Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis
Abstract
:1. Introduction
2. Related Works
3. Contribution
- (1)
- To the best of our knowledge, this is the first approach to develop a comprehensive deep learning-based framework for the classification of multiple GI diseases by considering deep spatiotemporal features. In contrast, most of the previous studies [6,7,8,9,10,11,12,13,14,15] considered the limited number of classes that are related to a specific type of GI portion.
- (2)
- We proposed a novel cascaded ResNet and LSTM-based framework in the medical domain to learn both spatial and temporal features for the different type of GI diseases. When compared to the previous methods based on handcrafted features and simple 2D-CNNs, our method can manage the large intraclass and low interclass variations among multiple classes more effectively.
- (3)
- We deeply analyzed the performance of our proposed method by selecting the multilevel spatial features for LSTM from the different layers of the ResNet network. Furthermore, the performance of multilevel spatial features was also analyzed by applying principal component analysis (PCA).
- (4)
- We compared the performance of the various state-of-the-art CNN models and different handcrafted feature-based approaches. Our analysis was more detailed, in contrast to previous studies [8,9], which provided only a limited performance analysis for a small number of classes related to a specific GI part.
- (5)
- Finally, we have ensure that our trained model and video indices of experimental endoscopic videos are publicly available through [18]; therefore, other researchers can evaluate and compare its performance.
4. Proposed Method
4.1. Overview of the Proposed Approach
4.2. Structure of Our Proposed Model
4.2.1. Spatial Features Extraction using a Convolutional Neural Network
4.2.2. Temporal Features Extraction by Long Short-term Memory Model
4.2.3. Classification
5. Experimental Setup and Performance Analysis
5.1. Dataset
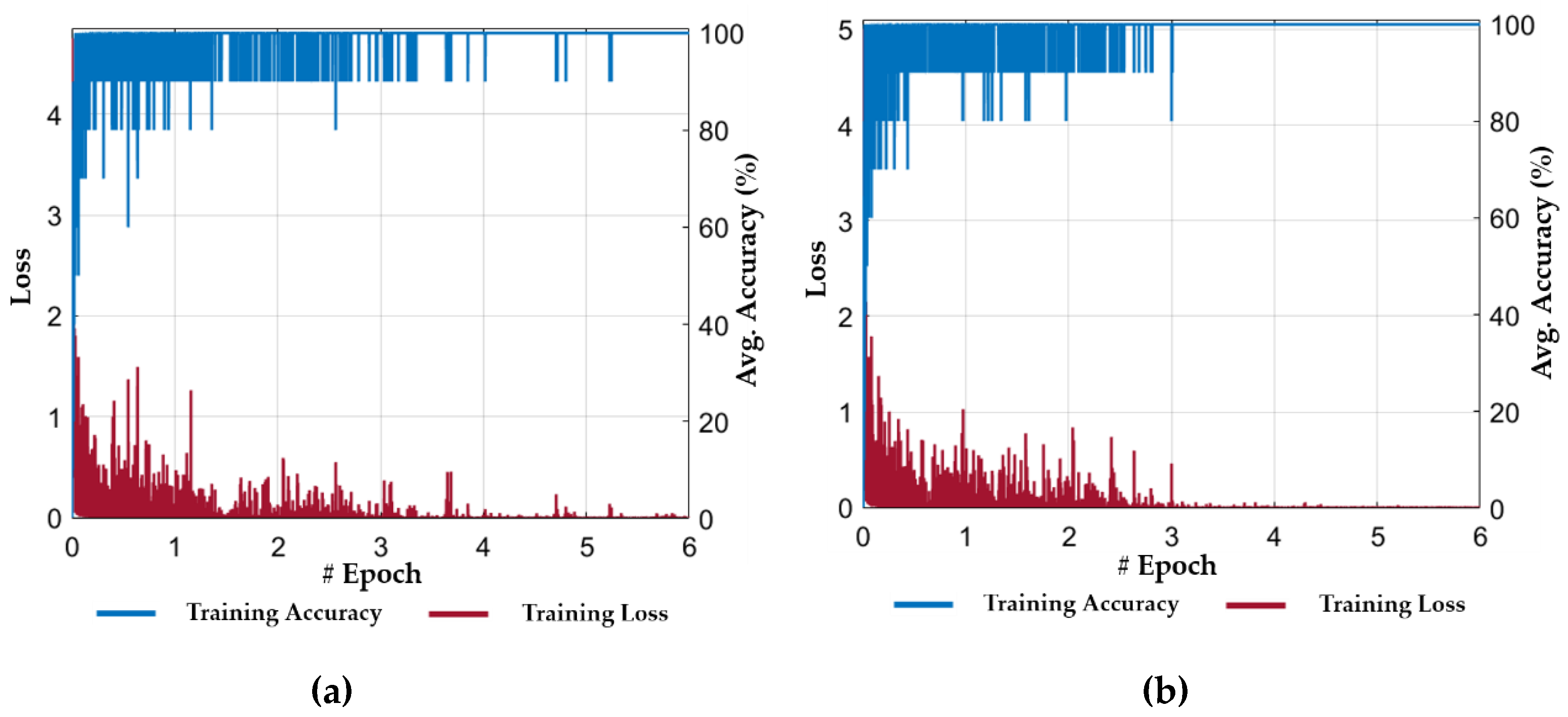
5.2. Experimental Setup and Training
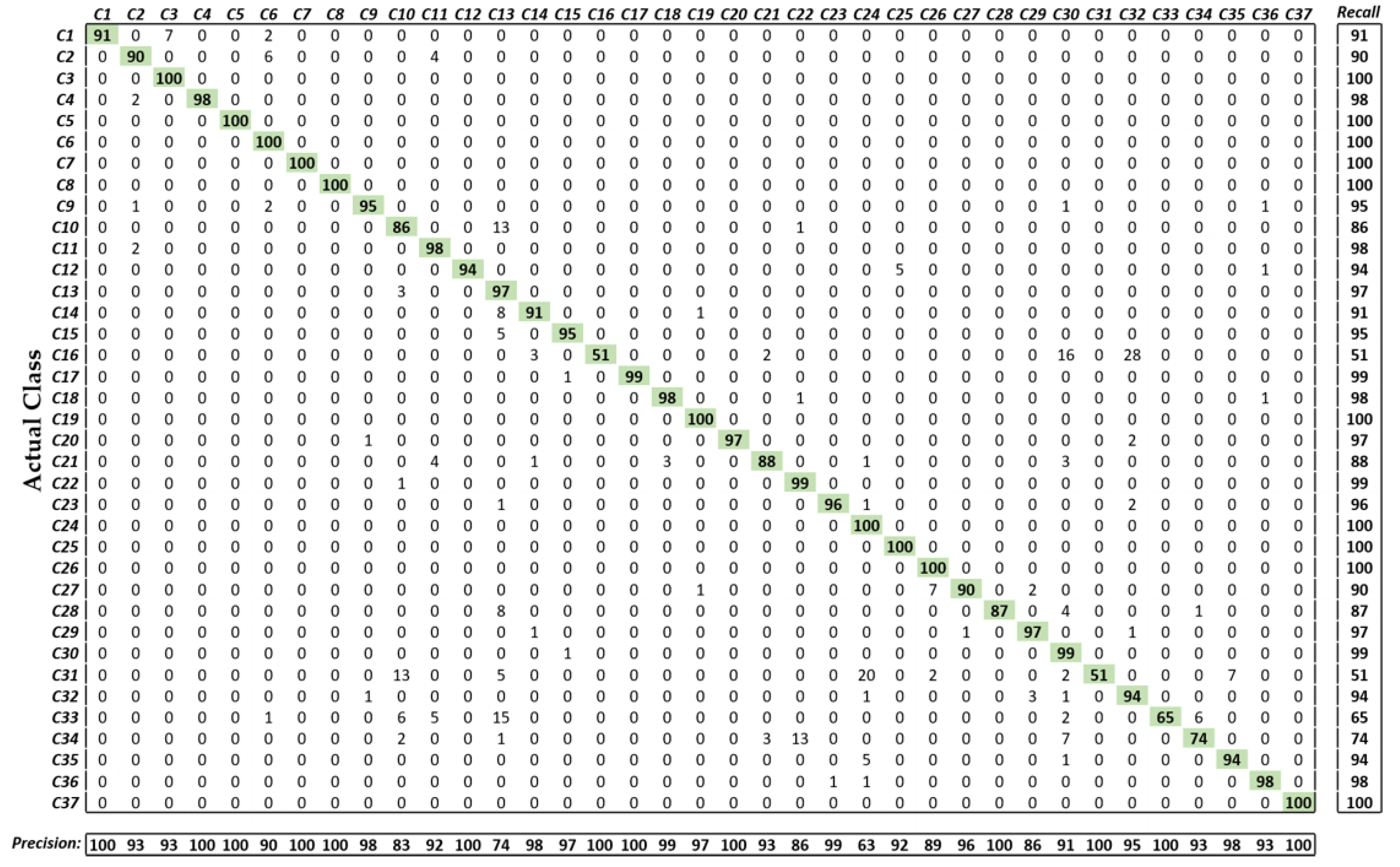
5.3. Evaluation of the Performance by Proposed Method
5.3.1. Performance Analysis Metric
5.3.2. Testing of the Proposed Method
5.3.3. Comparisons with Previous Methods
6. Discussion
7. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics. Ca-Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Vuong, Q.-H.; Ho, M.-T.; Vuong, T.-T.; La, V.-P.; Ho, M.-T.; Nghiem, K.-C.P.; Tran, B.X.; Giang, H.-H.; Giang, T.-V.; Latkin, C.; et al. Artificial intelligence vs. natural stupidity: Evaluating AI readiness for the vietnamese medical information system. J. Clin. Med. 2019, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.X.; Vu, G.T.; Ha, G.H.; Vuong, Q.