Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Population and Samples
2.3. Biopsy Samples
2.4. dd-cfDNA Measurement in Blood Samples
2.5. Statistical Analyses of dd-cfDNA and eGFR
3. Results
3.1. Patients and Blood Samples
3.2. dd-cfDNA and eGFR in Kidney Transplant Recipients
3.3. Performance Estimates for Discriminatory Ability of Tests
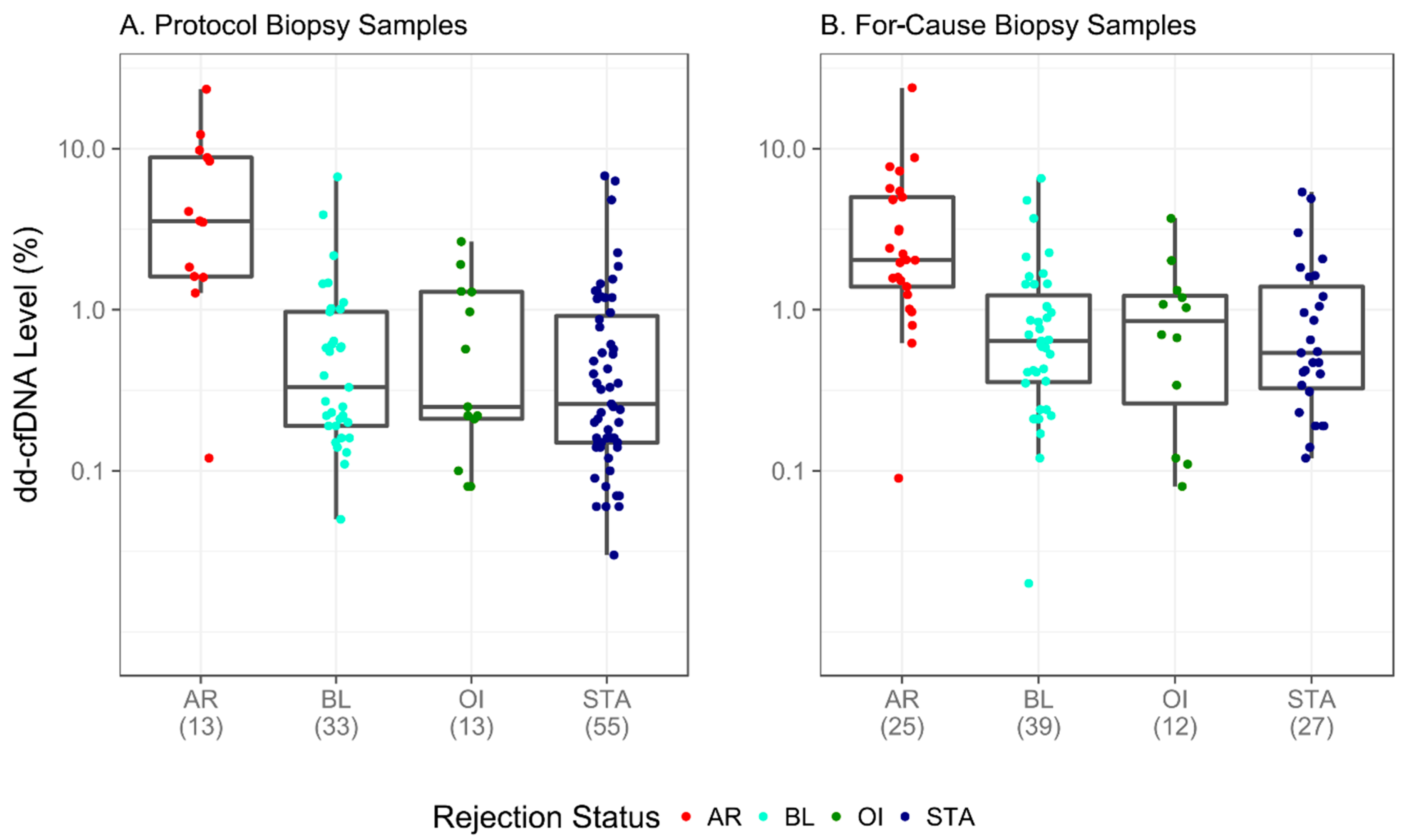
3.4. dd-cfDNA Performance in Unique Biopsy-Confirmed Subgroups
3.5. Relationship Between dd-cfDNA and Rejection Type
3.6. dd-cfDNA Levels by Donor Type
3.7. dd-cfDNA Variability over Time
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jacquemont, L.; Soulillou, J.P.; Degauque, N. Blood biomarkers of kidney transplant rejection, an endless search? Expert Rev. Mol. Diagn. 2017, 17, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Humar, A.; Kerr, S.; Hassoun, A.; Granger, D.; Suhr, B.; Matas, A. The association between acute rejection and chronic rejection in kidney transplantation. Transplant. Proc. 1999, 31, 1302–1303. [Google Scholar] [CrossRef]
- Nankivell, B.J.; Alexander, S.I. Rejection of the kidney allograft. N. Engl. J. Med. 2010, 363, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Sarwal, M.M. The proteogenomic path towards biomarker discovery. Pediatr. Transplant. 2008, 12, 737–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, D.J.; Kaplan, B.; Kirk, A.D. Biomarkers for kidney transplant rejection. Nat. Rev. Nephrol. 2014, 10, 215–225. [Google Scholar] [CrossRef]
- Nasr, M.; Sigdel, T.; Sarwal, M. Advances in diagnostics for transplant rejection. Expert Rev. Mol. Diagn. 2016, 16, 1121–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.Y.; Sarwal, M.M. Transplant genetics and genomics. Nat. Rev. Genet. 2017, 18, 309–326. [Google Scholar] [CrossRef]
- Loupy, A.; Haas, M.; Solez, K.; Racusen, L.; Glotz, D.; Seron, D.; Nankivell, B.J.; Colvin, R.B.; Afrouzian, M.; Akalin, E.; et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am. J. Transplant. 2017, 17, 28–41. [Google Scholar] [CrossRef]
- Snyder, T.M.; Khush, K.K.; Valantine, H.A.; Quake, S.R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl. Acad. Sci. USA 2011, 108, 6229–6234. [Google Scholar] [CrossRef] [Green Version]
- Beck, J.; Bierau, S.; Balzer, S.; Andag, R.; Kanzow, P.; Schmitz, J.; Gaedcke, J.; Moerer, O.; Slotta, J.E.; Walson, P.; et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin. Chem. 2013, 59, 1732–1741. [Google Scholar] [CrossRef]
