Design and Synthesis of Novel Aminoindazole-pyrrolo[2,3-b]pyridine Inhibitors of IKKα That Selectively Perturb Cellular Non-Canonical NF-κB Signalling
Abstract
1. Introduction
2. Results and Discussion
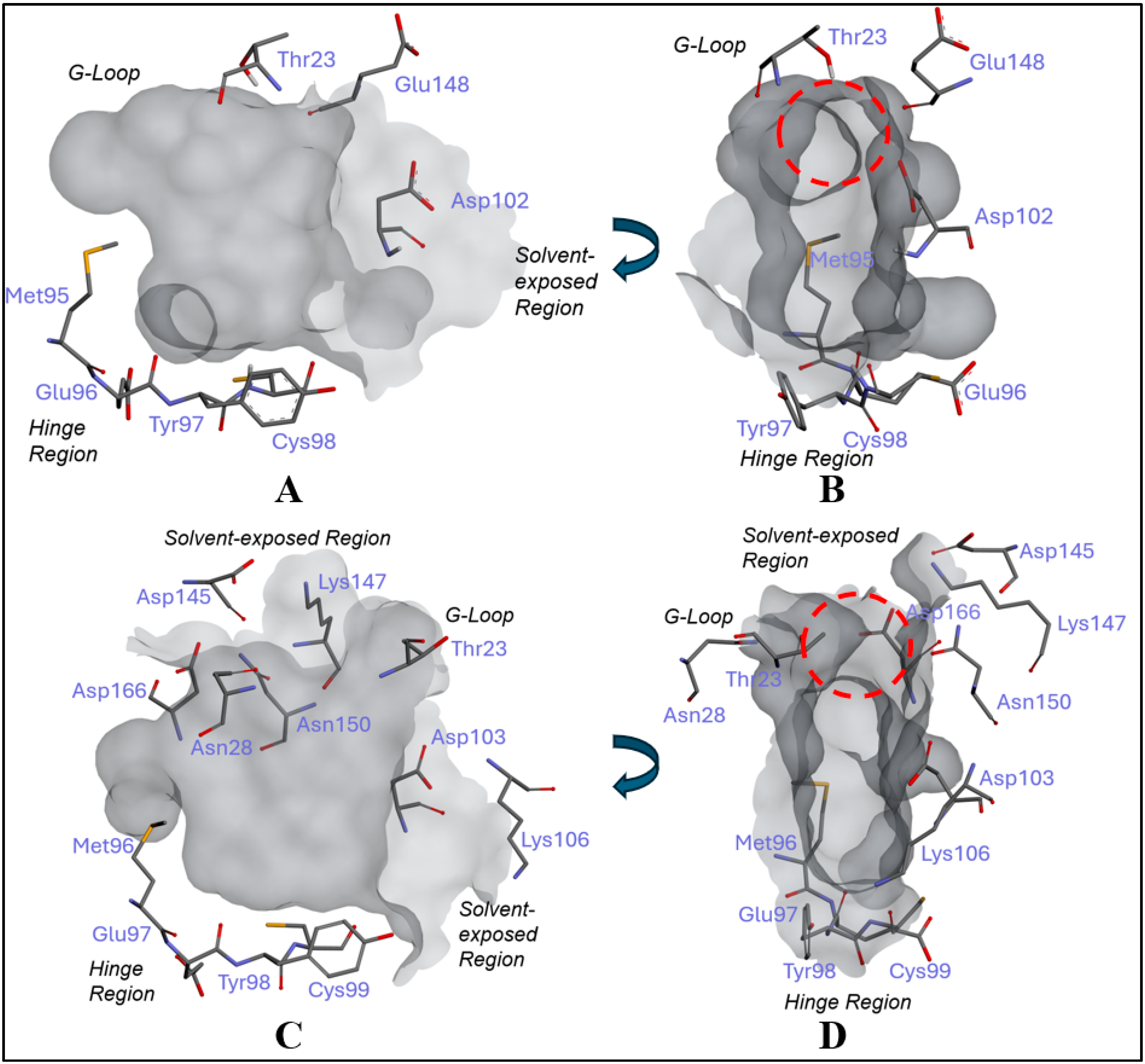
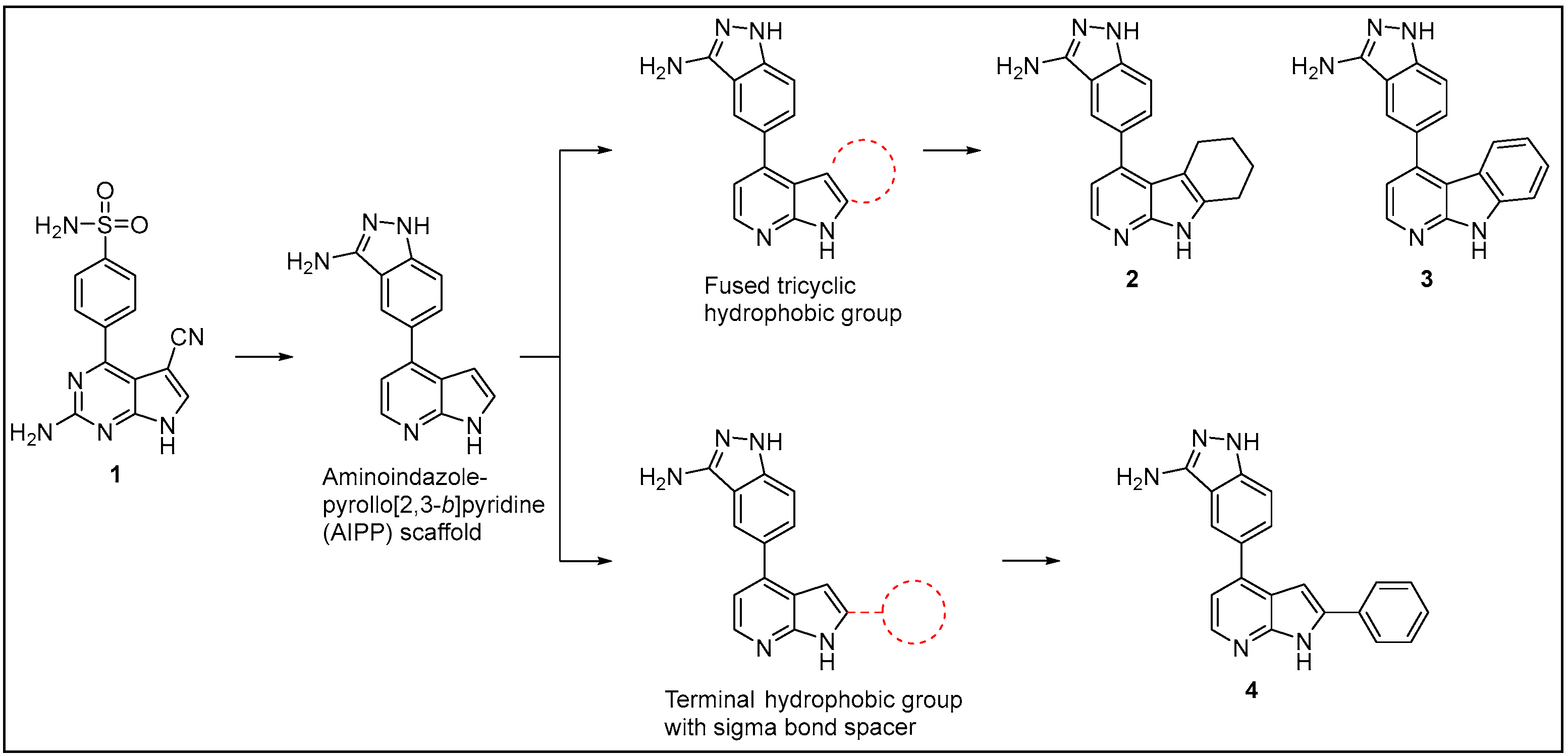
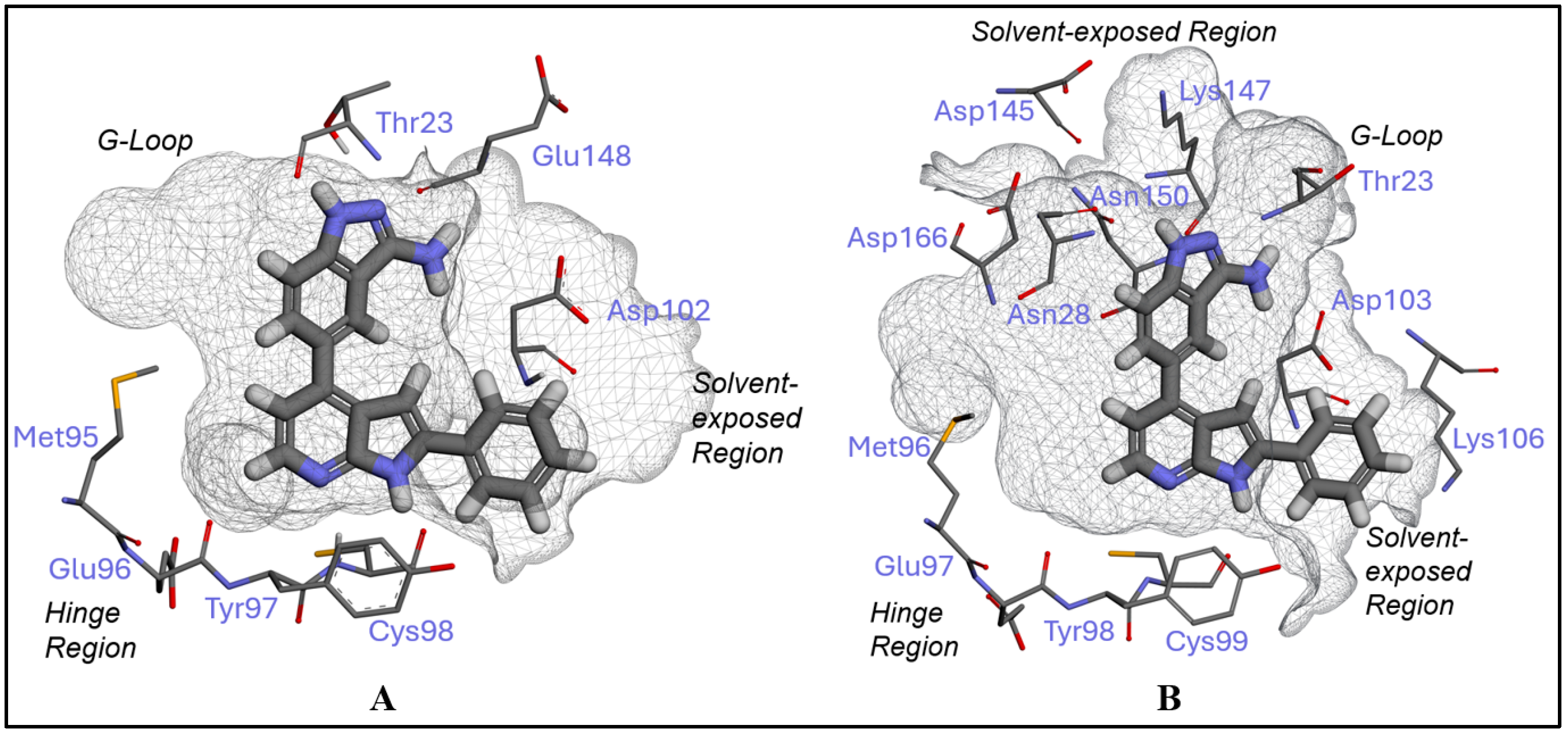
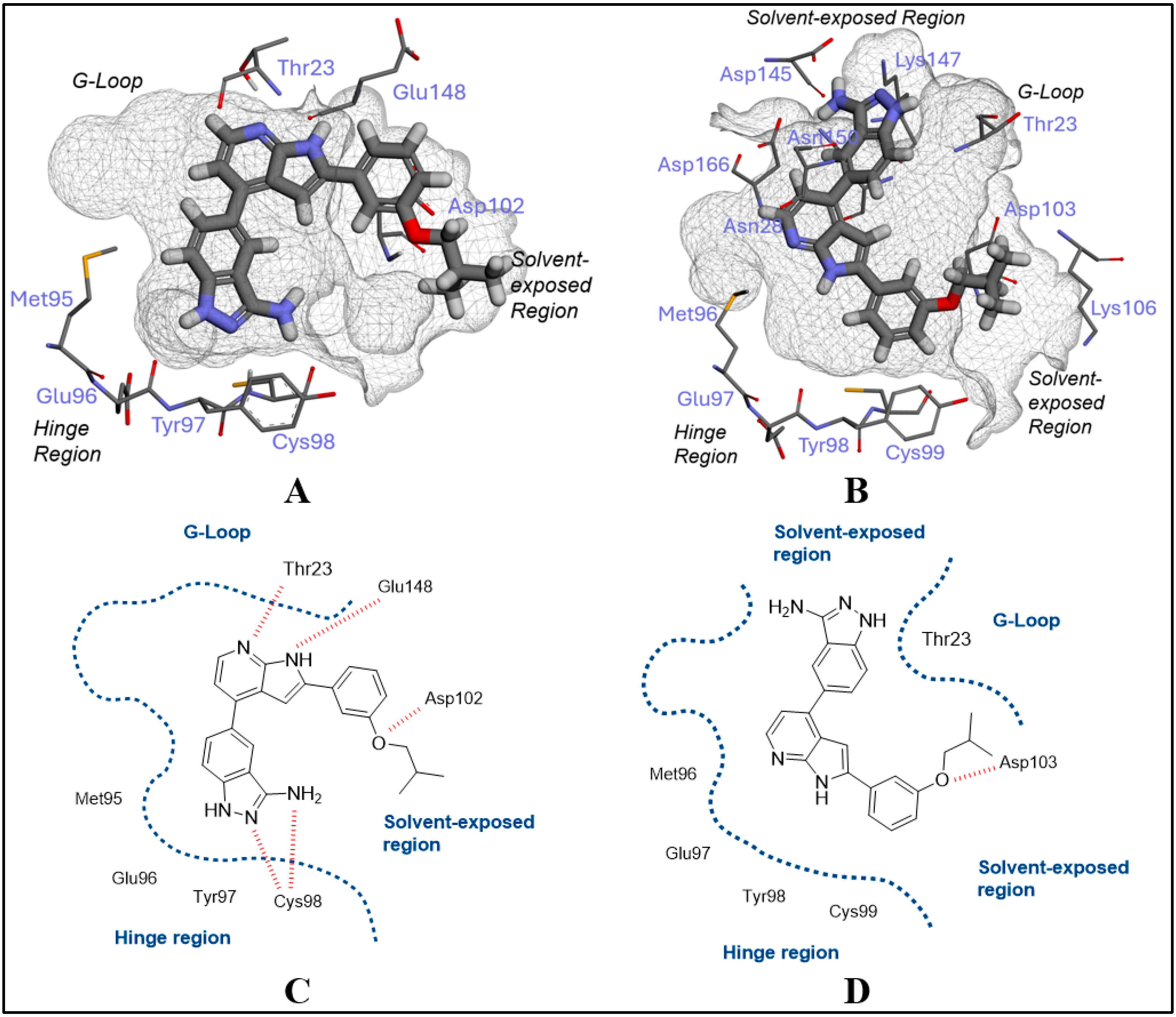
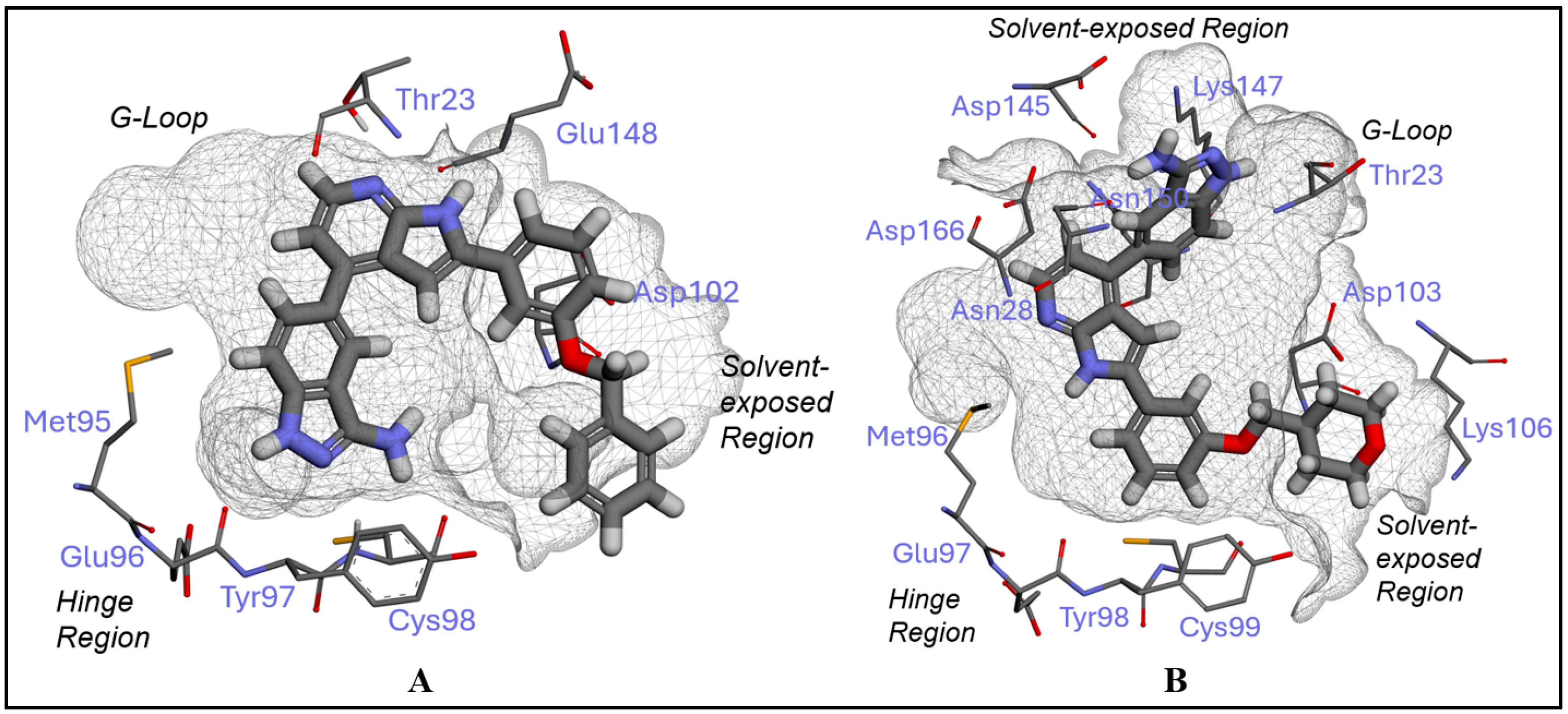
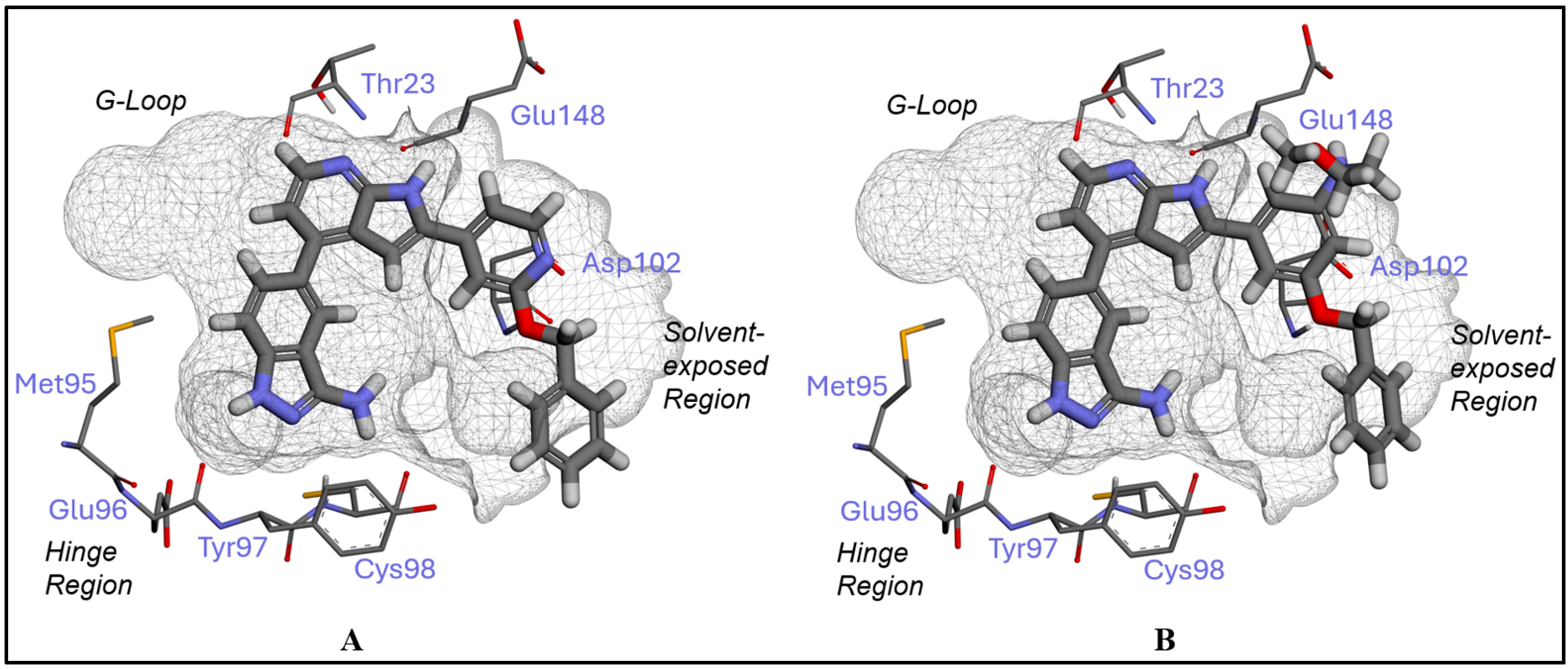
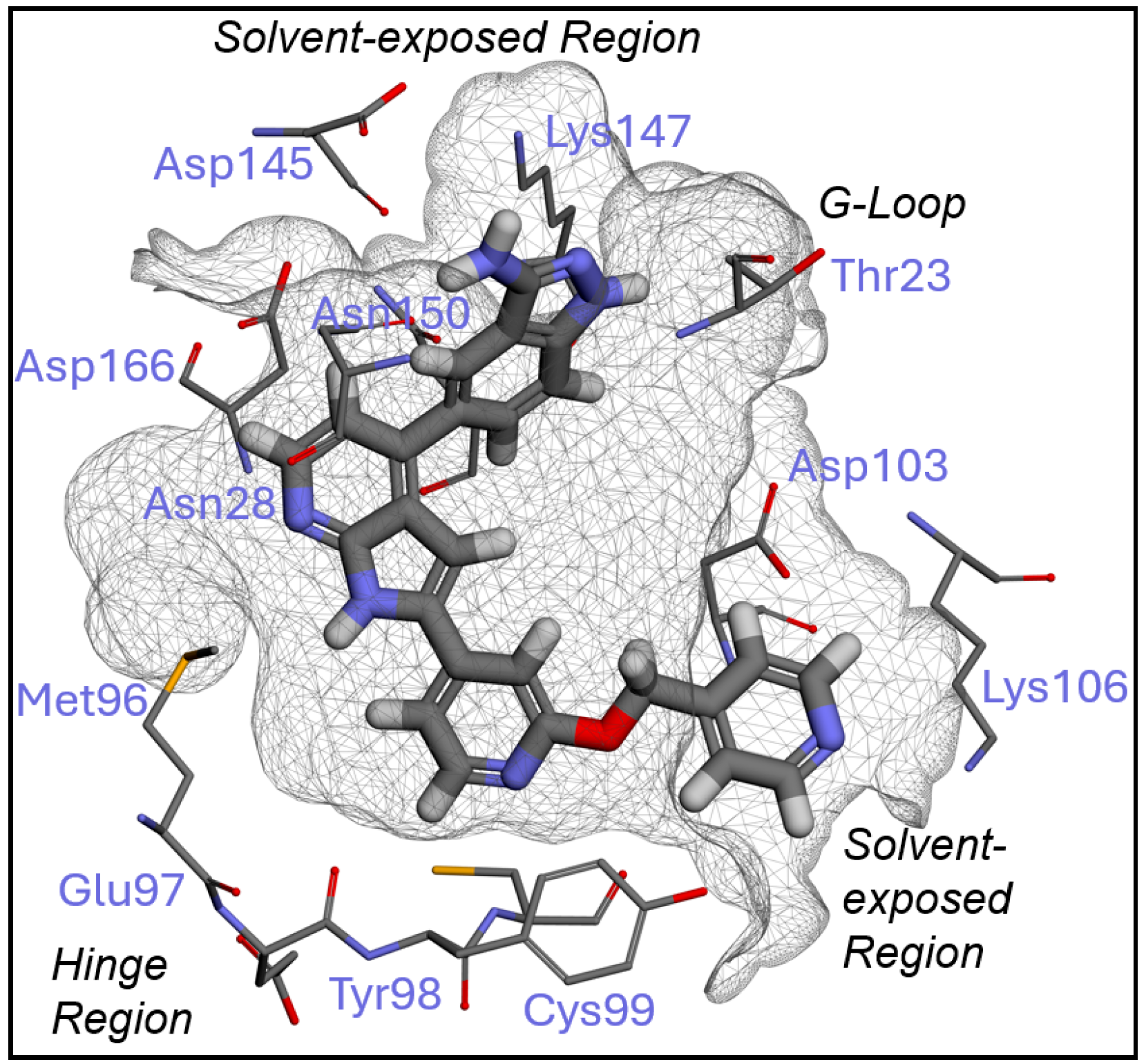
2.1. Inhibitor Design and Structure–Activity Relationship
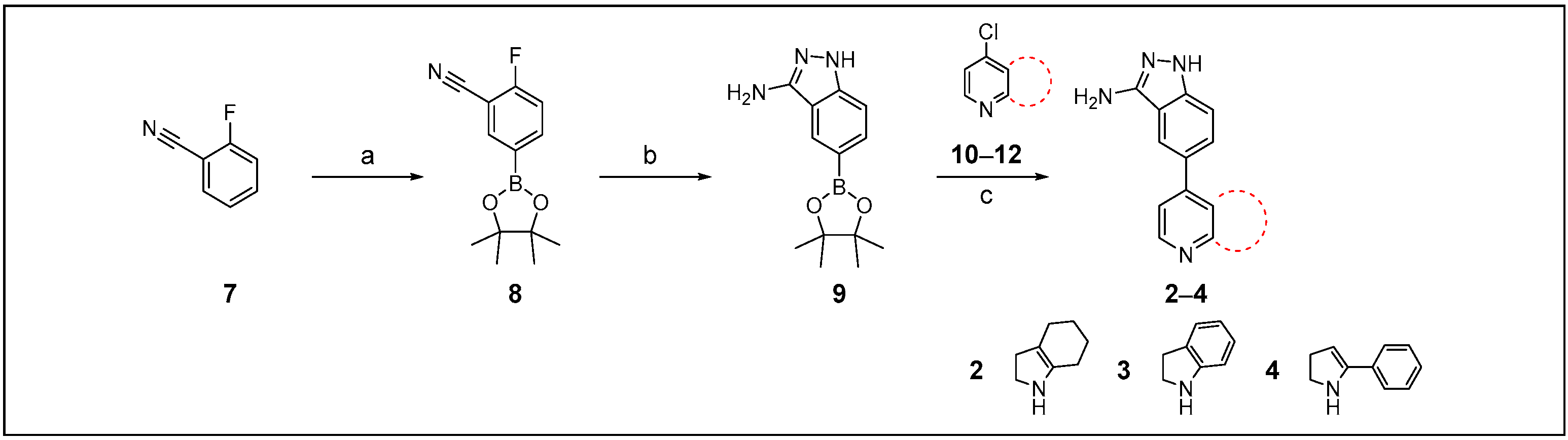
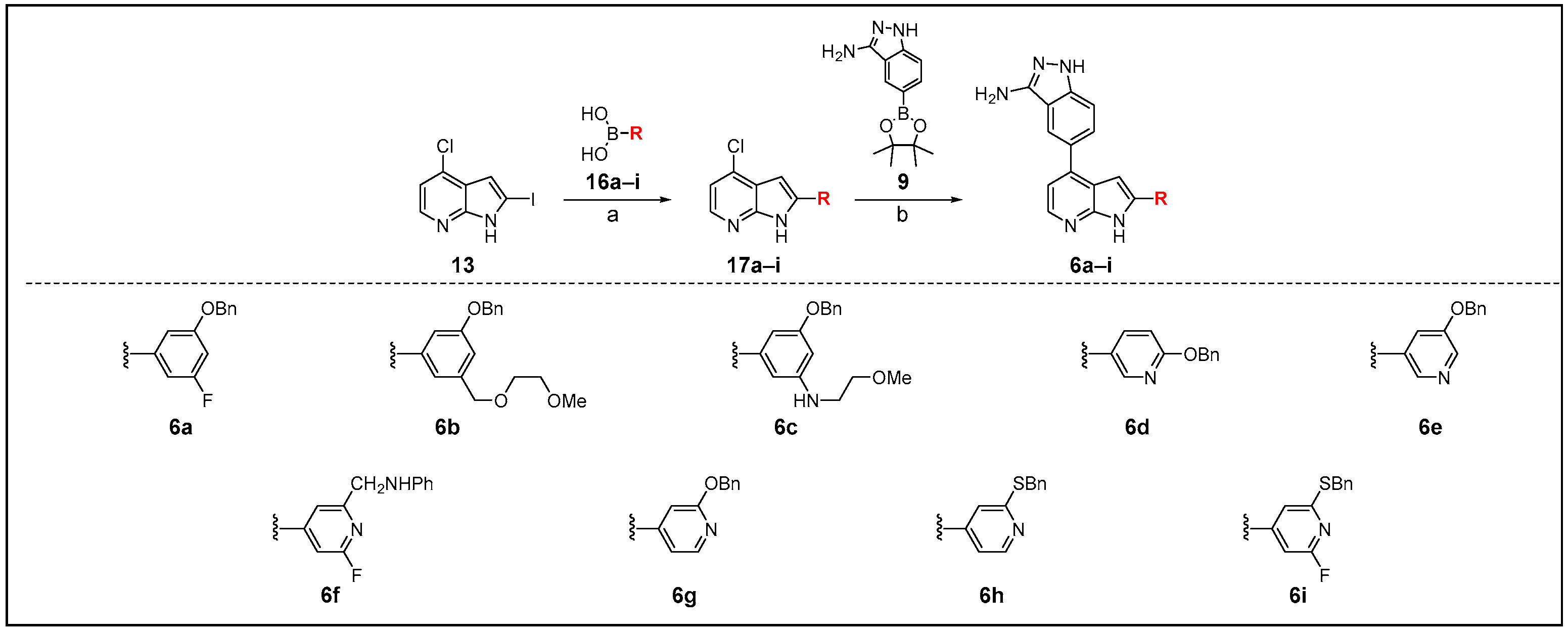
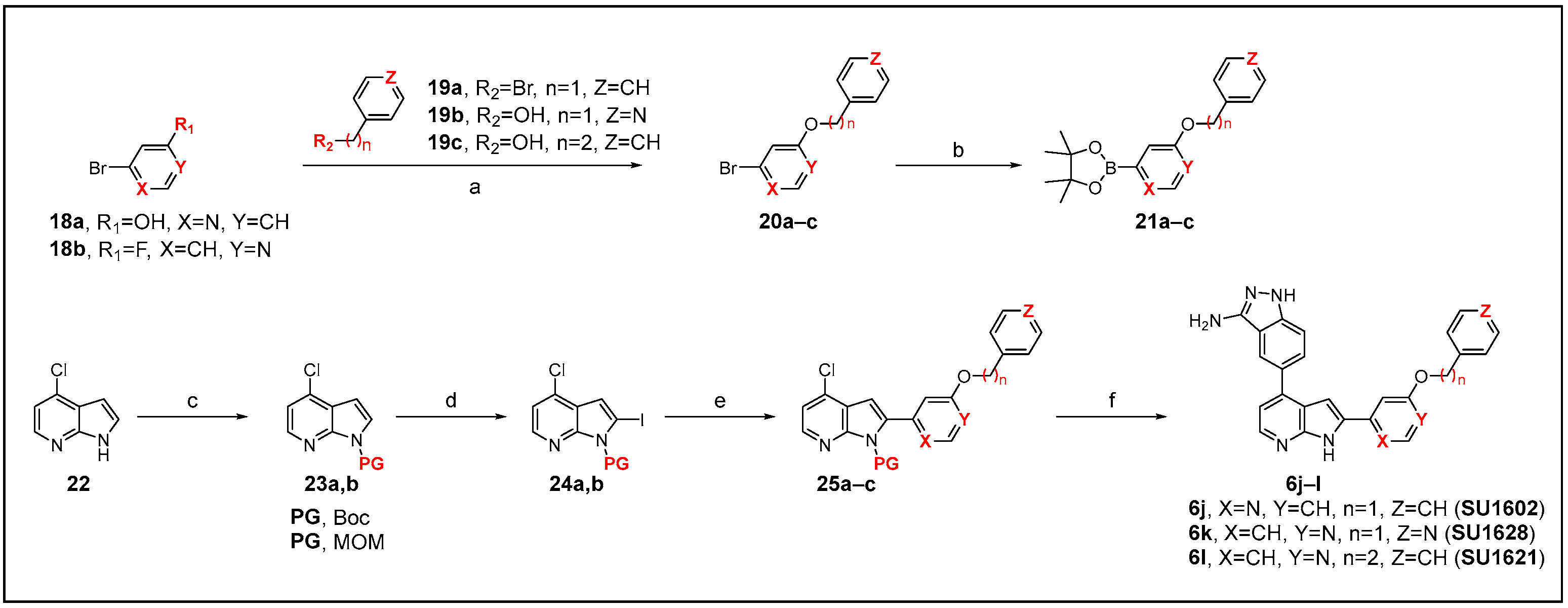
2.2. Chemistry
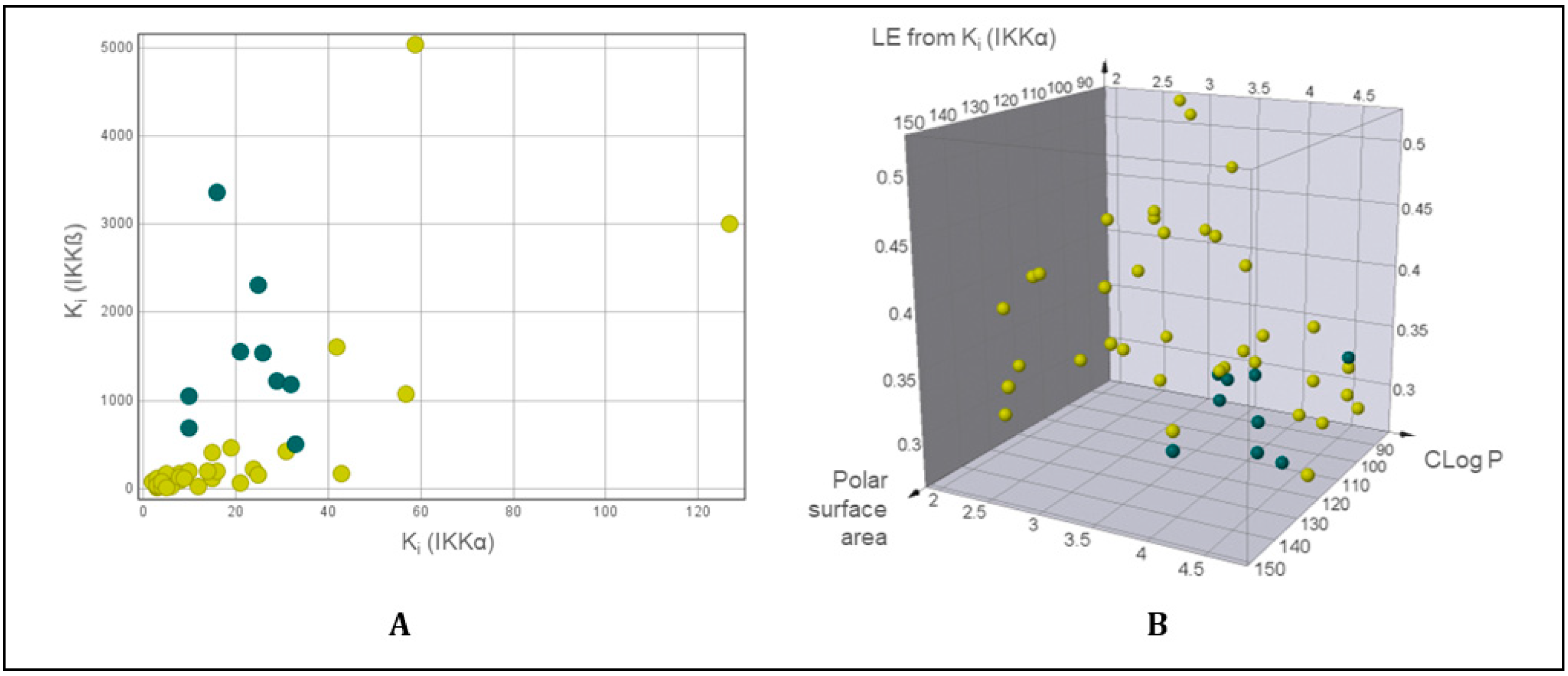
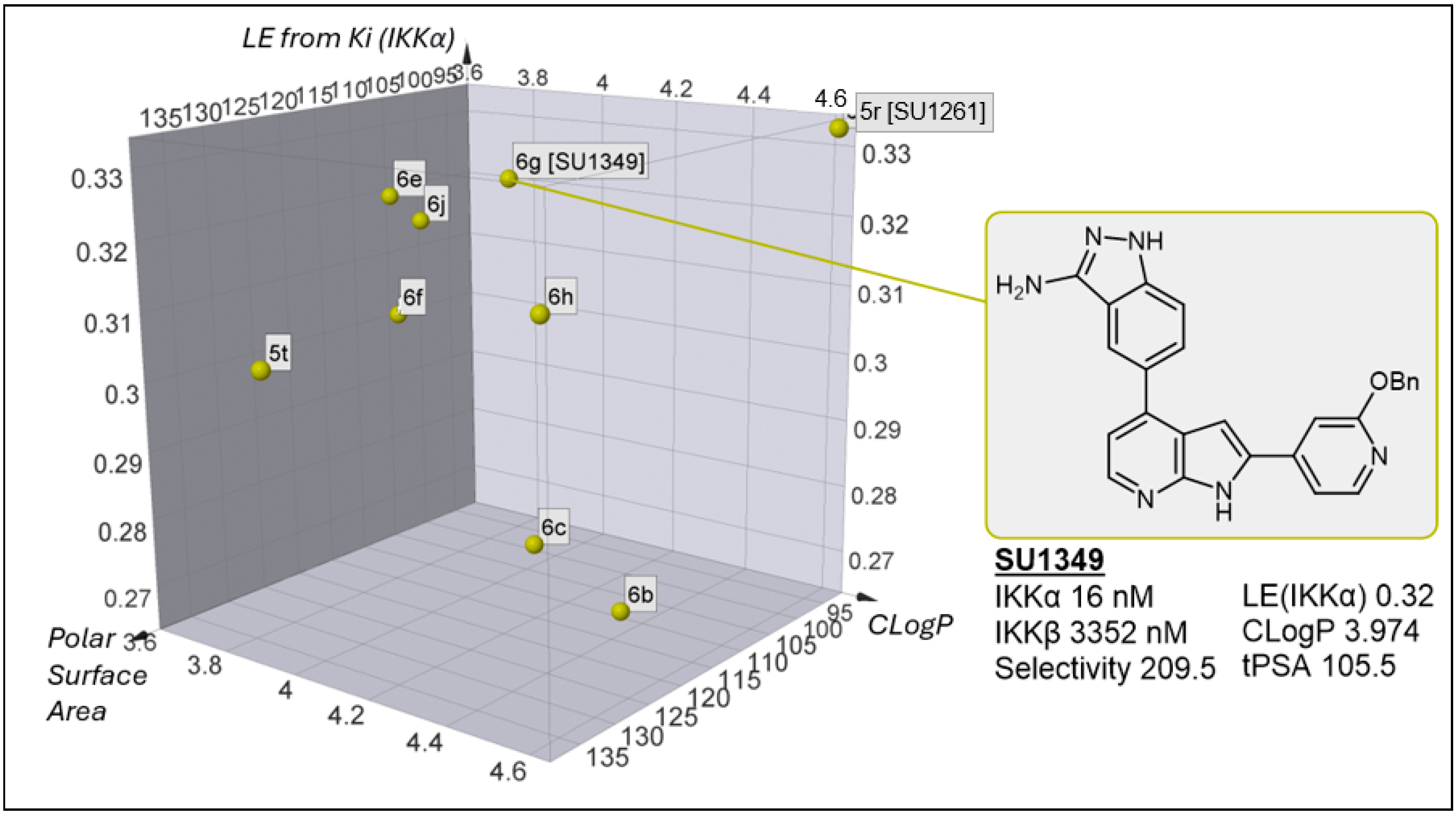
2.3. Physicochemical/DMPK Analysis
2.4. Kinome Profiling
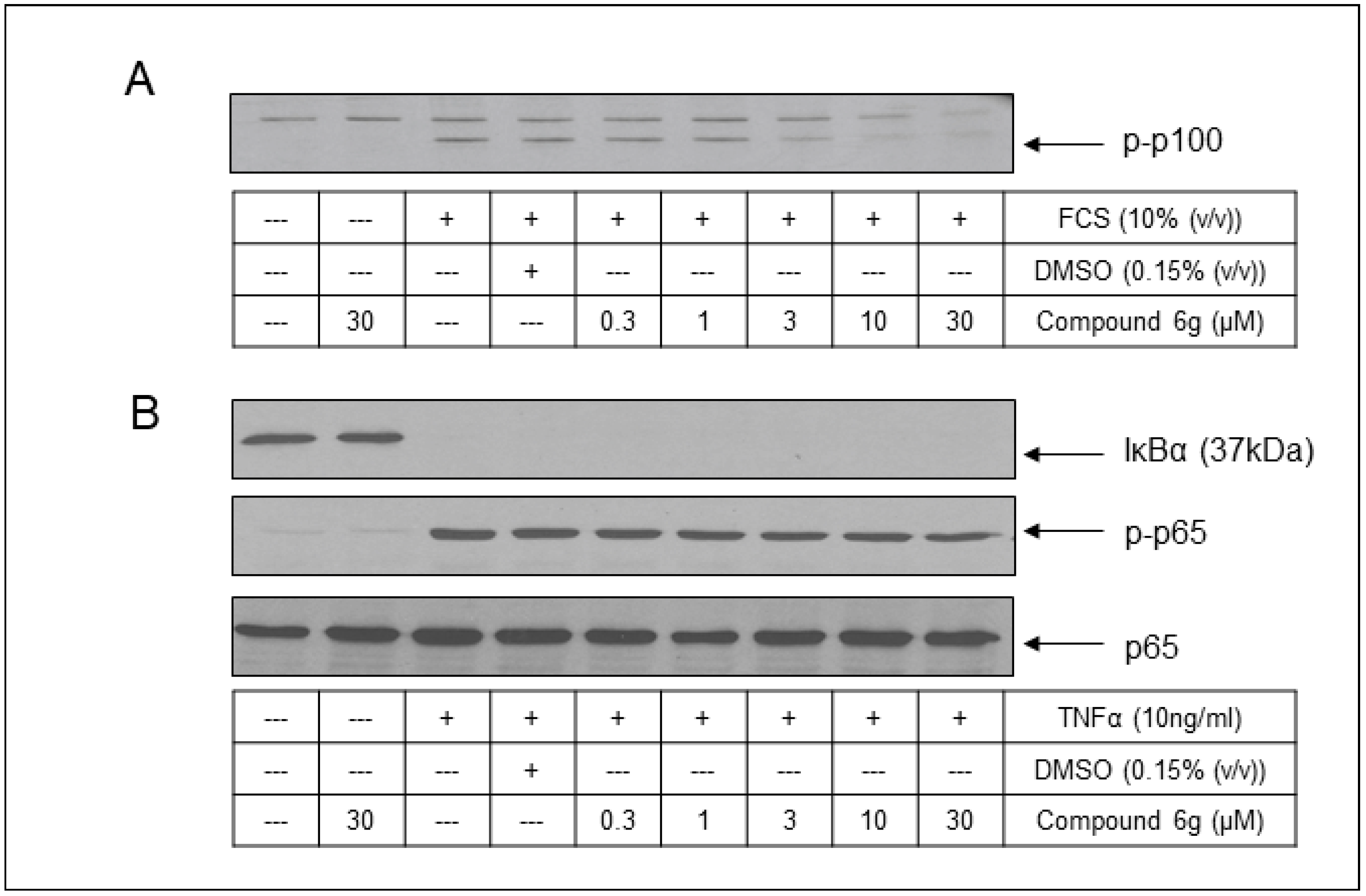
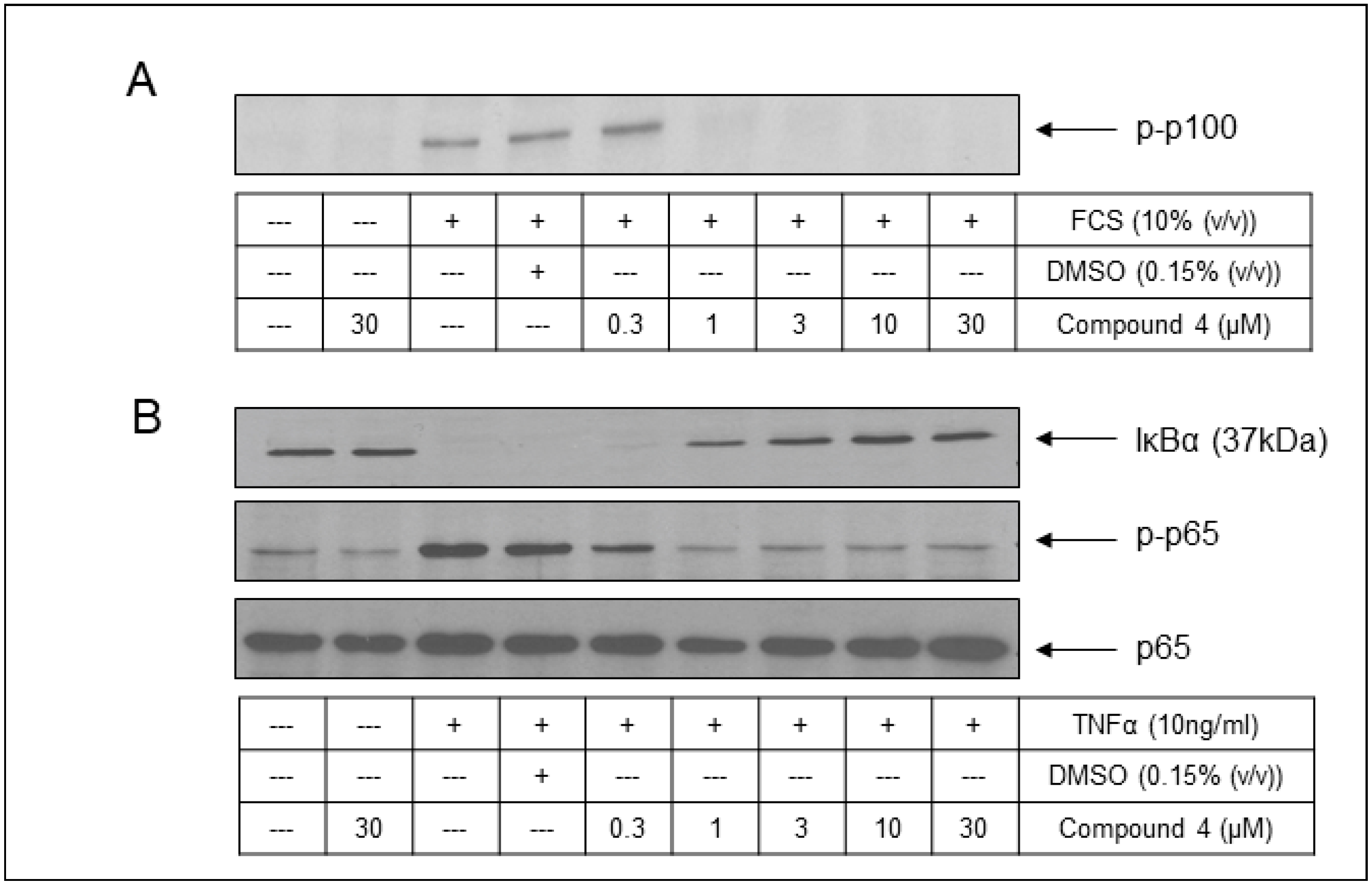
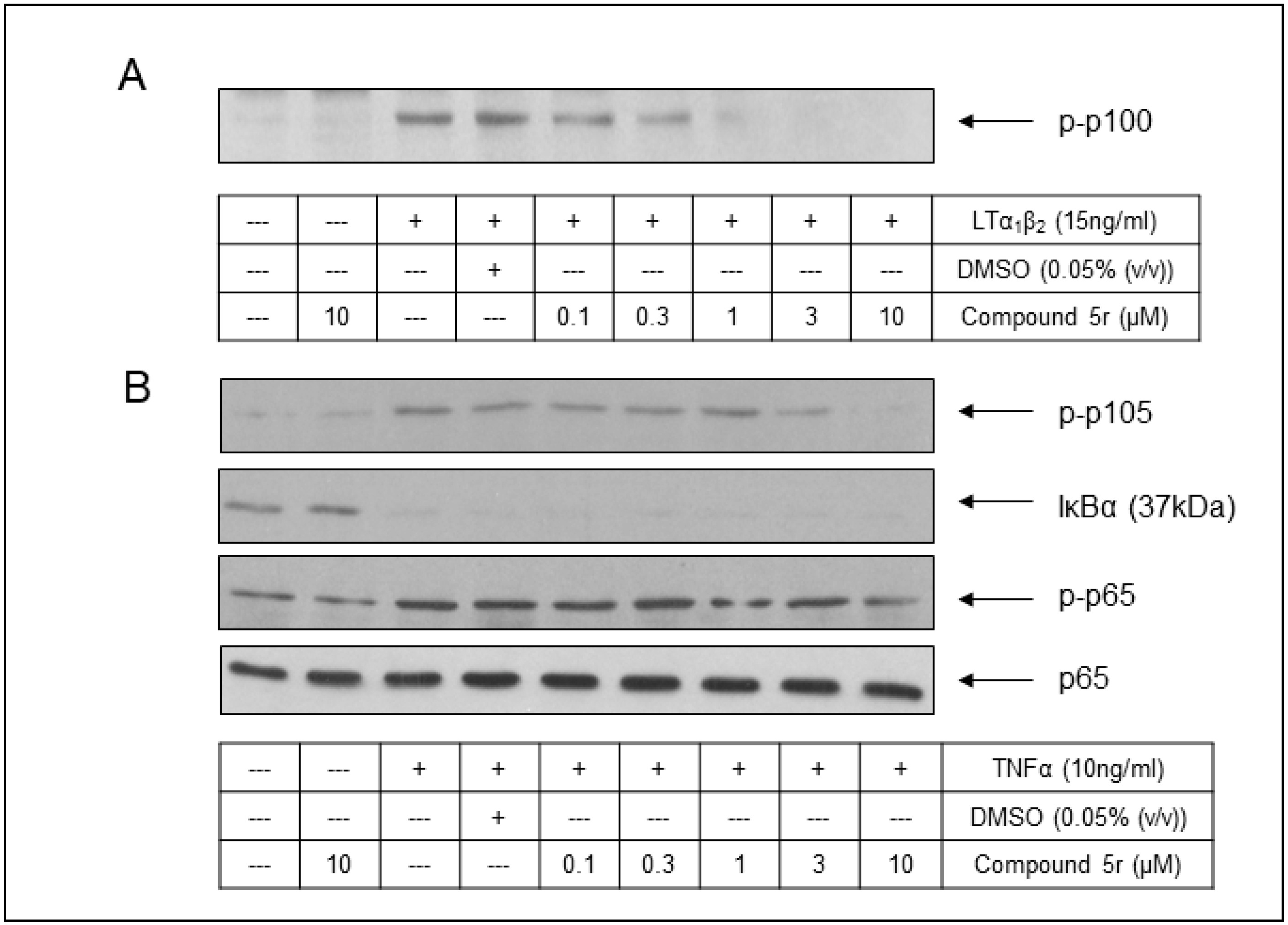
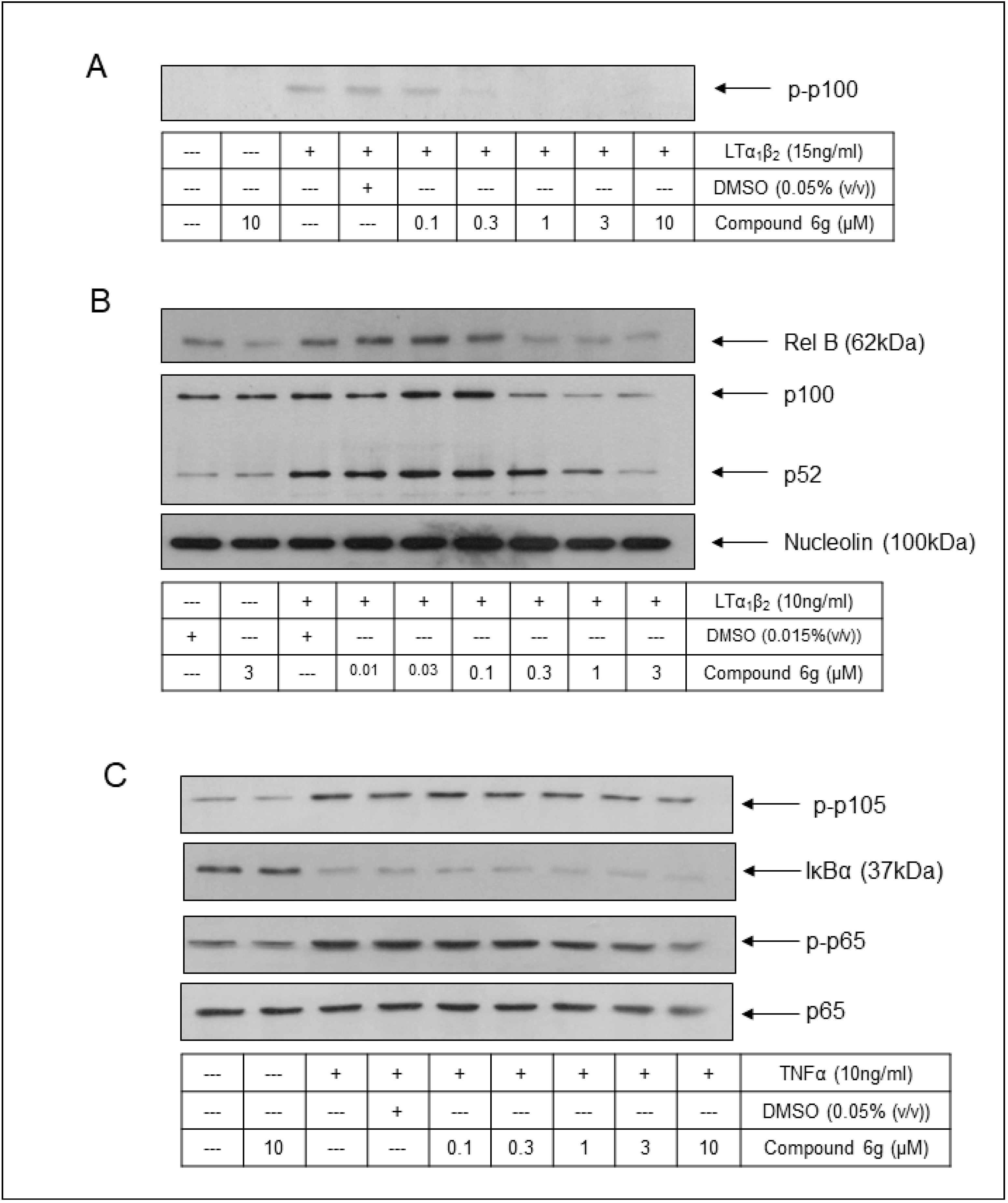
2.5. Cell-Based Assessment
2.6. Conclusions
3. Materials and Methods
3.1. Docking
3.2. Chemistry
3.3. Assessment of Kinase Activity of IKKα and IKKβ In Vitro
3.4. In Vitro DMPK Studies
3.5. Cell-Based Studies
3.5.1. Materials
3.5.2. Methodology
Cell Culture
Treatment of Cells, Preparation of Samples, and Western Blotting
Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karin, M. Nuclear Factor-ΚB in Cancer Development and Progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Ghosh, S. NF-Kappa B, an Evolutionarily Conserved Mediator of Immune and Inflammatory Responses. In Mechanisms of Lymphocyte Activation and Immune Regulation X: Innate Immunity; Gupta, S., Paul, W., Steinman, R., Eds.; Springer: Greer, SC, USA, 2005; Volume 560, pp. 41–45. [Google Scholar]