-H.; Ho, M.-T.; Vuong, T.-T.; La, V.-P.; Ho, M.-T.; Nghiem, K.-C.P.; Nguyen, H.L.T.; et al. Global evolution of research in artificial intelligence in health and medicine: A bibliometric study. J. Clin. Med. 2019, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Owais, M.; Arsalan, M.; Choi, J.; Park, K.R. Effective diagnosis and treatment through content-based medical image retrieval (CBMIR) by using artificial intelligence. J. Clin. Med. 2019, 8, 462. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-H.; Liu, W.-X. Identifying degenerative brain disease using rough set classifier based on wavelet packet method. J. Clin. Med. 2018, 7, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Meng, M.Q.-H. Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 323–329. [Google Scholar] [PubMed]
- Seguí, S.; Drozdzal, M.; Pascual, G.; Radeva, P.; Malagelada, C.; Azpiroz, F.; Vitrià, J. Generic feature learning for wireless capsule endoscopy analysis. Comput. Biol. Med. 2016, 79, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Takiyama, H.; Ozawa, T.; Ishihara, S.; Fujishiro, M.; Shichijo, S.; Nomura, S.; Miura, M.; Tada, T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Shichijo, S.; Nomura, S.; Aoyama, K.; Nishikawa, Y.; Miura, M.; Shinagawa, T.; Takiyama, H.; Tanimoto, T.; Ishihara, S.; Matsuo, K.; et al. Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine 2017, 25, 106–111. [Google Scholar] [CrossRef]
- Zhang, R.; Zheng, Y.; Mak, T.W.C.; Yu, R.; Wong, S.H.; Lau, J.Y.W.; Poon, C.C.Y. Automatic detection and classification of colorectal polyps by transferring low-level CNN features from nonmedical domain. IEEE J. Biomed. Health Inf. 2017, 21, 41–47. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Integrating online and offline three-dimensional deep learning for automated polyp detection in colonoscopy videos. IEEE J. Biomed. Health Inf. 2017, 21, 65–75. [Google Scholar] [CrossRef] [PubMed]
- He, J.-Y.; Wu, X.; Jiang, Y.-G.; Peng, Q.; Jain, R. Hookworm detection in wireless capsule endoscopy images with deep learning. IEEE Trans. Image Process. 2018, 27, 2379–2392. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J.; et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Perez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Han, G.; Li, B.N.; Lin, Z.; Ciaccio, E.J.; Green, P.H.; Qin, J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput. Biol. Med. 2017, 85, 1–6. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Dongguk, CNN and LSTM Models for the Classification of Multiple Gastrointestinal (GI) Diseases, and Video Indices of Experimental Endoscopic Videos. Available online: http://dm.dgu.edu/link.html (accessed on 15 June 2019).
