The Effects of Artificial Intelligence Assistance on the Radiologists’ Assessment of Lung Nodules on CT Scans: A Systematic Review
Abstract
1. Introduction
2. Methods
2.1. Eligibility Criteria
2.2. Search Strategy
2.3. Study Selection
2.4. Data Extraction and Synthesis
2.5. Quality Assessment
3. Results
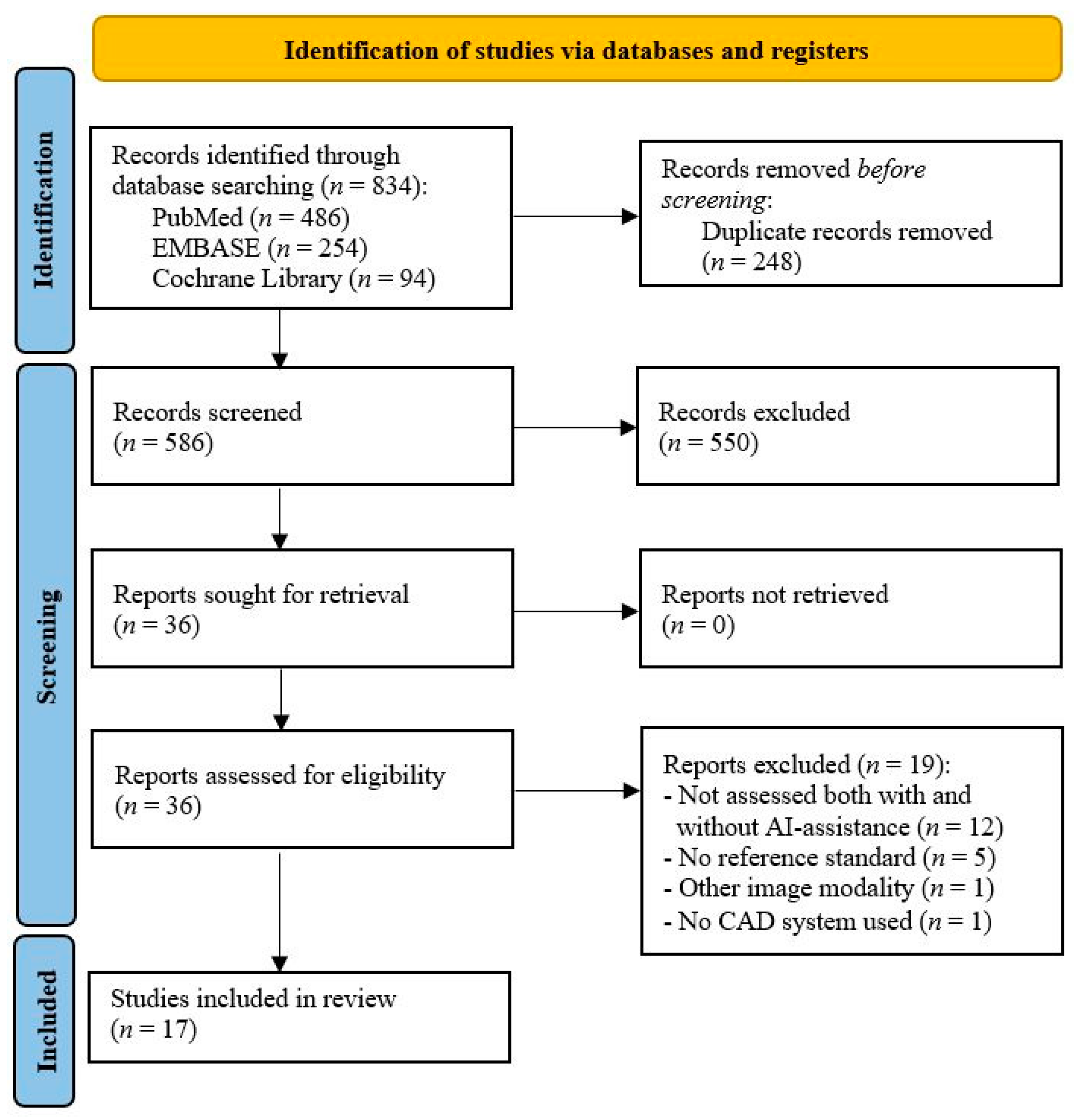
3.1. Study Selection
3.2. Study Characteristics
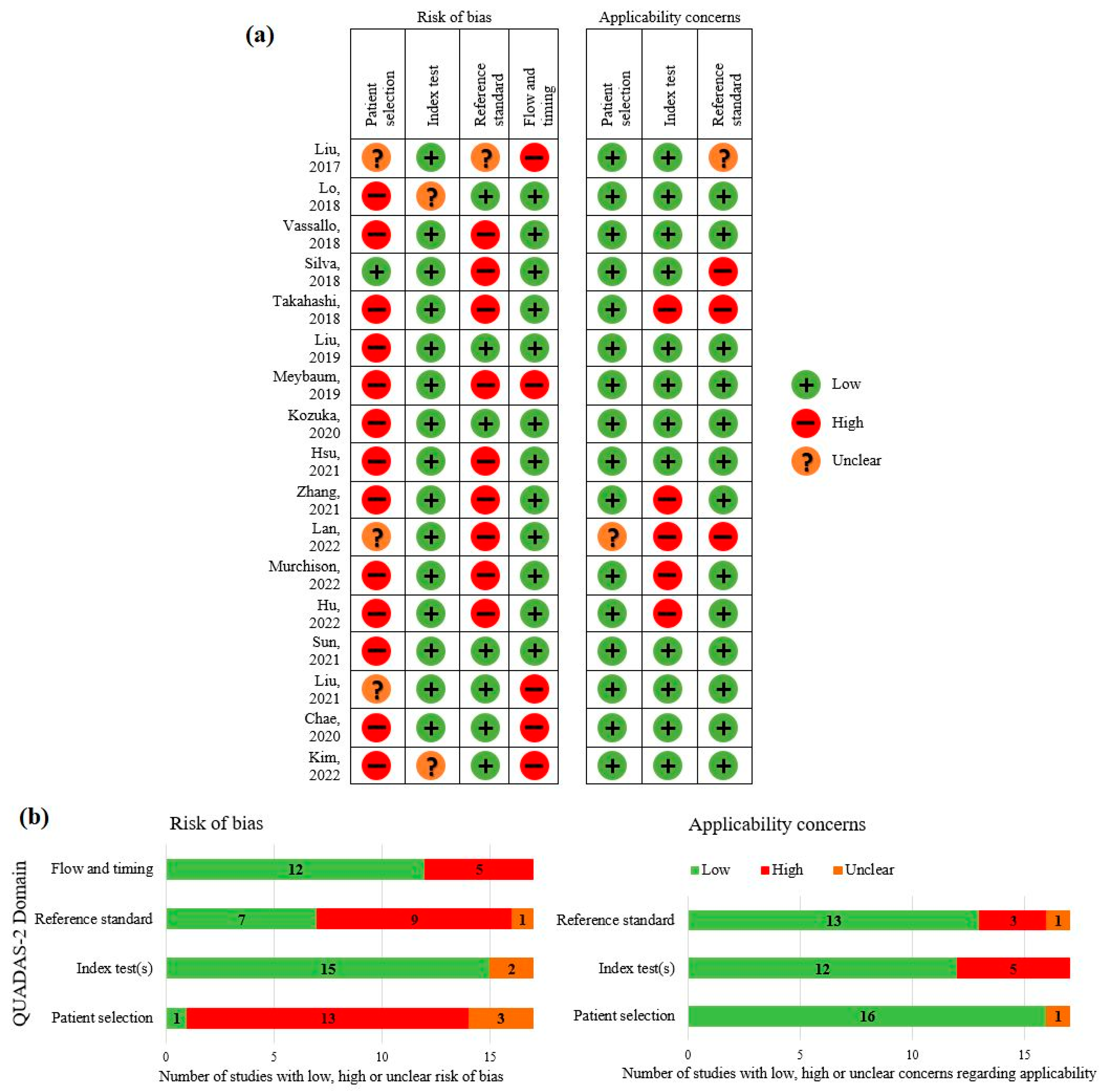
3.3. Quality Assessment
3.4. Data Analysis
3.4.1. Detection
3.4.2. Malignancy Prediction
3.4.3. Radiologists’ Workflow
4. Discussion
4.1. Detection and Malignancy Prediction: Performances of Radiologists
4.2. Reading Time
4.3. Radiologists’ Workflow
4.4. Shortcomings in Research on AI Assistance for Lung Nodule Assessment
4.5. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Chi, J.; Liu, J.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Lu, X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput. Biol. Med. 2021, 137, 104806. [Google Scholar] [CrossRef]
- Gu, D.; Liu, G.; Xue, Z. On the performance of lung nodule detection, segmentation and classification. Comput. Med. Imaging Graph. 2021, 89, 101886. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, F.; Cao, W.; Qin, C.; Dong, X.; Yang, Z.; Zheng, Y.; Luo, Z.; Zhao, L.; Yu, Y.; et al. Lung cancer risk prediction models based on pulmonary nodules: A systematic review. Thorac. Cancer 2022, 13, 664–677. [Google Scholar] [CrossRef]
- Li, R.; Xiao, C.; Huang, Y.; Hassan, H.; Huang, B. Deep Learning Applications in Computed Tomography Images for Pulmonary Nodule Detection and Diagnosis: A Review. Diagnostics 2022, 12, 298. [Google Scholar] [CrossRef]
- Wu, P.; Sun, X.; Zhao, Z.; Wang, H.; Pan, S.; Schuller, B. Classification of Lung Nodules Based on Deep Residual Networks and Migration Learning. Comput. Intell. Neurosci. 2020, 2020, 8975078. [Google Scholar] [CrossRef]
- Zhang, K.; Wei, Z.; Nie, Y.; Shen, H.; Wang, X.; Wang, J.; Yang, F.; Chen, K. Comprehensive Analysis of Clinical Logistic and Machine Learning-Based Models for the Evaluation of Pulmonary Nodules. JTO Clin. Res. Rep. 2022, 3, 100299. [Google Scholar] [CrossRef]
- Ali, I.; Muzammil, M.; Haq, I.U.; Khaliq, A.A.; Abdullah, S. Efficient lung nodule classification using transferable texture convolutional neural network. IEEE Access 2020, 8, 175859–175870. [Google Scholar] [CrossRef]
- Venkadesh, K.V.; Setio, A.A.A.; Schreuder, A.; Scholten, E.T.; Chung, K.; Wille, M.M.W.; Saghir, Z.; van Ginneken, B.; Prokop, M.; Jacobs, C. Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT. Radiology 2021, 300, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, K.G.; Schalekamp, S.; Rutten, M.J.C.M.; Van Ginneken, B.; De Rooij, M. Artificial intelligence in radiology: 100 commercially available products and their scientific evidence. Eur. Radiol. 2021, 31, 3797–3804. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Liu, J.K.; Jiang, H.Y.; Gao, M.D.; He, C.G.; Wang, Y.; Wang, P.; Ma, H.; Li, Y. An Assisted Diagnosis System for Detection of Early Pulmonary Nodule in Computed Tomography Images. J. Med. Syst. 2017, 41, 30. [Google Scholar] [CrossRef]
- Lo, S.C.B.; Freedman, M.T.; Gillis, L.B.; White, C.S.; Mun, S.K. Computer-aided detection of lung nodules on CT with a computerized pulmonary vessel suppressed function. Am. J. Roentgenol. 2018, 210, 480–488. [Google Scholar] [CrossRef]
- Vassallo, L.; Traverso, A.; Agnello, M.; Bracco, C.; Campanella, D.; Chiara, G.; Fantacci, M.E.; Lopez Torres, E.; Manca, A.; Saletta, M.; et al. A cloud-based computer-aided detection system improves identification of lung nodules on computed tomography scans of patients with extra-thoracic malignancies. Eur. Radiol. 2019, 29, 144–152. [Google Scholar] [CrossRef]
- Silva, M.; Schaefer-Prokop, C.M.; Jacobs, C.; Capretti, G.; Ciompi, F.; Van Ginneken, B.; Pastorino, U.; Sverzellati, N. Detection of Subsolid Nodules in Lung Cancer Screening: Complementary Sensitivity of Visual Reading and Computer-Aided Diagnosis. Investig. Radiol. 2018, 53, 441–449. [Google Scholar] [CrossRef]
