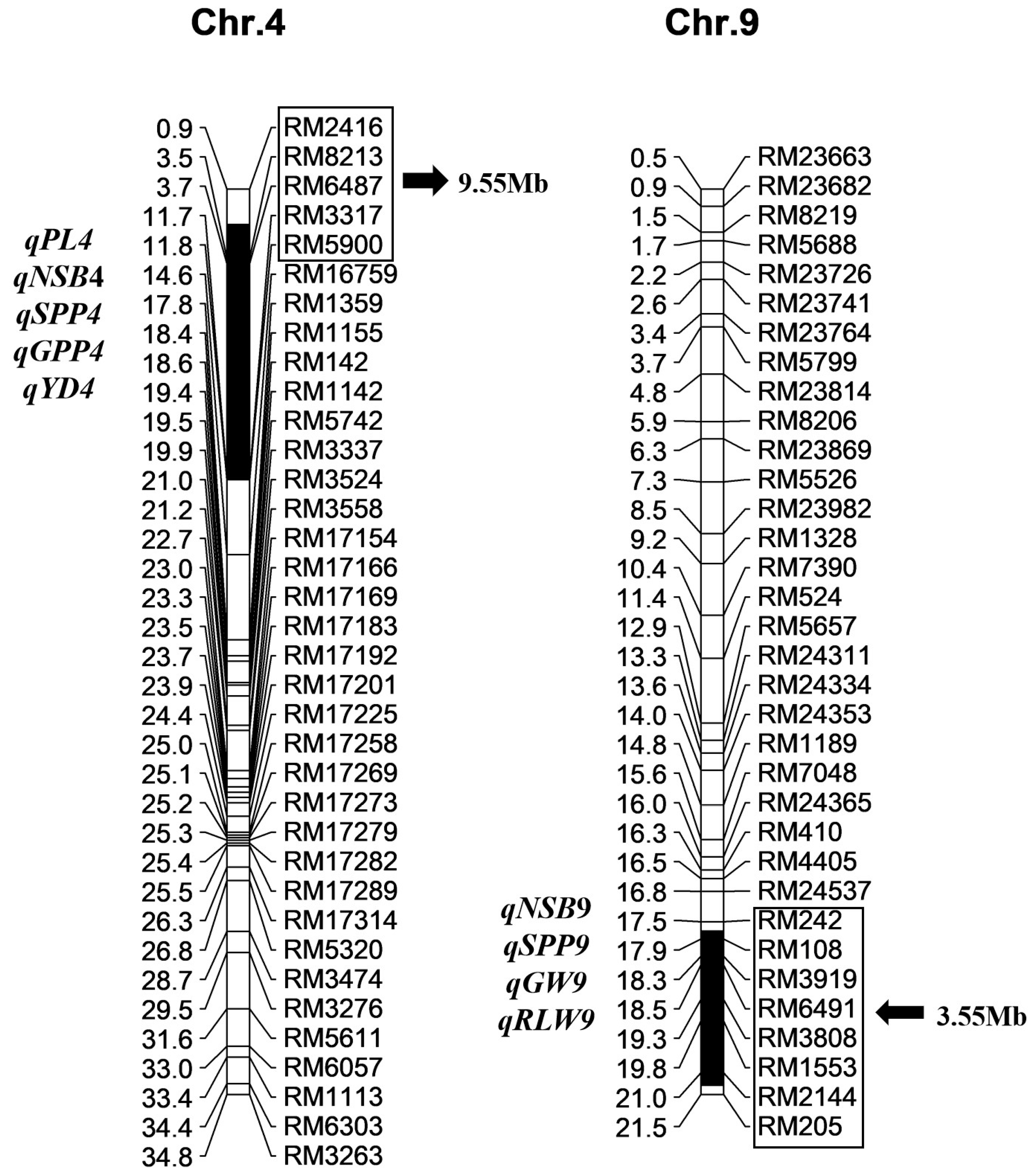

| Traits. | QTL | Chr. | Linked Marker | Additive Effect | Var. (%) | p-Value |
|---|---|---|---|---|---|---|
| Panicle length (cm) | qPL4 | 4 | RM3317 | −0.51 | 12.13 | 0.0338 |
| Number of secondary branches | qNSB4 | 4 | RM3317 | −2.84 | 13.07 | 0.0104 |
| qNSB9 | 9 | RM2144 | −2.51 | 10.27 | 0.0375 | |
| Spikelet number per panicle | qSPP4 | 4 | RM3317 | −10.75 | 8.05 | 0.0429 |
| qSPP9 | 9 | RM2144 | −11.78 | 9.67 | 0.0429 | |
| Grain number per panicle | qGPP4 | 4 | RM3317 | −11.92 | 10.51 | 0.0202 |
| Grain width (mm) | qGW9 | 9 | RM2144 | 0.04 | 11.60 | 0.0201 |
| Ratio of length to width | qRLW9 | 9 | RM2144 | −0.04 | 17.96 | 0.0064 |
| Yield (g) | qYD4 | 4 | RM3317 | −3.93 | 12.96 | 0.0096 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, K.; Zhang, H.; Wu, J.; Zhao, Z.; Wang, D.; Xu, G.; Yu, J.; Ling, Y.; Zhao, F. Development of Single-Segment Substitution Lines and Fine-Mapping of qSPP4 for Spikelets Per Panicle and qGW9 for Grain Width Based on Rice Dual-Segment Substitution Line Z783. Int. J. Mol. Sci. 2023, 24, 17305. https://doi.org/10.3390/ijms242417305
Deng K, Zhang H, Wu J, Zhao Z, Wang D, Xu G, Yu J, Ling Y, Zhao F. Development of Single-Segment Substitution Lines and Fine-Mapping of qSPP4 for Spikelets Per Panicle and qGW9 for Grain Width Based on Rice Dual-Segment Substitution Line Z783. International Journal of Molecular Sciences. 2023; 24(24):17305. https://doi.org/10.3390/ijms242417305
Chicago/Turabian StyleDeng, Keli, Han Zhang, Jiayi Wu, Zhuowen Zhao, Dachuang Wang, Guangyi Xu, Jinjin Yu, Yinghua Ling, and Fangming Zhao. 2023. "Development of Single-Segment Substitution Lines and Fine-Mapping of qSPP4 for Spikelets Per Panicle and qGW9 for Grain Width Based on Rice Dual-Segment Substitution Line Z783" International Journal of Molecular Sciences 24, no. 24: 17305. https://doi.org/10.3390/ijms242417305