- Prescott, J.A.; Cook, S.J. Targeting IKKβ in Cancer: Challenges and Opportunities for the Therapeutic Utilisation of IKKβ Inhibitors. Cells 2018, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Edwards, J.; Pepper, C.; Mackay, S. Inhibitory-ΚB Kinase (IKK) α and Nuclear Factor-ΚB (NFκB)-Inducing Kinase (NIK) as Anti-Cancer Drug Targets. Cells 2018, 7, 176. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Karin, M. NF-ΚB, Inflammation, Immunity and Cancer: Coming of Age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.; Vargas, J.; Hoffmann, A. Signaling via the NFκB System. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C. The Non-Canonical NF-ΚB Pathway in Immunity and Inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-ΚB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef] [PubMed]
- Häcker, H.; Karin, M. Regulation and Function of IKK and IKK-Related Kinases. Sci. STKE 2006, 2006. [Google Scholar] [CrossRef]
- Xiao, G.; Rabson, A.B.; Young, W.; Qing, G.; Qu, Z. Alternative Pathways of NF-ΚB Activation: A Double-Edged Sword in Health and Disease. Cytokine Growth Factor Rev. 2006, 17, 281–293. [Google Scholar] [CrossRef]
- Sizemore, N.; Lerner, N.; Dombrowski, N.; Sakurai, H.; Stark, G.R. Distinct Roles of the IκB Kinase α and β Subunits in Liberating Nuclear Factor ΚB (NF-ΚB) from IκB and in Phosphorylating the P65 Subunit of NF-ΚB. J. Biol. Chem. 2002, 277, 3863–3869. [Google Scholar] [CrossRef]
- Karin, M.; Greten, F.R. NF-ΚB: Linking Inflammation and Immunity to Cancer Development and Progression. Nat. Rev. Immunol. 2005, 5, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Baud, V.; Oga, T.; Kim, K.I.; Yoshida, K.; Karin, M. IKKα Controls Formation of the Epidermis Independently of NF-ΚB. Nature 2001, 410, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Park, K.J.; Krishnan, V.; O’Malley, B.W.; Yamamoto, Y.; Gaynor, R.B. Formation of an IKKα-Dependent Transcription Complex Is Required for Estrogen Receptor-Mediated Gene Activation. Mol. Cell 2005, 18, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Nong, Y.; Morgan, J.G.; Gangurde, P.; Bielecki, A.; DaSilva, J.; Keaveney, M.; Cheng, H.; Fraser, C.; Schopf, L.; et