- Takahashi, E.A.; Koo, C.W.; White, D.B.; Lindell, R.M.; Sykes, A.M.G.; Levin, D.L.; Kuzo, R.S.; Wolf, M.; Bogoni, L.; Carter, R.E.; et al. Prospective pilot evaluation of radiologists and computer-aided pulmonary nodule detection on ultra-low-dose CT with tin filtration. J. Thorac. Imaging 2018, 33, 396–401. [Google Scholar] [CrossRef]
- Liu, K.; Li, Q.; Ma, J.; Zhou, Z.; Sun, M.; Deng, Y.; Tu, W.; Wang, Y.; Fan, L.; Xia, C.; et al. Evaluating a Fully Automated Pulmonary Nodule Detection Approach and Its Impact on Radiologist Performance. Radiol. Artif. Intell. 2019, 1, e180084. [Google Scholar] [CrossRef]
- Meybaum, C.; Graff, M.; Fallenberg, E.M.; Leschber, G.; Wormanns, D. Contribution of CAD to the Sensitivity for Detecting Lung Metastases on Thin-Section CT—A Prospective Study with Surgical and Histopathological Correlation. RoFo Fortschr. Geb. Rontgenstrahlen Bildgeb. Verfahr. 2020, 192, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, T.; Matsukubo, Y.; Kadoba, T.; Oda, T.; Suzuki, A.; Hyodo, T.; Im, S.W.; Kaida, H.; Yagyu, Y.; Tsurusaki, M.; et al. Efficiency of a computer-aided diagnosis (CAD) system with deep learning in detection of pulmonary nodules on 1-mm-thick images of computed tomography. Jpn. J. Radiol. 2020, 38, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.H.; Ko, K.H.; Chou, Y.C.; Wu, Y.C.; Chiu, S.H.; Chang, C.K.; Chang, W.C. Performance and reading time of lung nodule identification on multidetector CT with or without an artificial intelligence-powered computer-aided detection system. Clin. Radiol. 2021, 76, e23–e626. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, B.; Zhang, L.; Greuter, M.J.W.; de Bock, G.H.; Zhang, H.; Xie, X. Lung Nodule Detectability of Artificial Intelligence-assisted CT Image Reading in Lung Cancer Screening. Curr. Med. Imaging 2021, 18, 327–334. [Google Scholar] [CrossRef]
- Lan, C.C.; Hsieh, M.S.; Hsiao, J.K.; Wu, C.W.; Yang, H.H.; Chen, Y.; Hsieh, P.C.; Tzeng, I.S.; Wu, Y.K. Deep Learning-based Artificial Intelligence Improves Accuracy of Error-prone Lung Nodules. Int. J. Med. Sci. 2022, 19, 490–498. [Google Scholar] [CrossRef]
- Murchison, J.T.; Ritchie, G.; Senyszak, D.; Nijwening, J.H.; van Veenendaal, G.; Wakkie, J.; van Beek, E.J.R. Validation of a deep learning computer aided system for CT based lung nodule detection, classification, and growth rate estimation in a routine clinical population. PLoS ONE 2022, 17, e0266799. [Google Scholar] [CrossRef]
- Hu, Q.; Wang, S.; Chen, C.; Kang, S.; Sun, Z.; Wang, Y.; Xiang, M.; Guan, H.; Liu, Y.; Xia, L. Comparison of two reader modes of computer-aided diagnosis in lung nodules on low-dose chest CT scan. J. Innov. Opt. Health Sci. 2022, 15, 2250013. [Google Scholar] [CrossRef]
- Sun, K.; Chen, S.; Zhao, J.; Wang, B.; Yang, Y.; Wang, Y.; Wu, C.; Sun, X. Convolutional Neural Network-Based Diagnostic Model for a Solid, Indeterminate Solitary Pulmonary Nodule or Mass on Computed Tomography. Front. Oncol. 2021, 11, 792062. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, L.; Han, X.; Ji, H.; Liu, L.; He, W. Estimation of malignancy of pulmonary nodules at CT scans: Effect of computer-aided diagnosis on diagnostic performance of radiologists. Asia Pac. J. Clin. Oncol. 2021, 17, 216–221. [Google Scholar] [CrossRef]
- Chae, K.J.; Jin, G.Y.; Ko, S.B.; Wang, Y.; Zhang, H.; Choi, E.J.; Choi, H. Deep Learning for the Classification of Small (≤2 cm) Pulmonary Nodules on CT Imaging: A Preliminary Study. Acad. Radiol. 2020, 27, e55–e63. [Google Scholar] [CrossRef]