- Karargyris, A.; Bourbakis, N. Detection of small bowel polyps and ulcers in wireless capsule endoscopy videos. IEEE Trans. Biomed. Eng. 2011, 58, 2777–2786. [Google Scholar] [CrossRef]
- Li, B.; Meng, M.Q.-H. Automatic polyp detection for wireless capsule endoscopy images. Expert Syst. Appl. 2012, 39, 10952–10958. [Google Scholar] [CrossRef]
- Yuan, Y.; Meng, M.Q.-H. Polyp classification based on bag of features and saliency in wireless capsule endoscopy. In Proceedings of the IEEE International Conference on Robotics and Automation, Hong Kong, China, 31 May–7 June 2014; pp. 3930–3935. [Google Scholar]
- Yuan, Y.; Li, B.; Meng, M.Q.-H. Improved bag of feature for automatic polyp detection in wireless capsule endoscopy images. IEEE Trans. Autom. Sci. Eng. 2016, 13, 529–535. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360v4. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Raychaudhuri, S. Introduction to Monte Carlo simulation. In Proceedings of the IEEE Winter Simulation Conference, Miami, FL, USA, 7–10 December 2008; pp. 91–100. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 630–645. [Google Scholar]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef] [Green Version]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Heaton, J. Artificial Intelligence for Humans; Deep Learning and Neural Networks; Heaton Research, Inc.: St. Louis, MO, USA, 2015; Volume 3. [Google Scholar]
- Gastrolab—The Gastrointestinal Site. Available online: http://www.gastrolab.net/ni.htm (accessed on 1 February 2019).
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; de Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.-T.; Lux, M.; Schmidt, P.T.; et al. KVASIR: A multi-class image dataset for computer aided gastrointestinal disease detection. In Proceedings of the 8th ACM Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169. [Google Scholar]
- Peng, X.; Tang, Z.; Yang, F.; Feris, R.S.; Metaxas, D. Jointly optimize data augmentation and network training: Adversarial data augmentation in human pose estimation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2226–2234. [Google Scholar]
- Wong, S.C.; Gatt, A.; Stamatescu, V.; McDonnell, M.D. Understanding data augmentation for classification: When to warp? In Proceedings of the IEEE International Conference on Digital Image Computing: Techniques and Applications, Gold Coast, Australia, 30 November–2 December 2016; pp. 1–6. [Google Scholar]
- MATLAB R2018b. Available online: https://www.mathworks.com/products/matlab.html (accessed on 1 February 2019).
- Deep Learning Toolbox. Available online: https://in.mathworks.com/products/deep-learning.html (accessed on 1 July 2019).
- Intel® Core i7-3770K Processor. Available online: https://ark.intel.com/content/www/us/en/ark/products/65523/intel-core-i7-3770k-processor-8m-cache-up-to-3-90-ghz.html (accessed on 1 February 2019).