al. A Selective Small Molecule IκB Kinase β Inhibitor Blocks Nuclear Factor ΚB-Mediated Inflammatory Responses in Human Fibroblast-Like Synoviocytes, Chondrocytes, and Mast Cells. J. Pharmacol. Exp. Ther. 2006, 317, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.R.; Pattoli, M.A.; Gregor, K.R.; Brassil, P.J.; MacMaster, J.F.; McIntyre, K.W.; Yang, X.; Iotzova, V.S.; Clarke, W.; Strnad, J.; et al. BMS-345541 Is a Highly Selective Inhibitor of IκB Kinase That Binds at an Allosteric Site of the Enzyme and Blocks NF-ΚB-Dependent Transcription in Mice. J. Biol. Chem. 2003, 278, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Lee, J.Y.; Seo, J.H.; Lee, H.Y.; Ryu, S.Y.; Ahn, B.W.; Lee, C.K.; Hwang, B.Y.; Han, S.B.; Kim, Y. Artemisolide Is a Typical Inhibitor of IκB Kinase β Targeting Cysteine-179 Residue and down-Regulates NF-ΚB-Dependent TNF-α Expression in LPS-Activated Macrophages. Biochem. Biophys. Res. Commun. 2007, 361, 593–598. [Google Scholar] [CrossRef]
- Podolin, P.L.; Callahan, J.F.; Bolognese, B.J.; Li, Y.H.; Carlson, K.; Davis, T.G.; Mellor, G.W.; Evans, C.; Roshak, A.K. Attenuation of Murine Collagen-Induced Arthritis by a Novel, Potent, Selective Small Molecule Inhibitor of IκB Kinase 2, TPCA-1 (2-[(Aminocarbonyl)Amino]-5-(4-Fluorophenyl)-3-Thiophenecarboxamide), Occurs via Reduction of Proinflammatory Cytokines and Antigen-Induced T Cell Proliferation. J. Pharmacol. Exp. Ther. 2005, 312, 373–381. [Google Scholar] [CrossRef]
- Strnad, J.; Burke, J.R. IκB Kinase Inhibitors for Treating Autoimmune and Inflammatory Disorders: Potential and Challenges. Trends Pharmacol. Sci. 2007, 28, 142–148. [Google Scholar] [CrossRef]
- Gamble, C.; McIntosh, K.; Scott, R.; Ho, K.H.; Plevin, R.; Paul, A. Inhibitory Kappa B Kinases as Targets for Pharmacological Regulation. Br. J. Pharmacol. 2012, 165, 802–819. [Google Scholar] [CrossRef]