- Kim, R.Y.; Oke, J.L.; Pickup, L.C.; Munden, R.F.; Dotson, T.L.; Bellinger, C.R.; Cohen, A.; Simoff, M.J.; Massion, P.P.; Filippini, C.; et al. Artificial Intelligence Tool for Assessment of Indeterminate Pulmonary Nodules Detected with CT. Radiology 2022, 304, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Pehrson, L.M.; Lauridsen, C.A.; Tøttrup, L.; Fraccaro, M.; Elliott, D.; Zając, H.D.; Darkner, S.; Carlsen, J.F.; Nielsen, M.B. The Added Effect of Artificial Intelligence on Physicians’ Performance in Detecting Thoracic Pathologies on CT and Chest X-ray: A Systematic Review. Diagnostics 2021, 11, 2206. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.J.Y.; Leong, L.C.H.; Teo, L.L.S.; Ong, C.C.; Cheah, F.K.; Tham, W.P.; Salahudeen, H.M.M.; Lee, C.H.; Kaw, G.J.L.; Tee, A.K.H.; et al. A practical and adaptive approach to lung cancer screening: A review of international evidence and position on CT lung cancer screening in the Singaporean population by the College of Radiologists Singapore. Singap. Med. J. 2019, 60, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.J.; Mondal, P.; Penz, E.; Tyan, C.C.; Lim, H.; Babyn, P. Development and Cost Analysis of a Lung Nodule Management Strategy Combining Artificial Intelligence and Lung-RADS for Baseline Lung Cancer Screening. J. Am. Coll. Radiol. 2021, 18, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Nagendran, M.; Chen, Y.; Lovejoy, C.A.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.A.; Collins, G.S.; Maruthappu, M. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies in medical imaging. BMJ 2020, 368, m689. [Google Scholar] [CrossRef]
- Recht, M.P.; Dewey, M.; Dreyer, K.; Langlotz, C.; Niessen, W.; Prainsack, B.; Smith, J.J. Integrating artificial intelligence into the clinical practice of radiology: Challenges and recommendations. Eur. Radiol. 2020, 30, 3576–3584. [Google Scholar] [CrossRef]
- Sounderajah, V.; Ashrafian, H.; Golub, R.M.; Shetty, S.; De Fauw, J.; Hooft, L.; Moons, K.; Collins, G.; Moher, D.; Bossuyt, P.M.; et al. Developing a reporting guideline for artificial intelligence-centred diagnostic test accuracy studies: The STARD-AI protocol. BMJ Open 2021, 11, e047709. [Google Scholar] [CrossRef]
- Collins, G.S.; Dhiman, P.; Andaur Navarro, C.L.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021, 11, e048008. [Google Scholar] [CrossRef]
- Vasey, B.; Nagendran, M.; Campbell, B.; Clifton, D.A.; Collins, G.S.; Denaxas, S.; Denniston, A.K.; Faes, L.; Geerts, B.; Ibrahim, M.; et al. Reporting guideline for the early stage clinical evaluation of decision support systems driven by artificial intelligence: DECIDE-AI. BMJ 2022, 28, 924–933. [Google Scholar] [CrossRef]
| First Author, Year of Publication, Country | Population | Exclusion Criteria | Algorithm/ Software | Use of Assistance | CT Scans | N Scans | N Scans with Nodules | N Nodules | Nodule Sizes | Nodule Types | Reference Standard (Years of Experience) | N Observers (Years of Experience) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liu, 2017 [15], China | Participants of screening | - | Sparse non-negative matrix factorization model | SR | SDCT - Single-center | 180 | 80 | 96 | 4–10 mm | Various types | Biopsy, resection or consensus by radiologists | 6 (2 resident radiologists (<5), 2 secondary chest radiologists (8–12), 2 senior chest radiologists (>15)) |
| Lo, 2018 [16], Unites States | Participants of screening | - | VIS/CADe | CR | LDCT No contrast Multi-center | 324 | 108 | 178 | ≥5 mm | Solid, part-solid, GGO | Detected by at least 2/3 radiologists | 12 radiologists (6–26) |
| Vassallo, 2018 [17], Italy | Patients with an extra-thoracic primary tumor |
| M5L lung CAD on-demand | SR | SDCT With and without contrast Single-center | 225 (28 with contrast) | 75 | 215 | ≥3 mm | Solid, part-solid, sub-solid, calcified | Consensus by 2 radiologists (>15) who assessed all nodules detected by at least 1 radiologist or/and CAD | 3 (1 resident radiologist (3), 2 radiologists (20, 35)) |
| Silva, 2018 [18], Italy | Participants of screening | - | CIRRUS Lung screening (prototype version of Veolity) | CR | LDCT No contrast Multi-center | 2303 | 155 | 194 | ≥5 mm | Sub-solid (including non-solid, part-solid) | Detected by either or both radiologists or CAD | AI-assisted: consensus by 2 radiologists (8, 11) Radiologists only: 1 out of 7 radiologists (4–20) |
| Takahashi, 2018 [19], United States | Patients with clinically indicated CT scan |
| Syngo.CT Lung CAD (Siemens Health Care) | SR | Ultra LDCT No contrast Single-center | 55 | 30 | 45 | ≥5 mm | Solid, part-solid, GGN | Assessed by 1 radiologist (>20) using SDCT (acquired consecutively) | 2 out of 4 radiologists (5–21) |