- GeForce GTX 1070. Available online: https://www.geforce.com/hardware/desktop-gpus/geforce-gtx-1070/specifications (accessed on 1 February 2019).
- Bottou, L. Stochastic gradient descent tricks. In Neural Networks: Tricks of the Trade; Springer: Berlin, Germany, 2012; pp. 421–436. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Hossin, M.; Sulaiman, M.N. A review on evaluation metrics for data classification evaluations. Int. J. Data Min. Knowl. Manag. Process. 2015, 5, 1–11. [Google Scholar]
- Cover, T.M.; Hart, P.E. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Ilin, A.; Raiko, T. Practical approaches to principal component analysis in the presence of missing values. J. Mach. Learn. Res. 2010, 11, 1957–2000. [Google Scholar]
- Precision and Recall. Available online: https://en.wikipedia.org/wiki/Precision_and_recall (accessed on 21 May 2019).
- Student’s T-test. Available online: https://en.wikipedia.org/wiki/Student%27s_t-test (accessed on 20 March 2019).
- Cohen, J. A power primer. Psychol. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Cuthill, I.C. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol. Rev. 2007, 82, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanyam, M.; Maheshwari, R.P.; Balasubramanian, R. Local maximum edge binary patterns: A new descriptor for image retrieval and object tracking. Signal Process. 2012, 92, 1467–1479. [Google Scholar] [CrossRef]
- Velmurugan, K.; Baboo, S.S. Image retrieval using Harris corners and histogram of oriented gradients. Int. J. Comput. Appl. 2011, 24, 6–10. [Google Scholar]
- Nguyen, D.T.; Pham, T.D.; Baek, N.R.; Park, K.R. Combining deep and handcrafted image features for presentation attack detection in face recognition systems using visible-light camera sensors. Sensors 2018, 18, 699. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Hsu, C.-W.; Lin, C.-J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 2002, 13, 415–425. [Google Scholar] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]




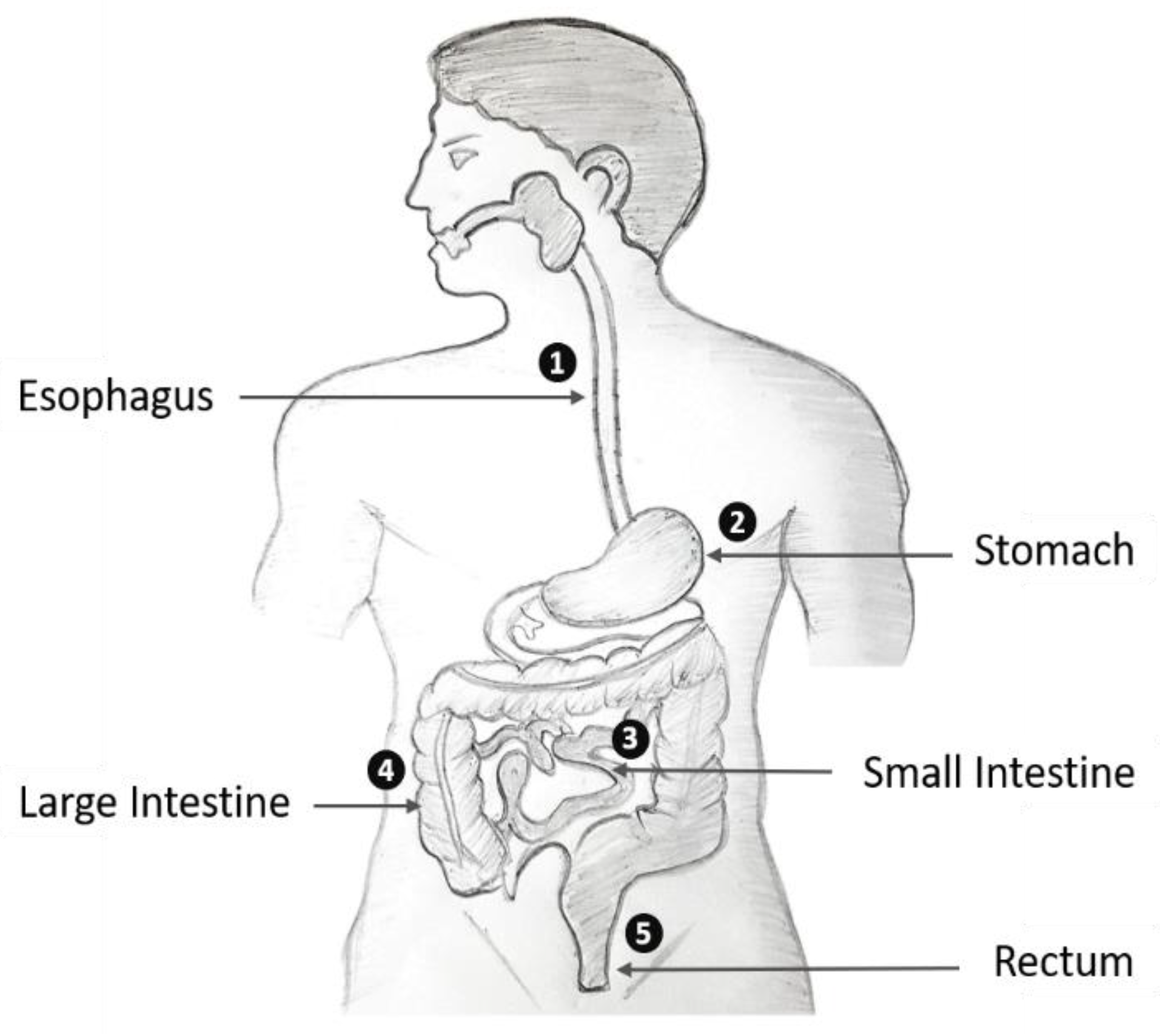
















| Endoscopy Type | Method | Purpose | No. of Classes | Strength | Weakness |
|---|---|---|---|---|---|
| CE | Log Gabor filter, SUSAN edge detection and SVM [19] | Small bowel polyps and ulcers detection | 2 | Computationally efficient | Limited dataset and number of classes Low detection performance |
| CE | Texture features (ULBP, wavelet) + SVM [20] | Polyp detection in GI tract | 2 | Robust to illumination change and scale invariant | Limited dataset and number of classes |
| CE | Texture features (LBP, wavelet) + SVM [6] | Tumor recognition in the digestive tract | 2 | Invariant to illumination change Extract multiresolution features | Limited dataset and number of classes |
| CE | Texture features (SIFT, Saliency) + SVM [21] | Polyp classification | 2 | Extract scale invariant features | Limited dataset and number of classes |
| CE | Texture features (SIFT, HoG, LBP, CLBP, ULBP) + SVM, FLDA [22] | Polyp Detection | 2 | Extract scale invariant features High classification performance | Limited dataset and number of classes |