- De Vlaminck, I.; Valantine, H.A.; Snyder, T.M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; Bernstein, D.; Weisshaar, D.; et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med. 2014, 6, 241ra77. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Vitalone, M.J.; Tran, T.Q.; Dai, H.; Hsieh, S.C.; Salvatierra, O.; Sarwal, M.M. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 2013, 96, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.R.; Thorne, A.; Faro, M.L.L. Donor-specific Cell-Free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation 2018. [Google Scholar] [CrossRef] [PubMed]
- Bloom, R.D.; Bromberg, J.S.; Poggio, E.D.; Bunnapradist, S.; Langone, A.J.; Sood, P.; Matas, A.J.; Mehta, S.; Mannon, R.B.; Sharfuddin, A.; et al. Cell-Free DNA and Active Rejection in Kidney Allografts. J. Am. Soc. Nephrol. 2017, 28, 2221–2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dar, P.; Curnow, K.J.; Gross, S.J.; Hall, M.P.; Stosic, M.; Demko, Z.; Zimmermann, B.; Hill, M.; Sigurjonsson, S.; Ryan, A.; et al. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing. Am. J. Obstet. Gynecol. 2014, 211, e1–e17. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.; Hunkapiller, N.; Banjevic, M.; Vankayalapati, N.; Fong, N.; Jinnett, K.N.; Demko, Z.; Zimmermann, B.; Sigurjonsson, S.; Gross, S.J.; et al. Validation of an Enhanced Version of a Single-Nucleotide Polymorphism-Based Noninvasive Prenatal Test for Detection of Fetal Aneuploidies. Fetal Diagn. Ther. 2016, 40, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Pergament, E.; Cuckle, H.; Zimmermann, B.; Banjevic, M.; Sigurjonsson, S.; Ryan, A.; Hall, M.P.; Dodd, M.; Lacroute, P.; Stosic, M.; et al. Single-nucleotide polymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet. Gynecol. 2014, 124 Pt 1, 210–218. [Google Scholar] [CrossRef]
- Haas, M.; Loupy, A.; Lefaucheur, C.; Roufosse, C.; Glotz, D.; Seron, D.; Nankivell, B.J.; Halloran, P.F.; Colvin, R.B.; Akalin, E.; et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am. J. Transplant. 2018, 18, 293–307. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Xie, W.; Wang, H.; Jin, J.; Wu, J.; He, Q. C4d as a significant predictor for humoral rejection in renal allografts. Clin. Transplant. 2005, 19, 785–791. [Google Scholar]