| Liu, 2019 [20], China | Patients who underwent a clinical scan and participants for screening |
| InferRead CT Lung (Infervision) | CR | LDCT and SDCT - Multi-center | 123 for per-nodule analysis, 148 for per-patient analysis | - | - | >3 mm | Solid | Assessment by 2 radiologists (±10) with access to original radiology report, differences checked by 3rd radiologist (±10) | 2 radiologists (±10) |
| Meybaum, 2019 [21], Germany | Patients who underwent surgery for lung metastases |
| LMS 6.0, Median Technologies, Valbonne, France | SR | SDCT - Single-center | 95 | - | 646 | - | - | Surgical findings during pulmonary metastasectomy | 1 out of 4 radiologists (6–20) |
| Kozuka, 2020 [22], Japan | Patients with suspected lung cancer |
| InferRead CT Lung (Infervision) | CR | SDCT No contrast Single-center | 117 | 111 | 743 | ≥3 mm | Solid, part-solid, calcified, GGN | Assessment by 2 radiologists, differences checked by 3rd radiologist (6, 12, 26) | 2 radiologists (1, 5) |
| Hsu, 2021 [23], Taiwan | Patients who underwent a CT scan with proved small pulmonary nodules (≤10 mm), stable nodules for at least 2 years, incidental small nodules on screening |
| ClearRead CT | CR and SR | LDCT and SDCT No contrast Single-center | 150 (57 LDCT, 93 SDCT) | 98 | 340 | ≤10 mm | Solid, part-solid, GGN | Consensus by 2 radiologists (>15), first assessed without pre-knowledge, subsequently reviewed all nodules detected by any radiologist or/and AI | 6 (3 resident radiologists (1–2), 3 radiologists (5, 10, 25)) |
| Zhang, 2021 [24], China | Asymptomatic participants of screening |
| InferRead CT Lung (Infervision) | CR | LDCT No contrast Single-center | 860 | 250 solid; 13 part-solid; 111 non-solid (at least 1 nodule of that type) | - | ≥3 mm | Solid, part-solid, non-solid | Assessment by 2 radiologists (20, 31) who assessed all scans with pre-knowledge of nodules detected by radiologists or AI | AI-assisted: Consensus by a resident radiologist (5) supervised by a radiologist (20) Radiologists only: 1/14 resident radiologists (2–5) supervised by 1/15 radiologists (10–30) |
| Lan, 2022 [25], Taiwan | Patients who underwent a CT scan | - | ResNet 18 | CR | SDCT - Single-center | 60 | - | 266 | - | Solid, part-solid, GGO | Detected by AI and at least 3/5 doctors | 5 (2 assessed with AI, 3 without AI): 3 chest physicians, 1 chest surgeon, 1 radiologist, (all >10 years of experience in reading chest CT scans) |
| Murchison, 2022 [26], United Kingdom | (Previous) smokers and/or radiological evidence of pulmonary emphysema |
| Veye Lung Nodules (Aidence) | CR | SDCT With and without contrast Single-center | 273 (22 with contrast) | - | 269 | 5–30 mm | Solid, sub-solid | Detected by both radiologists or detected by 1 radiologists and confirmed by third radiologist | 1 out of 2 radiologists (≥9) |
| Hu, 2022 [27], China | Patients with an extra-thoracic primary tumor |
| InferRead CT Lung (Infervision) | CR and SR | LDCT No contrast Single-center | 117 | - | 650 | - | Solid, sub-solid calcified | Consensus by 2 radiologists (>20) who assessed all scans with access to CAD outcomes | 9 (3 radiologists (5–10), 3 resident radiologists (2–3), 3 interns (<1)) |
| First Author, Year of Publication, Country | Population | Exclusion Criteria | Algorithm/ Software | Use of Assistance | CT Scans | N Scans | N Scans with Nodules | N Nodules | Nodule Sizes | Nodule Types | Assessment Observers | Output AI | Reference Standard (Years of Experience) | N Observers (Years of Experience) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sun, 2021 [28], China | Patients with a primary intrapulmonary lesion |
| ResNet-101 | CR | SDCT - Single-center | 47 | 47 | - | >8 mm | Solid intermediate solitary pulmonary nodules | Benign or malignant | Both benign and malignant probability in percentages | Histopathologic confirmed | Consensus by 2 radiologists (4, 9) and in cases of discrepancies a third radiologist (>29) |