| CE | CNN [7] | Small intestine movement characterization | 6 | High classification performance | Limited number of classes |
| CE | CNN [15] | Celiac disease classification | 2 | High sensitivity and specificity | Limited dataset and number of classes |
| CE | CNN [12] | Hookworm detection | 2 | Edge extraction network results in better performance | Limited number of classes |
| EGD | CNN [9] | H. pylori infection detection | 9 | Comparable performance of second CNN with the clinical diagnosis reference standard | CAD performance should be enhanced. A limited number of classes |
| EGD | CNN [8] | Anatomical classification of GI images | 6 | High classification performance Computationally efficient | Limited number of classes Only used for anatomical classification |
| EGD | CNN-based SSD detector [13] | Gastric cancer detection | 2 | High sensitivity Computationally efficient | Overall low positive prediction value Limited dataset and number of classes |
| Colonoscopy | CNN [10] | Colorectal polyp detection and classification | 3 | High detection performance | Limited dataset and number of classes Low classification performance |
| Colonoscopy | CNN [14] | Real-time colorectal polyp type analysis | 4 | High accuracy and sensitivity | Limited number of classes Low specificity |
| Colonoscopy | Online and offline 3D-CNN [11] | Detection of colorectal polyps | 2 | Computationally efficient | CAD performance should be enhanced. |
| EGD, Colonoscopy, Sigmoidoscopy, Rectoscopy | CNN (ResNet) + LSTM (Proposed) | Classification of multiple GI diseases | 37 | Computationally efficient High classification performance | Cascaded training of CNN and LSTM requires more time |
| Layer Name | Feature Map Size | Filters | Kernel Size | Stride | #Padding | Total Learnable Parameters |
|---|---|---|---|---|---|---|
| Image input layer | n/a | n/a | n/a | n/a | n/a | |
| Conv1 | 64 | 2 | 3 | 9600 | ||
| Max pooling | 1 | 2 | 1 | |||
| Conv2-1–Conv2-2 (Identity Mapping) | 64 64 | 1 1 | 1 1 | 74,112 | ||
| Conv3-1–Conv3-2 (Identity Mapping) | 64 64 | 1 1 | 1 1 | 74,112 | ||
| Conv4-1–Conv4-2 (1 × 1 Convolutional Mapping) | 128 128 128 | 2 1 2 | 1 1 0 | 230,528 | ||
| Conv5-1–Conv5-2 (Identity Mapping) | 128 128 | 1 1 | 1 1 | 295,680 | ||
| Conv6-1–Conv6-3 (1 × 1 Convolutional Mapping) | 256 256 256 | 2 1 2 | 1 1 0 | 919,808 | ||
| Conv7-1–Conv7-2 (Identity Mapping) | 256 256 | 1 1 | 1 1 | 1,181,184 | ||
| Conv8-1–Conv8-3 (1 × 1 Convolutional Mapping) | 512 512 512 | 2 1 2 | 1 1 0 | 3,674,624 | ||
| Conv9-1–Conv9-2 (Identity Mapping) | 512 512 | 1 1 | 1 1 | 4,721,664 | ||
| Avg pooling | 1 | 7 | 0 | |||
| FC layer | 37 | 18,981 | ||||
| Softmax | 37 | |||||
| Classification layer | 37 | |||||
| Total number of learnable parameters: 11,200,293 | ||||||
| Layer Name | Feature Map Size | Total Learnable |
|---|---|---|
| Sequence input layer | ||
| LSTM | 600 | 1,951,200 |
| Dropout | 600 | |
| FC layer | 37 | 22,237 |
| Softmax | 37 | |
| Classification layer | 37 | |