- Crespo, M.; Pascual, M.; Tolkoff-Rubin, N.; Mauiyyedi, S.; Collins, A.B.; Fitzpatrick, D.; Farrell, M.L.; Williams, W.W.; Delmonico, F.L.; Cosimi, A.B.; et al. Acute humoral rejection in renal allograft recipients: I. Incidence, serology and clinical characteristics. Transplantation 2001, 71, 652–658. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, B.; Hill, M.; Gemelos, G.; Demko, Z.; Banjevic, M.; Baner, J.; Ryan, A.; Sigurjonsson, S.; Chopra, N.; Dodd, M.; et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat. Diagn. 2012, 32, 1233–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Dunn, O.J. Multiple Comparisons Using Rank Sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Levey, A.S.; Bosch, J.P.; Lewis, J.B.; Greene, T.; Rogers, N.; Roth, D. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Modification of Diet in Renal Disease Study Group. Ann. Intern. Med. 1999, 130, 461–470. [Google Scholar] [CrossRef]
- Schwartz, G.J.; Munoz, A.; Schneider, M.F.; Mak, R.H.; Kaskel, F.; Warady, B.A.; Furth, S.L. New equations to estimate GFR in children with CKD. J. Am. Soc. Nephrol. 2009, 20, 629–637. [Google Scholar] [CrossRef]
- National Institute of Diabetes and Digestive and Kidney Diseases. Chronic Kidney Disease Tests & Diagnosis. Available online: https://www.niddk.nih.gov/health-information/kidney-disease/chronic-kidney-disease-ckd/tests-diagnosis (accessed on 31 August 2018).
- Efron, B.; Tibshirani, R. Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Stat. Sci. 1986, 1, 54–75. [Google Scholar] [CrossRef]
- Laird, N.M.; Ware, J.H. Random-effects models for longitudinal data. Biometrics 1982, 38, 963–974. [Google Scholar] [CrossRef]
- Kenward, M.G.; Roger, J.H. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 1997, 53, 983–997. [Google Scholar] [CrossRef]







| Phenotype Characteristic | Active Rejection (38 Samples) | Non-Rejection | |||
|---|---|---|---|---|---|
| Stable (82 Samples) | Borderline AR (72 Samples) | Other Injury (25 Samples) b | Combined (179 Samples) | ||
| Recipient age, year * (p-value < 0.0001) | |||||
| (0, 18) | 0 (0) | 44 (53.7) | 1 (1.4) | 4 (16.0) | 49 (27.4) |
| (18, 40) | 10 (26.3) | 32 (39.0) | 18 (18.0) | 8 (32.0) | 58 (32.4) |
| (40, 80) | 28 (73.7) | 6 (7.3) | 53 (73.6) | 13 (52.0) | 72 (40.2) |
| Mean ± SD | 47.91 ± 14.31 | 20.04 ± 11.97 | 47.88 ± 13.24 | 44.75 ± 23.73 | 34.65 ± 19.87 |
| Median | 49.13 | 19.96 | 47.46 | 40.97 | 31.33 |
| Range | 23–76 | 3–70 | 5–74 | 3–80 | 3–80 |
| Male/female, no. (%) (p-value = 0.5988) | |||||
| Male | 17 (44.7) | 48 (58.5) | 40 (55.6) | 15 (60) | 103 (57.5) |
| Female | 21 (55.3) | 34 (41.5) | 32 (44.4) | 10 (40) | 76 (42.5) |