| Liu, 2021 [29], China | Patients with lung nodules | - | ResNet-50 | SR | SDCT - Multi-center | - | - | 168 (112 malignant, 56 benign) | 6–30 mm | - | 5 categories | Number between 0 and 1 | Histopathologic confirmed or benign based on stability on CT during 2 years or disappeared | 6 (4 radiologists (8, 12, and two >15), 2 resident radiologists (<5)) |
| Chae, 2020 [30], South Korea | Patients with difficult-to-diagnose lung nodules |
| CT-lungNET | SR | SDCT No contrast Single-center | 60 | 60 | 60 (30 malignant, 30 benign) | 5–20 mm | - | 4 categories | Percentages | Histopathologic confirmed or benign based on stability on CT during >1 year | 8 (2 third-year medical students, 2 physicians (non-radiology) (1), 2 resident radiologists (2), 2 radiologists (3, 5)) |
| Kim, 2022 [31], United Kingdom | Patients with indeterminate lung nodules |
| Virtual Nodule Clinic (Optellum) | SR | LDCT (n = 124) and SDCT (n = 176) No contrast Multi-center | 300 | 300 (150 malignant, 150 benign) | 300 (150 malignant, 150 benign) | 5–30 mm | Solid, mixed, part-solid | Score between 0 and 100 | Number between 1 and 10 (decile scale) | Histopathologic confirmed or benign based on stability on CT during 2 years or disappeared | 12 (6 pulmonologists (2 in thoracic oncology), 6 radiologists (2 in thoracic radiology)) |
| First Author, Year of Publication | Per-Nodule/Per-Patient (Mean) Sensitivity 95% CI TP/(TP + FN) | Per-Patient (Mean) Specificity 95% CI TN/(TN + FP) | (Mean) AUC 95% CI | Mean FP Per Scan | Reporting Time Mean Time Per Scan ± SD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AI | Observers | AI-Assisted | AI | Observers | AI-Assisted | Observers | AI-Assisted | AI | Observers | AI-Assisted | Observers | AI-Assisted | Difference | |
| Liu, 2017 [15] | Per-nodule | - | - | - | - | - | 0.09 | 0.016 | - | - | - | - | ||
| 92.7% 89/96 | 87.3% 503/576 | 96.9% 558/576 | ||||||||||||
| Lo, 2018 [16] | Per-nodule | - | 89.9% 85.9–93.9% 194/216 | 84.4% 80.4–88.4% 182/216 | 0.584 0.518–0.650 | 0.692 0.626–0.758 | 0.58 | 0.17 | 0.28 | 132.3 s | 98.0 s | −26% | ||
| 82.0% 146/178 | 60.1% 53.5–66.7% 107/178 | 72.5% 65.9–79.1% 129/178 | ||||||||||||
| Vassallo, 2018 [17] | Per-nodule | - | 85% 71–79% 382/450 | 82% 79–86% 369/450 | - | - | 3.8 | 0.13 | 0.16 | 296 ± 80 s | 329 ± 83 s | +11% * | ||
| 85% 82–91% 183/215 | 65% 61–69% 421/645 | 88% 86–91% 570/645 | ||||||||||||
| Per-patient | ||||||||||||||
| - | 75% 71–79% 169/225 | 82% 79–86% 186/225 | ||||||||||||
| Silva, 2018 [18] | Per-nodule | - | - | - | - | - | - | - | 0.26 | - | - | - | ||
| - | 37.1% 72/194 | 90.2% 175/194 | ||||||||||||
| Per-patient | ||||||||||||||
| - | 34.2% 53/155 | 88.4% 137/155 | ||||||||||||
| Takahashi, 2018 [19] | Per-nodule | - | - | - | - | - | 2 | 0.45 | - | - | - | - | ||
| 71% 57–82% 32/45 | ~60% | 70.8% 63/89 | ||||||||||||
| Liu, 2019 [20] | Per-nodule | - | - | - | Per-patient | - | - | - | ±15 min | ±5–10 min | - | |||
| - | - | Improved, shown by radar plot | 0.660 | 0.778 | ||||||||||
| Meybaum, 2019 [21] | Per-nodule | - | - | - | - | - | - | - | - | - | - | - | ||
| 53.6% 49.7–57.4% 346/646 | 67.5% 63.9–71.1% 436/646 | 77.9% 74.7–81.1% 503/646 | ||||||||||||
| Kozuka, 2020 [22] | Per-nodule | 83.3% 35.9–99.6% 5/6 | 91.7% 61.5–99.8% 11/12 | 83.3% 51.6–97.9% 10/12 | - | - | 3.25 | 1.11 | 2.97 | 3.1 min | 2.8 min | −11.3% | ||
| 70.3% 66.8–73.5% 522/743 | 20.9% 18.8–23.0% 310/1486 | 38.0% 35.5–40.5% 564/1486 | ||||||||||||
| Per-patient | ||||||||||||||
| 95.5% 89.8–98.5% 106/111 | 68% 61.4–74.1% 151/222 | 85.1% 79.8–89.5% 189/222 | ||||||||||||
| Hsu, 2021 [23] | Per-nodule | 87% 45/52 | 80% 78–81% 250/312 | CR: 83% 82–85% 259/312 SR: 84% 82–85% 262/312 | 0.72 0.70–0.74 | CR: 0.82 0.80–0.83 SR: 0.83 0.81–0.85 | 0.67 (diameter threshold > 5 mm) | - | - | CR: 156 ± 34 s SR: 156 ± 34 s | CR: 124 ± 25 s SR: 197 ± 46 s | CR: −21% SR: +26% * | ||
| 86% 291/340 | 64% 62–66% 1306/2040 | CR: 80% 79–82% 1632/2040 SR: 82% 80–84% 1673/2040 | ||||||||||||
| Zhang, 2021 [24] | Per-patient | - | 100% | ~99% | - | - | - | - | - | - | - | - | ||
| - | 43.3% 162/374 | 98.9% 370/374 | ||||||||||||
| Lan, 2022 [25] | Per-nodule | - | - | - | - | - | - | 0.634 | 0.122 | - | - | - | ||