| Total learnable parameters: 1,973,437 | ||
| Gastrointestinal Tract | Class Name (Normal/Disease Cases) | Training Set (Frames) | Testing Set (Frames) | Total | |
|---|---|---|---|---|---|
| Anatomical District | Subcategory | ||||
| Esophagus | Larynx | C1: Normal | 387 | 387 | 774 |
| Upper part | C2: Normal | 625 | 625 | 1250 | |
| C3: Esophageal candidiasis | 419 | 419 | 838 | ||
| C4: Esophageal papillomatosis | 272 | 272 | 544 | ||
| Lower part (z-line) | C5: Normal | 250 | 250 | 500 | |
| Stomach | Cardia | C6: Hiatal hernia | 648 | 648 | 1296 |
| Fundus | C7: Atrophic gastritis | 241 | 241 | 482 | |
| C8: Atrophic and xanthoma gastritis | 255 | 254 | 509 | ||
| Body | C9: Benign hyperplastic polyps | 1070 | 1070 | 2140 | |
| C10: Adenocarcinoma (Cancer) | 955 | 955 | 1910 | ||
| Pylorus | C11: Normal | 1275 | 1275 | 2550 | |
| Small Intestine | Duodenum | C12: Normal | 423 | 423 | 846 |
| C13: Ulcer | 1345 | 1345 | 2690 | ||
| C14: Papilla vateri | 702 | 702 | 1404 | ||
| Terminal Ileum | C15: Crohn’s disease | 840 | 840 | 1680 | |
| Ileocecal | C16: Severe Crohn’s disease | 278 | 278 | 556 | |
| Ileocecal valve | C17: Crohn’s disease | 838 | 838 | 1676 | |
| Large Intestine | Caecum | C18: Adenocarcinoma (Cancer) | 1301 | 1301 | 2602 |
| C19: Melanosis coli | 342 | 342 | 684 | ||
| C20: Caecal angiectasia | 403 | 404 | 807 | ||
| C21: Appendix aperture | 694 | 694 | 1388 | ||
| Ascending/ Transverse/Descending Colon | C22: Adenocarcinoma (Cancer) | 1293 | 1293 | 2586 | |
| C23: Melanosis coli | 603 | 604 | 1207 | ||
| C24: Other types of polyps | 250 | 250 | 500 | ||
| C25: Dyed resection margins | 250 | 250 | 500 | ||
| C26: Dyed lifted polyps | 250 | 250 | 500 | ||
| C27: Melanosis coli and tuber adenoma | 243 | 243 | 486 | ||
| C28: Inflammatory polyposis | 382 | 382 | 764 | ||
| C29: Normal | 500 | 500 | 1000 | ||
| Sigmoid Colon | C30: Tuber adenoma | 2212 | 2212 | 4424 | |
| C31: Polypoid cancer | 282 | 282 | 564 | ||
| Rectosigmoid | C32: Ulcerative colitis | 2071 | 2071 | 4142 | |
| Rectum | C33: Severe Crohn’s disease | 1074 | 1074 | 2148 | |
| C34: Adenocarcinoma (Cancer) | 1362 | 1362 | 2724 | ||
| C35: Tuber adenoma | 1069 | 1069 | 2138 | ||
| C36: Normal | 420 | 420 | 840 | ||
| C37: A focal radiation injury | 411 | 411 | 822 | ||
| Model | Number of Training Epochs | Initial Learning Rate | Momentum | L2-Regularization | Learning Rate Drop Factor | Mini-Batch Size |
|---|---|---|---|---|---|---|
| ResNet18 | 8 | 0.001 | 0.9 | 0.0001 | 0.1 | 10 |
| LSTM | 10 | 0.0001 | 0.9 | 0.0001 | 0.1 | 50 |
| Layer Name | Feature Dim. | ResNet18 [23] | Proposed | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Accuracy ± Std | F1 score ± Std | mAP ± Std | mAR ± Std | Accuracy ± Std | F1 score ± Std | mAP ± Std | mAR ± Std | ||
| Conv6-2 | 50,176 | 75.86 ± 4.03 | 78.62 ± 1.28 | 81.64 ± 0.35 | 75.85 ± 2.69 | 87.15 ± 1.02 | 87.61 ± 0.04 | 88.85 ± 0.81 | 86.40 ± 0.85 |
| Conv7-2 | 50,176 | 77.13 ± 3.61 | 79.61 ± 0.73 | 82.42 ± 0.76 | 77.02 ± 2.02 | 88.02 ± 2.78 | 88.94 ± 1.18 | 91.20 ± 0.12 | 86.81 ± 2.36 |