- Pasparakis, M. Regulation of Tissue Homeostasis by NF-ΚB Signalling: Implications for Inflammatory Diseases. Nat. Rev. Immunol. 2009, 9, 778–788. [Google Scholar] [CrossRef]
- Li, Q.; Van Antwerp, D.; Mercurio, F.; Lee, K.-F.; Verma, I.M. Severe Liver Degeneration in Mice Lacking the IκB Kinase 2 Gene. Science 1999, 284, 321–325. [Google Scholar] [CrossRef]
- Cao, Y.; Bonizzi, G.; Seagroves, T.N.; Greten, F.R.; Johnson, R.; Schmidt, E.V.; Karin, M. IKKα Provides an Essential Link between RANK Signaling and Cyclin D1 Expression during Mammary Gland Development. Cell 2001, 107, 763–775. [Google Scholar] [CrossRef]
- Anthony, N.G.; Baiget, J.; Berretta, G.; Boyd, M.; Breen, D.; Edwards, J.; Gamble, C.; Gray, A.I.; Harvey, A.L.; Hatziieremia, S.; et al. Inhibitory Kappa B Kinase α (IKKα) Inhibitors That Recapitulate Their Selectivity in Cells against Isoform-Related Biomarkers. J. Med. Chem. 2017, 60, 7043–7066. [Google Scholar] [CrossRef]
- Llona-Minguez, S.; Baiget, J.; Mackay, S.P. Small-Molecule Inhibitors of IκB Kinase (IKK) and IKK-Related Kinases. Pharm. Pat. Anal. 2013, 2, 481–498. [Google Scholar] [CrossRef]
- Asamitsu, K.; Yamaguchi, T.; Nakata, K.; Hibi, Y.; Victoriano, A.F.B.; Imai, K.; Onozaki, K.; Kitade, Y.; Okamoto, T. Inhibition of Human Immunodeficiency Virus Type 1 Replication by Blocking IκB Kinase with Noraristeromycin. J. Biochem. 2008, 144, 581–589. [Google Scholar] [CrossRef]
- Shukla, S.; Shankar, E.; Fu, P.; MacLennan, G.T.; Gupta, S. Suppression of NF-ΚB and NF-ΚB-Regulated Gene Expression by Apigenin through IκBα and IKK Pathway in TRAMP Mice. PLoS ONE 2015, 10, e0138710. [Google Scholar] [CrossRef]
- Shukla, S.; Kanwal, R.; Shankar, E.; Datt, M.; Chance, M.R.; Fu, P.; MacLennan, G.T.; Gupta, S.; Shukla, S.; Kanwal, R.; et al. Apigenin Blocks IKKα Activation and Suppresses Prostate Cancer Progression. Oncotarget 2015, 6, 31216–31232. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Zhu, H.Y.; Zhang, X.H.; Du, Z.Y.; Xu, Y.J.; Yu, X.D. Apigenin Inhibits Proliferation and Induces Apoptosis in Human Multiple Myeloma Cells through Targeting the Trinity of CK2, Cdc37 and Hsp90. Mol. Cancer 2011, 10, 104. [Google Scholar] [CrossRef]