| - | 63.1% 52.0–74.1% 504/798 | 69.8% 50.9–88.6% 371/532 | ||||||||||||
| Murchison, 2022 [26] | Per-nodule | - | - | - | - | - | - | 0.11 | 0.16 | - | - | - | ||
| - | 71.9% 66.0–77.0% 193/269 | 80.3% 75.2–85.0% 216/269 | ||||||||||||
| Hu, 2022 [27] | Per-nodule | - | - | - | - | - | - | SDCT: 1.90 LDCT: 1.76 | SDCT: 2.89 LDCT: 2.63 | - | - | - | ||
| - | SDCT: 45.35% 2694/5940 LDCT: 42.11% 2463/5850 | SDCT: 57.77% 3432/5940 LDCT: 56.70% 3317/5850 | ||||||||||||
| - | 44.27% 1197/2704 | CR: 64.76% 1751/2704 SR: 66.86% 1808/2704 | - | - | - | - | - | - | 2.67 | CR: 4.09 SR: 4.11 | CR: 235 ± 162 s SR: 235 ± 162 s | CR: 165 ± 133 s SR: 294 ± 153 s | CR: −30% SR: +25% * | |
| First Author, Year of Publication | Malignancy Risk Threshold | Per-Nodule | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (Mean) Sensitivity 95% CI TP/(TP + FN) | (Mean) Specificity 95% CI TN/(TN + FP) | (Mean) AUC 95% CI | ||||||||
| AI | Observers | AI-Assisted | AI | Observers | AI-Assisted | Observers | AI-Assisted | Difference | ||
| Sun, 2021 [28] | - | 86% | 82% | 89% | 79% | 42% | 84% | 0.91 | - | - |
| 73–93% | 69–90% | 77–95% | 65–88% | 29–56% | 71–92% | 0.83–0.99 | ||||
| Liu, 2021 [29] | 58% (Youden Index) | 93.8% | - | - | 83.9% | - | - | - | 0.913 | 0.938 |
| Chae, 2020 [30] | - | 60% | 70% | 65% | 87% | 69% | 85% | 0.85 | ~0.72 | ~0.79 |
| 0.74–0.93 | ||||||||||
| Kim, 2022 [31] | 5% | - | 94.1% | 97.9% | - | 37.4% | 42.3% | - | 0.82 | 0.89 |
| 90.8–97.4% | 96.0–99.7% | 27.2–47.6% | 31.3–53.3% | |||||||
| 1693/1800 | 1762/1800 | 674/1800 | 761/1800 | |||||||
| 65% | - | 52.6% | 63.1% | - | 87.3% | 89.9% | 0.77–0.86 | 0.85–0.92 | ||
| 41.8–63.3% | 53.7–72.5% | 81.0–93.6% | 83.3–96.6% | |||||||
| 946/1800 | 1136/1800 | 1572/1800 | 1619/1800 | |||||||
| First Author, Year of Publication | Unassisted Reading | AI-Assisted Reading | Comparison with Reference Standard | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Provided Information | Functionalities Workstation | How Nodule Positions were Marked/Scans were Scored | Provided Information | How Images were Transferred to the AI Tool | Training for Familiarization with the AI Tool | How AI Results were Shown | What Radiologists Could do with the AI Results | Provided Information | Information Provided | |
| Liu, 2017 [15] | Observers interpreted the original CT images using the PACS workstation. Observers marked the detected nodule on the images using a drawing tool internally installed in the workstation. | N | B | Before the observation, we introduced the CAD system to the observers to make sure they can use it. All observers were told the accuracy of the CAD technique before the performance study. The CAD output image was displayed on a monitor. The observers made a refined decision after taking CAD output into consideration. However, if they make a change, they must record it. | N | B | B | N | - | N |
| Lo, 2018 [16] | A workstation with standard interpretation functions (i.e., zoom, pan, magnified glass, maximum intensity projection, sagittal or coronal viewing, lung or soft-tissue window setting, window or level control and other functions) was used. Radiologists marked the center of each nodule. | Y | B | Fifteen training cases were performed to familiarize with the workstation and its interpretation functions. Detected nodules by AI were marked with a score that indicated the likelihood that a nodule was present. If the score was greater than a selected threshold value, the nodules were shown as boundary contours on both the standard CT scan (left) and vessel-suppressed image (right). These images were coupled. Nodule’s density, maximum and minimum diameters, and volume were provided. The same process as during unassisted reading was performed. | N | Y | Y | N | If a mark of the radiologist was within the ellipsoid range of the nodule defined by the reference standard, the mark was considered as TP, otherwise as FP. | Y |