| Ethnicity, no. (%) (p-value = 1) | |||||
| Hispanic or Latino | 13 (34.2) | 28 (34.1) | 24 (33.3) | 10 (40) | 62 (34.6) |
| Not Hispanic or Latino | 25 (65.8) | 54 (65.9) | 48 (66.7) | 15 (60) | 117 (65.4) |
| Race groups, no. (%) (p-value = 0.4695) | |||||
| White or Caucasian | 10 (26.6) | 42 (51.2) | 16 (22.2) | 6 (24) | 64 (35.8) |
| Black or African American | 6 (15.8) | 7 (8.5) | 14 (19.4) | 4 (16) | 25 (14.0) |
| Asian or Pacific Islander | 8 (21.1) | 4 (4.9) | 15 (20.8) | 4 (16) | 23 (12.8) |
| Other/Not reported | 14 (36.8) | 29 (35.4) | 27 (37.8) | 11 (44.0) | 67 (37.4) |
| Recipient weight, kg (p-value = 0.6039) | |||||
| Mean ± SD | 76.22 ± 19.7 | 70.9 ± 8.8 | 79.18 ± 18.7 | 78.33 ± 17.1 | 78.1 ± 17.6 |
| Median | 72.5 | 73.0 | 78.0 | 76.0 | 76.0 |
| Range | 45–119 | 52–81 | 46–134 | 47–109 | 46–134 |
| Unknown | 6 | 72 | 7 | 7 | 86 |
| DSA positive, no. (%) (p-value = 0.1928) | |||||
| Yes | 15 (39.5) | 0 (0) | 18 (25) | 2 (8) | 20 (11.2) |
| No | 21 (55.3) | 0 (0) | 48 (66.7) | 3 (12) | 51 (28.5) |
| Not recorded | 2 (5.3) | 82 (100) | 6 (8.3) | 20 (80) | 108 (60.3) |
| Indication for renal transplantation, no. (%) (p-value = 0.4869) | |||||
| Glomerulonephritis | 5 (13.2) | 6 (7.3) | 4 (5.6) | 1 (4) | 11 (6.1) |
| Focal segmental glomerulosclerosis | 5 (13.2) | 5 (6.1) | 6 (8.3) | 2 (8) | 13 (7.3) |
| Diabetes mellitus | 5 (13.2) | 3 (3.7) | 15 (20.8) | 5 (20) | 23 (12.8) |
| Thin basement membrane nephropathy | 0 (0) | 0 (0) | 2 (2.8) | 0 (0) | 2 (1.1) |
| Polycystic kidney disease | 3 (7.9) | 2 (2.4) | 7 (9.7) | 1 (4) | 10 (5.6) |
| Solitary kidney | 0 (0) | 0 (0) | 3 (4.2) | 0 (0) | 3 (1.7) |
| Hypertension | 4 (10.5) | 2 (2.4) | 13 (18.1) | 3 (12) | 18 (10.1) |
| IgA nephropathy | 3 (7.9) | 0 (0) | 7 (9.7) | 1 (4) | 8 (4.5) |
| Lupus nephritis | 2 (5.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0.0) |
| ANCA—vasculitis | 1 (2.6) | 0 (0) | 2 (2.8) | 0 (0) | 2 (1.1) |
| Other/Unknown | 10 (26.3) | 64 (78.1) | 13 (18.1) | 12 (48) | 89 (49.7) |
| Donor source *, no. (%) (p-value < 0.0001) | |||||
| Living related | 1 (2.8) | 2 (2.4) | 9 (12.5) | 3 (12) | 14 (7.8) |
| Living unrelated | 2 (5.3) | 50 (61) | 18 (25) | 7 (28) | 75 (41.9) |
| Deceased unrelated | 35 (92.1) | 30 (36.6) | 45 (62.5) | 15 (60) | 90 (50.3) |
| Parameter | Active Rejection | Non-Rejection | |||
|---|---|---|---|---|---|
| Stable | Borderline AR | Other Injury | Combined | ||
| dd-cfDNA | |||||