- Shukla, S.; Bhaskaran, N.; Babcook, M.A.; Fu, P.; MacLennan, G.T.; Gupta, S. Apigenin Inhibits Prostate Cancer Progression in TRAMP Mice via Targeting PI3K/Akt/FoxO Pathway. Carcinogenesis 2014, 35, 452–460. [Google Scholar] [CrossRef]
- Palayoor, S.T.; Youmell, M.Y.; Calderwood, S.K.; Coleman, C.N.; Price, B.D. Constitutive Activation of IκB Kinase α and NF-ΚB in Prostate Cancer Cells Is Inhibited by Ibuprofen. Oncogene 1999, 18, 7389–7394. [Google Scholar] [CrossRef]
- Ammirante, M.; Luo, J.L.; Grivennikov, S.; Nedospasov, S.; Karin, M. B-Cell-Derived Lymphotoxin Promotes Castration-Resistant Prostate Cancer. Nature 2010, 464, 302–305. [Google Scholar] [CrossRef]
- Luo, J.L.; Tan, W.; Ricono, J.M.; Korchynskyi, O.; Zhang, M.; Gonias, S.L.; Cheresh, D.A.; Karin, M. Nuclear Cytokine-Activated IKKα Controls Prostate Cancer Metastasis by Repressing Maspin. Nature 2007, 446, 690–694. [Google Scholar] [CrossRef]
- Margalef, P.; Fernández-Majada, V.; Villanueva, A.; Garcia-Carbonell, R.; Iglesias, M.; López, L.; Martínez-Iniesta, M.; Villà-Freixa, J.; Mulero, M.C.; Andreu, M.; et al. A Truncated Form of IKKα Is Responsible for Specific Nuclear IKK Activity in Colorectal Cancer. Cell Rep. 2012, 2, 840–854. [Google Scholar] [CrossRef]
- Margalef, P.; Colomer, C.; Villanueva, A.; Montagut, C.; Iglesias, M.; Bellosillo, B.; Salazar, R.; Martínez-Iniesta, M.; Bigas, A.; Espinosa, L. BRAF-Induced Tumorigenesis Is IKKα-Dependent but NF-ΚB-Independent. Sci. Signal 2015, 8. [Google Scholar] [CrossRef]
- Hao, L.; Rizzo, P.; Osipo, C.; Pannuti, A.; Wyatt, D.; Cheung, L.W.K.; Sonenshein, G.; Osborne, B.A.; Miele, L. Notch-1 Activates Estrogen Receptor-α-Dependent Transcription via IKKα in Breast Cancer Cells. Oncogene 2009, 29, 201–213. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, J.L.; Karin, M. IκB Kinase α Kinase Activity Is Required for Self-Renewal of ErbB2/Her2-Transformed Mammary Tumor-Initiating Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 15852. [Google Scholar] [CrossRef]