| Vassallo, 2018 [17] | CT images were stored on picture archiving and communication system (PACS). Maximum-intensity projection volume-rendering technique was used. The radiologists searched freely for lesions and measured the longest diameter of each nodule using a lung window on the plane where the lesion was more conspicuous, provided the nodule coordinates. All information was stored on an online web-form. | B | Y | The AI-detected nodules became accessible through a dedicated web-form. Radiologists could classify unmatched findings as FP, TP or irrelevant (<3 mm). | B | N | N | Y | If the Euclidean distance between the 3D centers of the mark of the radiologist and the AI-detected nodule was within the nodule diameter, the mark was considered TP. This was an automatic process. | Y |
| Silva, 2018 [18] | - | N | N | Radiologists reviewed the marks of the AI-detected nodules by selecting marks reflecting SSNs and discarding FPs. | N | N | N | Y | - | N |
| Takahashi, 2018 [19] | Radiologists marked nodules using a specially configured workstation (Mayo Biomedical Imaging Resource). | N | N | The CAD system is a fully automated computer-assisted tool that can identify pulmonary nodules regardless of density. The CAD markings were shown on the CT images in addition to their corresponding unassisted detections. They characterized each CAD marking as TP or FP. | N | N | B | Y | - | N |
| Liu, 2019 [20] | Radiologists marked nodules by square bounding boxes with the nodule at the center. | N | Y | Output of the model was the detection marked with a square bounding box. The nodule type and the model’s confidence in its prediction were also included. Radiologists used the AI assistance and reported the nodule type and confidence level. | N | N | Y | N | - | N |
| Meybaum, 2019 [21] | Radiologists used the user interface of the CAD system for CT reading, which provides original images and sliding thin-slab maximum-intensity projection. They labeled all nodules and made a screenshot of each nodule. | B | B | The CT images were transferred to the CAD system, and the CAD analysis was sent to the PACS system. The CAD findings were given. TP findings were labeled, and FPs were rejected. The total number of nodules per CT scan was recorded in a comment field. The radiologist filled out a study documentation sheet containing lesion label, lung segment, lesion size and lung scheme in three views, on which the radiologist marked the approximate position of all lesions. | Y | N | N | Y | - | N |
| Kozuka, 2020 [22] | Radiologists marked each nodule using a workstation with general interpretation functions. | B | N | The CAD system had functions to display marks, density, major axis and the volume of the detected nodules. Object bounding boxes and scores were provided. | N | N | Y | N | - | N |
| Hsu, 2021 [23] | - | N | N | Radiologists were trained in how to operate the procedures with several training images. The displays of the CAD results were combined to make the final decision. The CAD system renders a boundary contour on detected nodules, which is classified as actionable and provides a report table with nodule density, maximum and minimum diameters, and volume to the radiologists. | N | Y | Y | N | - | N |
| Zhang, 2021 [24] | - | N | N | The CT images were automatically transmitted to the AI server for segmentation and detection of lung nodules. Radiologists read the results on their image reading terminals. The system automatically depicts a suspected nodule with a bounding box and reveals its characteristics, including its components (solid/part-solid/non-solid), diameter and volume. | Y | N | Y | N | - | N |
| Lan, 2022 [25] | - | N | N | - | N | N | N | N | - | N |
| Murchison, 2022 [26] | Actionable nodules were marked manually. | N | N | Radiologists received training on the annotation tasks and annotation tool, with written instructions available throughout. Radiologists classified a CAD prompt as either TP or FP. Any actionable nodules identified on aided scans, which had not been detected by CAD, were also recorded. | N | Y | N | Y | - | N |