| Number of samples (%) | 38 (17.5) | 82 (37.8) | 72 (33.2) | 25 (11.5) | 179 (82.5) |
| Mean (SD) | 4.64 (5.45) | 0.90 (1.36) | 0.95 (1.31) | 0.89 (0.91) | 0.92 (1.28) |
| Median (range) | 2.32 (0.1–23.9) | 0.4 (0.03–6.8) | 0.58 (0.02–6.7) | 0.67 (0.08–3.69) | 0.47 (0.04–6.78) |
| eGFR | |||||
| Number of samples (%) | 38 (17.5) | 82 (37.8) | 72 (33.2) | 25 (11.5) | 179 (82.5) |
| Score mean (SD) | 49.0 (22.4) | 99.5 (16.1) | 55.9 (21.4) | 63.8 (29.0) | 77.0 (8.45) |
| Score median (range) | 45.67 (8.0–100.4) | 104.5 (47.4–131.1) | 55.99 (6.4–109.4) | 57.4 (25.0–116.9) | 76.06 (6.4–131.1) |
| Rejection Status | Biopsy Reason | Total | Median | Low | High | Mean | SD |
|---|---|---|---|---|---|---|---|
| AR | For-cause | 25 | 2.04 | 0.09 | 23.9 | 3.85 | 4.81 |
| Protocol | 13 | 3.56 | 0.12 | 23.4 | 6.16 | 6.44 | |
| BL | For-cause | 39 | 0.64 | 0.02 | 6.54 | 1.07 | 1.32 |
| Protocol | 33 | 0.33 | 0.05 | 6.69 | 0.82 | 1.30 | |
| OI | For-cause | 12 | 0.865 | 0.08 | 3.69 | 1.03 | 1.02 |
| Protocol | 13 | 0.25 | 0.08 | 2.65 | 0.76 | 0.82 | |
| STA | For-cause | 27 | 0.54 | 0.12 | 5.38 | 1.12 | 1.36 |
| Protocol | 55 | 0.26 | 0.03 | 6.78 | 0.80 | 1.37 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sigdel, T.K.; Archila, F.A.; Constantin, T.; Prins, S.A.; Liberto, J.; Damm, I.; Towfighi, P.; Navarro, S.; Kirkizlar, E.; Demko, Z.P.; et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J. Clin. Med. 2019, 8, 19. https://doi.org/10.3390/jcm8010019
Sigdel TK, Archila FA, Constantin T, Prins SA, Liberto J, Damm I, Towfighi P, Navarro S, Kirkizlar E, Demko ZP, et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. Journal of Clinical Medicine. 2019; 8(1):19. https://doi.org/10.3390/jcm8010019
Chicago/Turabian StyleSigdel, Tara K., Felipe Acosta Archila, Tudor Constantin, Sarah A. Prins, Juliane Liberto, Izabella Damm, Parhom Towfighi, Samantha Navarro, Eser Kirkizlar, Zachary P. Demko, and et al. 2019. "Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR" Journal of Clinical Medicine 8, no. 1: 19. https://doi.org/10.3390/jcm8010019
APA StyleSigdel, T. K., Archila, F. A., Constantin, T., Prins, S. A., Liberto, J., Damm, I., Towfighi, P., Navarro, S., Kirkizlar, E., Demko, Z. P., Ryan, A., Sigurjonsson, S., Sarwal, R. D., Hseish, S. -C., Chan-On, C., Zimmermann, B., Billings, P. R., Moshkevich, S., & Sarwal, M. M. (2019). Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. Journal of Clinical Medicine, 8(1), 19. https://doi.org/10.3390/jcm8010019