- Liou, G.Y.; Döppler, H.; Necela, B.; Krishna, M.; Crawford, H.C.; Raimondo, M.; Storz, P. Macrophage-Secreted Cytokines Drive Pancreatic Acinar-to-Ductal Metaplasia through NF-ΚB and MMPs. J. Cell Biol. 2013, 202, 563. [Google Scholar] [CrossRef]
- Storz, P. Targeting the Alternative NF-ΚB Pathway in Pancreatic Cancer: A New Direction for Therapy? Expert. Rev. Anticancer Ther. 2013, 13, 501–504. [Google Scholar] [CrossRef][Green Version]
- Döppler, H.; Liou, G.Y.; Storz, P. Downregulation of TRAF2 Mediates NIK-Induced Pancreatic Cancer Cell Proliferation and Tumorigenicity. PLoS ONE 2013, 8, e53676. [Google Scholar] [CrossRef]
- Cohen, M.S.; Zhang, C.; Shokat, K.M.; Taunton, J. Biochemistry: Structural Bioinformatics-Based Design of Selective, Irreversible Kinase Inhibitors. Science 2005, 308, 1318–1321. [Google Scholar] [CrossRef]
- Liu, S.; Misquitta, Y.R.; Olland, A.; Johnson, M.A.; Kelleher, K.S.; Kriz, R.; Lin, L.L.; Stahl, M.; Mosyak, L. Crystal Structure of a Human IκB Kinase β Asymmetric Dimer. J. Biol. Chem. 2013, 288, 22758–22767. [Google Scholar] [CrossRef] [PubMed]
- Polley, S.; Passos, D.O.; Huang, D.B.; Mulero, M.C.; Mazumder, A.; Biswas, T.; Verma, I.M.; Lyumkis, D.; Ghosh, G. Structural Basis for the Activation of IKK1/α. Cell Rep. 2016, 17, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Christopher, J.; Jung, D.; Lackey, K. Lh-Indazole-3-Amine Compounds as Ikk1 Inhibitors. Patent No. WO2008132121A1, 11 June 2008. [Google Scholar]
- Ishiyama, T.; Takagi, J.; Hartwig, J.; Miyaura, N. A Stoichiometric Aromatic C—H Borylation Catalyzed by Iridium(I)/2,2′-Bipyridine Complexes at Room Temperature. Angew. Chem. Int. Ed. 2002, 41, 3056–3058. [Google Scholar] [CrossRef]