| Hu, 2022 [27] | - | N | N | Radiologists were trained on the CAD system. Radiologists were allowed to adjust the window level, zoom in and out, invert gray-scale and use maximum-intensity projection thick-slab images. Each by AI-identified nodule was marked on the image showing the nodule’s largest diameter. | N | Y | Y | N | - | N |
| Sun, 2021 [28] | - | N | N | Response heat maps were shown, in which red areas indicate that the model mainly extracted diagnostic characteristics from the region, while blue areas indicate that less discriminative features were found in that region. | N | N | B | N | - | N |
| Liu, 2021 [29] | Radiologists observed the CT scans with two clinical parameters (age and sex) to rate a diagnostic score. According to the likelihood of malignancy, the diagnosis score ranged from 1 to 5 (highly unlikely, moderately unlikely, indeterminate, moderately suspicious, highly suspicious). The nodules were indicated with red arrows. Radiologists could scroll through the slices, adjust the width and window level, and use the zoom function. | Y | Y | Radiologists were trained on the CAD system, with two training cases to learn how to use the system and learn the rating method. They were introduced to the performance of the CAD system. After initial suspicion rating, the CAD output was displayed on the monitor. Radiologists could update their suspicion. | N | Y | N | B | - | N |
| Chae, 2020 [30] | Radiologists reviewed the CT scans with the given positions of the centers of the nodules. They interpreted the probability of the malignancy of each nodule on a four-point scale (highly suggestive of benign nature (<25%), more likely benign than malignant (25–50%), more likely malignant than benign (50–75%), highly suggestive of malignant nature (>75%)). | N | Y | Each reviewer reclassified the previously determined category after considering the malignancy prediction rate of the AI system. | N | N | N | B | - | N |
| Kim, 2022 [31] | Radiologists loaded each of the scans into the software, which highlighted the nodule of interest. They scrolled through the entire set of images with axial views. They estimated the malignancy risk on a 100-point scale and selected a management recommendation (no action, CT follow-up after more than 6 months, CT follow-up after 6 weeks to 6 months, immediate imaging follow-up, nonsurgical biopsy or surgical resection). | N | Y | Radiologists had 1 hour of training with the CAD system and evaluated 17 example cases. The lung cancer prediction score was displayed, and readers were asked to provide an updated risk estimate and management recommendation. | N | Y | B | B | - | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ewals, L.J.S.; van der Wulp, K.; van den Borne, B.E.E.M.; Pluyter, J.R.; Jacobs, I.; Mavroeidis, D.; van der Sommen, F.; Nederend, J. The Effects of Artificial Intelligence Assistance on the Radiologists’ Assessment of Lung Nodules on CT Scans: A Systematic Review. J. Clin. Med. 2023, 12, 3536. https://doi.org/10.3390/jcm12103536
Ewals LJS, van der Wulp K, van den Borne BEEM, Pluyter JR, Jacobs I, Mavroeidis D, van der Sommen F, Nederend J. The Effects of Artificial Intelligence Assistance on the Radiologists’ Assessment of Lung Nodules on CT Scans: A Systematic Review. Journal of Clinical Medicine. 2023; 12(10):3536. https://doi.org/10.3390/jcm12103536
Chicago/Turabian StyleEwals, Lotte J. S., Kasper van der Wulp, Ben E. E. M. van den Borne, Jon R. Pluyter, Igor Jacobs, Dimitrios Mavroeidis, Fons van der Sommen, and Joost Nederend. 2023. "The Effects of Artificial Intelligence Assistance on the Radiologists’ Assessment of Lung Nodules on CT Scans: A Systematic Review" Journal of Clinical Medicine 12, no. 10: 3536. https://doi.org/10.3390/jcm12103536
APA StyleEwals, L. J. S., van der Wulp, K., van den Borne, B. E. E. M., Pluyter, J. R., Jacobs, I., Mavroeidis, D., van der Sommen, F., & Nederend, J. (2023). The Effects of Artificial Intelligence Assistance on the Radiologists’ Assessment of Lung Nodules on CT Scans: A Systematic Review. Journal of Clinical Medicine, 12(10), 3536. https://doi.org/10.3390/jcm12103536