| Conv8-2 | 25,088 | 84.39 ± 1.54 | 84.75 ± 0.69 | 85.92 ± 0.20 | 83.62 ± 1.15 | 89.07 ± 0.10 | 89.96 ± 0.88 | 91.24 ± 0.86 | 88.72 ± 0.91 |
| Conv9-2 | 25,088 | 87.10 ± 0.70 | 87.57 ± 0.47 | 88.19 ± 0.17 | 86.97 ± 1.09 | 89.39 ± 1.10 | 89.70 ± 1.69 | 90.24 ± 1.61 | 89.18 ± 1.76 |
| Avg. pooling | 512 | 89.95 ± 1.26 | 90.35 ± 1.74 | 90.72 ± 1.17 | 89.99 ± 2.29 | 92.57 ± 0.66 | 93.41 ± 0.12 | 94.58 ± 0.37 | 92.28 ± 0.58 |
| Method | ResNe18 [23] | Proposed | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy ± Std | F1 score ± Std | mAP ± Std | mAR ± Std | Accuracy ± Std | F1 score ± Std | mAP ± Std | mAR ± Std | |
| With PCA (No. of eigenvectors = 136) | 88.50 ± 1.01 | 90.16 ± 0.16 | 91.85 ± 0.11 | 88.52 ± 0.20 | 90.01 ± 0.17 | 91.82 ± 0.37 | 94.22 ± 0.40 | 89.54 ± 0.33 |
| Without PCA | 89.95 ± 1.26 | 90.35 ± 1.74 | 90.72 ± 1.17 | 89.99 ± 2.29 | 92.57 ± 0.66 | 93.41 ± 0.12 | 94.58 ± 0.37 | 92.28 ± 0.58 |
| Methods | Accuracy | F1 Score | mAP | mAR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold 1 | Fold 2 | Avg. ± Std | Fold 1 | Fold 2 | Avg. ± Std | Fold 1 | Fold 2 | Avg. ± Std | Fold 1 | Fold 2 | Avg. ± Std | |
| SqueezeNet [24] | 78.69 | 77.00 | 77.84 ± 1.19 | 77.53 | 75.95 | 76.74 ± 1.12 | 78.38 | 75.16 | 76.77 ± 2.27 | 76.70 | 76.76 | 76.73 ± 0.04 |
| AlexNet [16] | 79.19 | 80.97 | 80.08 ± 1.26 | 80.31 | 80.66 | 80.49 ± 0.24 | 80.55 | 80.85 | 80.70 ± 0.21 | 80.08 | 80.47 | 80.28 ± 0.27 |
| GoogLeNet [8,12,15,17] | 83.36 | 85.82 | 84.59 ± 1.74 | 84.99 | 85.29 | 85.14 ± 0.21 | 84.67 | 85.92 | 85.29 ± 0.89 | 85.32 | 84.66 | 84.99 ± 0.47 |
| VGG19 [25] | 84.81 | 85.49 | 85.15 ± 0.48 | 84.57 | 86.02 | 85.29 ± 1.03 | 85.48 | 86.27 | 85.88 ± 0.56 | 83.67 | 85.77 | 84.72 ± 1.48 |
| VGG16 [25] | 83.88 | 87.57 | 85.72 ± 2.61 | 84.84 | 86.77 | 85.80 ± 1.37 | 85.20 | 87.28 | 86.24 ± 1.47 | 84.48 | 86.26 | 85.37 ± 1.26 |
| InceptionV3 [14,26] | 87.23 | 88.61 | 87.92 ± 0.98 | 87.80 | 89.10 | 88.45 ± 0.92 | 86.50 | 89.24 | 87.87 ± 1.93 | 89.14 | 88.96 | 89.05 ± 0.13 |
| ResNet50 [23] | 88.94 | 90.17 | 89.55 ± 0.87 | 90.13 | 91.06 | 90.60 ± 0.66 | 89.59 | 91.82 | 90.70 ± 1.58 | 90.68 | 90.32 | 90.50 ± 0.26 |
| ResNet18 [23] | 90.84 | 89.06 | 89.95 ± 1.26 | 91.58 | 89.13 | 90.35 ± 1.74 | 91.55 | 89.89 | 90.72 ± 1.17 | 91.62 | 88.37 | 89.99 ± 2.29 |
| Proposed | 92.10 | 93.03 | 92.57 ± 0.66 | 93.49 | 93.33 | 93.41 ± 0.12 | 94.32 | 94.84 | 94.58 ± 0.37 | 92.68 | 91.87 | 92.28 ± 0.58 |
| CNN Models | Size (MB) | No. of Conv. Layers | No. of FC Layers | No. of LSTM Layers | Network Depth | Parameters (Millions) | Image Input Size |
|---|---|---|---|---|---|---|---|
| SqueezeNet [24] | 4.6 MB | 18 | 18 | 1.24 | 227-by-227 | ||
| AlexNet [16] | 227 MB | 5 | 3 | 8 | 61 | 227-by-227 | |
| GoogLeNet [8,12,15,17] | 27 MB | 21 | 1 | 22 | 7.0 | 224-by-224 | |
| VGG19 [25] | 535 MB | 16 | 3 | 19 | 144 | 224-by-224 | |
| VGG16 [25] | 515 MB | 13 | 3 | 16 | 138 | 224-by-224 | |
| InceptionV3 [14,26] | 89 MB | 47 | 1 | 48 | 23.9 | 299-by-299 | |
| ResNet50 [23] | 96 MB | 49 | 1 | 50 | 25.6 | 224-by-224 | |