- Miyaura, N.; Yamada, K.; Suzuki, A. A New Stereospecific Cross-Coupling by the Palladium-Catalyzed Reaction of 1-Alkenylboranes with 1-Alkenyl or 1-Alkynyl Halides. Tetrahedron Lett. 1979, 20, 3437–3440. [Google Scholar] [CrossRef]
- D’Alterio, M.C.; Casals-Cruañas, È.; Tzouras, N.V.; Talarico, G.; Nolan, S.P.; Poater, A. Mechanistic Aspects of the Palladium-Catalyzed Suzuki-Miyaura Cross-Coupling Reaction. Chem.—A Eur. J. 2021, 27, 13481–13493. [Google Scholar] [CrossRef]
- Ishiyama, T.; Murata, M.; Miyaura, N. Palladium(0)-Catalyzed Cross-Coupling Reaction of Alkoxydiboron with Haloarenes: A Direct Procedure for Arylboronic Esters. J. Org. Chem. 1995, 60, 7508–7510. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program for Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Hopkins, A.L.; Groom, C.R.; Alex, A. Ligand Efficiency: A Useful Metric for Lead Selection. Drug Discov. Today 2004, 9, 430–431. [Google Scholar] [CrossRef]
- Barré, B.; Perkins, N.D. A Cell Cycle Regulatory Network Controlling NF-ΚB Subunit Activity and Function. EMBO J. 2007, 26, 4841–4855. [Google Scholar] [CrossRef]
- McIntosh, K.; Khalaf, Y.H.; Craig, R.; West, C.; McCulloch, A.; Waghmare, A.; Lawson, C.; Chan, E.Y.W.; Mackay, S.; Paul, A.; et al. IL-1β Stimulates a Novel, IKKα -Dependent, NIK -Independent Activation of Non-Canonical NFκB Signalling. Cell Signal 2023, 107, 110684. [Google Scholar] [CrossRef]
- Madge, L.A.; Kluger, M.S.; Orange, J.S.; May, M.J. Lymphotoxin-A1β2 and LIGHT Induce Classical and Noncanonical NF-ΚB-Dependent Proinflammatory Gene Expression in Vascular Endothelial Cells. J. Immunol. 2008, 180, 3467. [Google Scholar] [CrossRef]
- Lang, V.; Janzen, J.; Fischer, G.Z.; Soneji, Y.; Beinke, S.; Salmeron, A.; Allen, H.; Hay, R.T.; Ben-Neriah, Y.; Ley, S.C. ΒTrCP-Mediated Proteolysis of NF-ΚB1 P105 Requires Phosphorylation of P105 Serines 927 and 932. Mol. Cell Biol. 2003, 23, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes. Available online: https://www.3ds.com/ (accessed on 18 June 2024).
- RCSB PDB. Available online: https://www.rcsb.org/ (accessed on 18 June 2024).
- RCSB PDB-5EBZ: Crystal Structure of Human IKK1. Available online: https://www.rcsb.org/structure/5ebz (accessed on 18 June 2024).
- RCSB PDB-4KIK: Human IkB Kinase Beta. Available online: https://www.rcsb.org/structure/4KIK (accessed on 18 June 2024).
- Brooks, B.R.; Brooks, C.L.; Mackerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Robertson, D.H.; Brooks, C.L.; Vieth, M. Detailed Analysis of Grid-Based Molecular Docking: A Case Study of CDOCKER—A CHARMm-Based MD Docking Algorithm. J. Comput. Chem. 2003, 24, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, C.J.; Ritchie, E.; Paul, A.; Plevin, R. IKKα and IKKβ Function in TNFα-Stimulated Adhesion Molecule Expression in Human Aortic Smooth Muscle Cells. Cell Signal 2007, 19, 75–80. [Google Scholar] [CrossRef]
- Liu, L.; Paul, A.; MacKenzie, C.J.; Bryant, C.; Graham, A.; Plevin, R. Nuclear Factor Kappa B Is Involved in Lipopolysaccharide-Stimulated Induction of Interferon Regulatory Factor-1 and GAS/GAF DNA-Binding in Human Umbilical Vein Endothelial Cells. Br. J. Pharmacol. 2001, 134, 1629–1638. [Google Scholar] [CrossRef][Green Version]



| Compound | Ki Values (nM) | |
|---|---|---|
| IKKα | IKKβ | |
| 2 | 2 | 77 |
| 3 | 3 | 5 |
| 4 | 3 | 15 |
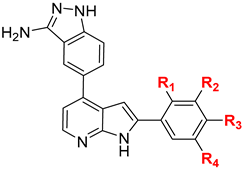 | ||||||
|---|---|---|---|---|---|---|
| Cmpd | R1 | R2 | R3 | R4 | Ki Values (nM) | |
| IKKα | IKKβ | |||||
| 5a | OH | H | H | H | 4 | 42 |
| 5b | OEt | H | H | H | 8 | 86 |
| 5c | H | NH2 | H | H | 5 | 28 |
| 5d | H | OH | H | H | 5 | 26 |
| 5e | H | OMe | H | H | 5 | 40 |
| 5f | H | OH | H | F | 4 | 42 |
| 5g | H | OMe | H | F | 3 | 32 |
| 5h | H | 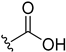 | H | H | 8 | 168 |
| 5i | H | 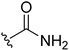 | H | H | 5 | 159 |
| 5j | H | 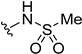 | H | H | 3 | 109 |
| 5k | H | 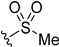 | H | H | 3 | 36 |
| 5l | H | 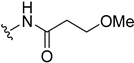 | H | H | 6 | 25 |
| 5m | H |  | H | H | 15 | 107 |
| 5n | H | 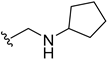 | H | H | 10 | 191 |
| 5o | H | 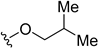 | H | H | 19 | 458 |
| 5p | H | 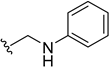 | H | H | 35 | 470 |
| 5q | OBn | H | H | H | 57 | 1064 |
| 5r [SU1261] | H | 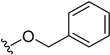 | H | H | 10 | 680 |
| 5s | H | H | OBn | H | 16 | 189 |
| 5t | H | NHTs | H | H | 10 | 1048 |
| 5u | H | H | NHTs | H | 4 | 67 |
| 5v | H | 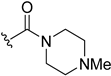 | H | H | 24 | 215 |
| 5w | H | 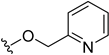 | H | H | 16 | 402 |
| 5x | H | 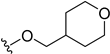 | H | H | 8 | 122 |
| 5y | H | 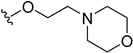 | H | H | 9 | 29 |
| 5z | H | 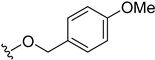 | H | H | 43 | 166 |
| 5aa | H | 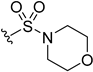 | H | H | 25 | 149 |
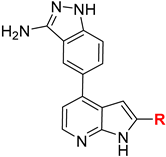 | |||
|---|---|---|---|
| Cmpd | R | Ki Values (nM) | |
| IKKα | IKKβ | ||
| 6a | 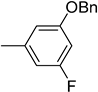 | 59 | 5029 |
| 6b | 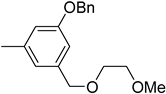 | 29 | 1215 |
| 6c | 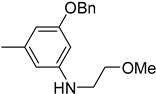 | 26 | 1532 |
| 6d | 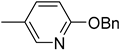 | 12 | 22 |
| 6e | 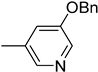 | 21 | 1550 |
| 6f | 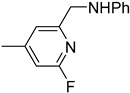 | 33 | 503 |
| 6g [SU1349] | 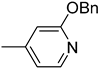 | 16 | 3352 |
| 6h | 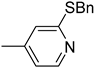 | 5 | 1180 |
| 6i | 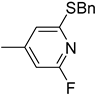 | 127 | 3000 |
| 6j | 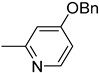 | 25 | 2300 |
| 6k | 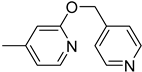 | 6 | 12 |
| 6l | 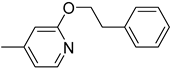 | 42 | 1600 |
| Cmpd | Solubility (μM) | Mouse Hepatocyte Clearance (μL/min/106 Cells) | Mouse Hepatocyte Half-Life (min) |
|---|---|---|---|
| 5r [SU1261] | <1 | 38 | 36 |
| 6b | 3.75 | N.D. | N.D. |
| 6c | 2 | N.D. | N.D. |
| 6e | 6.5 | N.D. | N.D. |
| 6f | 3.75 | N.D. | N.D. |
| 6g [SU1349] | 20 | 27 | 51 |
| 6h | 2 | N.D. | N.D. |
| 6j | 2 | N.D. | N.D. |
| 6k | >100 | 11 | 121 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riley, C.; Ammar, U.; Alsfouk, A.; Anthony, N.G.; Baiget, J.; Berretta, G.; Breen, D.; Huggan, J.; Lawson, C.; McIntosh, K.; et al. Design and Synthesis of Novel Aminoindazole-pyrrolo[2,3-b]pyridine Inhibitors of IKKα That Selectively Perturb Cellular Non-Canonical NF-κB Signalling. Molecules 2024, 29, 3515. https://doi.org/10.3390/molecules29153515
Riley C, Ammar U, Alsfouk A, Anthony NG, Baiget J, Berretta G, Breen D, Huggan J, Lawson C, McIntosh K, et al. Design and Synthesis of Novel Aminoindazole-pyrrolo[2,3-b]pyridine Inhibitors of IKKα That Selectively Perturb Cellular Non-Canonical NF-κB Signalling. Molecules. 2024; 29(15):3515. https://doi.org/10.3390/molecules29153515
Chicago/Turabian StyleRiley, Christopher, Usama Ammar, Aisha Alsfouk, Nahoum G. Anthony, Jessica Baiget, Giacomo Berretta, David Breen, Judith Huggan, Christopher Lawson, Kathryn McIntosh, and et al. 2024. "Design and Synthesis of Novel Aminoindazole-pyrrolo[2,3-b]pyridine Inhibitors of IKKα That Selectively Perturb Cellular Non-Canonical NF-κB Signalling" Molecules 29, no. 15: 3515. https://doi.org/10.3390/molecules29153515
APA StyleRiley, C., Ammar, U., Alsfouk, A., Anthony, N. G., Baiget, J., Berretta, G., Breen, D., Huggan, J., Lawson, C., McIntosh, K., Plevin, R., Suckling, C. J., Young, L. C., Paul, A., & Mackay, S. P. (2024). Design and Synthesis of Novel Aminoindazole-pyrrolo[2,3-b]pyridine Inhibitors of IKKα That Selectively Perturb Cellular Non-Canonical NF-κB Signalling. Molecules, 29(15), 3515. https://doi.org/10.3390/molecules29153515