| ResNet18 [23] | 44 MB | 17 | 1 | 18 | 11.7 | 224-by-224 | |
| Proposed | 48 MB | 17 | 1 | 1 | 19 | 13.17 | 224-by-224 |
| Method | Classifiers | Accuracy | F1 Score | mAP | mAR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold 1 | Fold 2 | Avg. ± Std | Fold 1 | Fold 2 | Avg. ± Std | Fold 1 | Fold 2 | Avg. ± Std | Fold 1 | Fold 2 | Avg. ± Std | ||
| LBP [49] | AdaBoostM2 | 36.90 | 34.57 | 35.74 ± 1.65 | 28.85 | 26.55 | 27.70 ± 1.63 | 36.90 | 34.57 | 35.74 ± 1.65 | 23.68 | 21.55 | 22.61 ± 1.51 |
| Multi-SVM | 45.53 | 42.15 | 43.84 ± 2.39 | 43.34 | 41.35 | 42.35 ± 1.41 | 44.05 | 41.94 | 42.99 ± 1.49 | 42.66 | 40.77 | 41.72 ± 1.34 | |
| RF | 57.37 | 56.84 | 57.10 ± 0.37 | 53.40 | 54.31 | 53.85 ± 0.64 | 54.53 | 55.06 | 54.79 ± 0.37 | 52.31 | 53.58 | 52.95 ± 0.90 | |
| KNN | 49.68 | 51.24 | 50.46 ± 1.10 | 46.28 | 48.44 | 47.36 ± 1.53 | 45.73 | 47.99 | 46.86 ± 1.59 | 46.84 | 48.90 | 47.87 ± 1.46 | |
| HoG [50] | AdaBoostM2 | 40.28 | 38.41 | 39.35 ± 1.33 | 33.04 | 32.68 | 32.86 ± 0.25 | 40.28 | 38.41 | 39.35 ± 1.33 | 28.00 | 28.44 | 28.22 ± 0.31 |
| Multi-SVM | 47.96 | 51.73 | 49.84 ± 2.67 | 51.95 | 55.66 | 53.80 ± 2.63 | 68.13 | 66.64 | 67.39 ± 1.05 | 41.97 | 47.79 | 44.88 ± 4.11 | |
| RF | 60.10 | 62.72 | 61.41 ± 1.85 | 61.73 | 64.66 | 63.19 ± 2.07 | 68.03 | 69.29 | 68.66 ± 0.89 | 56.49 | 60.61 | 58.55 ± 2.91 | |
| KNN | 50.14 | 56.26 | 53.20 ± 4.33 | 52.22 | 57.13 | 54.68 ± 3.47 | 57.37 | 59.45 | 58.41 ± 1.47 | 47.93 | 54.98 | 51.45 ± 4.99 | |
| MLBP [51] | AdaBoostM2 | 46.42 | 41.62 | 44.02 ± 3.40 | 40.04 | 34.85 | 37.45 ± 3.67 | 46.42 | 41.62 | 44.02 ± 3.40 | 35.20 | 29.98 | 32.59 ± 3.69 |
| Multi-SVM | 56.18 | 54.76 | 55.47 ± 1.00 | 53.72 | 52.49 | 53.10 ± 0.87 | 55.70 | 53.81 | 54.75 ± 1.33 | 51.87 | 51.23 | 51.55 ± 0.45 | |
| RF | 61.56 | 61.24 | 61.40 ± 0.22 | 56.98 | 58.16 | 57.57 ± 0.84 | 58.41 | 59.75 | 59.08 ± 0.95 | 55.62 | 56.65 | 56.13 ± 0.73 | |
| KNN | 54.38 | 56.43 | 55.40 ± 1.45 | 50.90 | 53.49 | 52.20 ± 1.83 | 50.92 | 53.21 | 52.06 ± 1.61 | 50.88 | 53.78 | 52.33 ± 2.05 | |
| Proposed | 92.10 | 93.03 | 92.57 ± 0.66 | 93.49 | 93.33 | 93.41 ± 0.12 | 94.32 | 94.84 | 94.58 ± 0.37 | 92.68 | 91.87 | 92.28 ± 0.58 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Owais, M.; Arsalan, M.; Choi, J.; Mahmood, T.; Park, K.R. Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis. J. Clin. Med. 2019, 8, 986. https://doi.org/10.3390/jcm8070986
Owais M, Arsalan M, Choi J, Mahmood T, Park KR. Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis. Journal of Clinical Medicine. 2019; 8(7):986. https://doi.org/10.3390/jcm8070986
Chicago/Turabian StyleOwais, Muhammad, Muhammad Arsalan, Jiho Choi, Tahir Mahmood, and Kang Ryoung Park. 2019. "Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis" Journal of Clinical Medicine 8, no. 7: 986. https://doi.org/10.3390/jcm8070986
APA StyleOwais, M., Arsalan, M., Choi, J., Mahmood, T., & Park, K. R. (2019). Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis. Journal of Clinical Medicine, 8(7), 986. https://doi.org/10.3390/jcm8070986






