Cellular Automata on Graphs: Topological Properties of ER Graphs Evolved towards Low-Entropy Dynamics
Abstract
:1. Introduction
2. Methods and Previous Results
2.1. Cellular Automata on Graphs
2.2. Entropy Measures
2.3. Previous Results
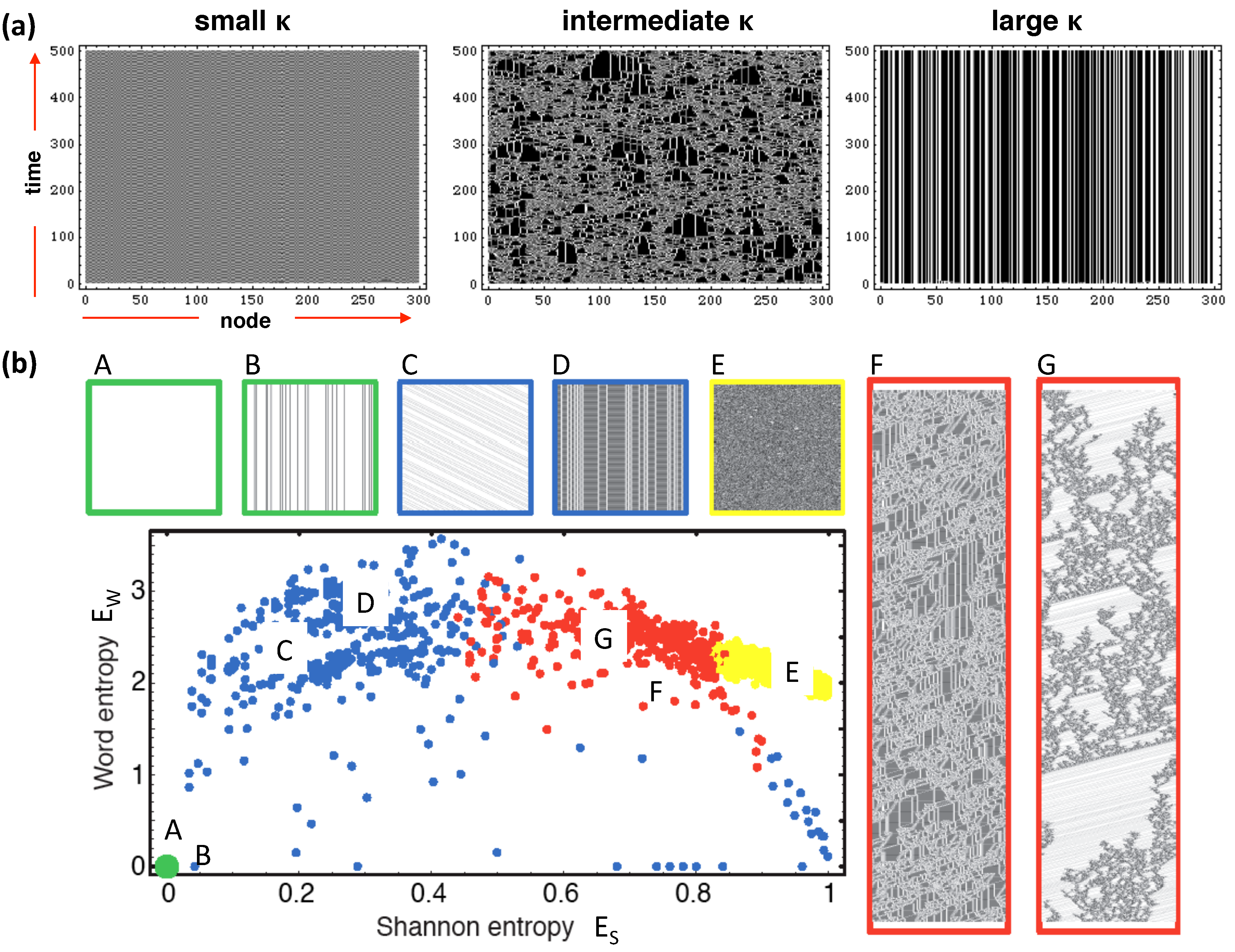
2.4. Networks
2.5. Rule Sensitivity
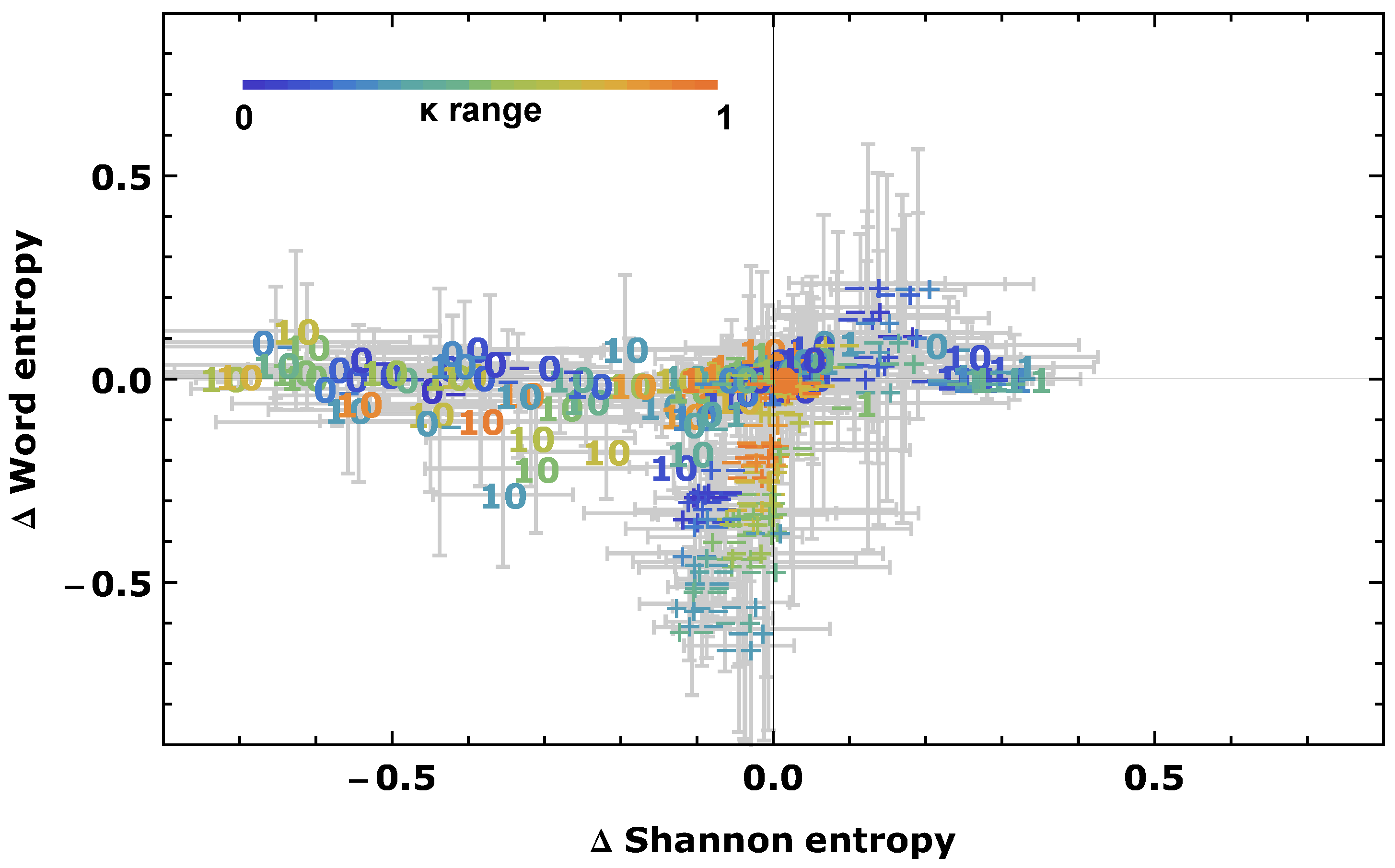
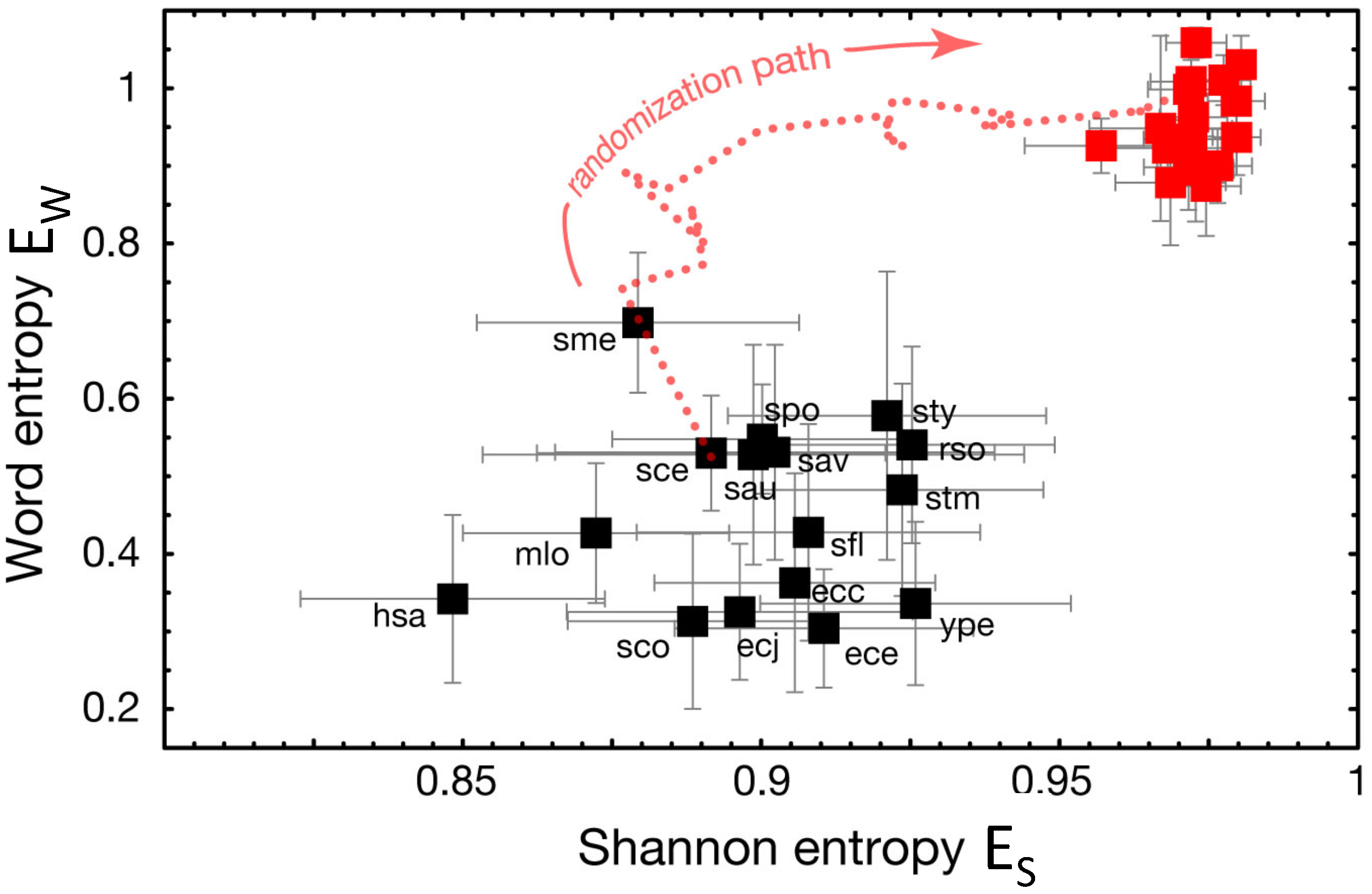
2.6. Simulated Annealing
- Generate a directed ER graph with 100 nodes and about 200 links ( 0.02).
- Implement the cellular automaton defined in Equation (1) with , 200 timesteps and 100 different initial conditions.
- Evaluate the (node-)averaged Shannon and word entropies as defined in Equations (2) and (3).
- Apply a single rewiring step by randomly choosing a directed link and connect either the start or the end of the link to a randomly chosen node. Ensure that the new link does not yet exist and that the resulting graph is weakly connected (i.e., the undirected version of the graph is connected). Repeat steps 2–3 for the new graph.
- Accept the new graph (labeled i) and discard graph , if the entropy differences . If , accept the new graph with probability . Here, we choose a linear cooling scheme where s denotes the number of evolution steps (5000) and denotes the initial temperature, which we choose as 0.01. If graph i is accepted, continue with step 2, otherwise choose a new graph i with step 4.
3. Results
3.1. Evolved Networks
3.2. Topological Analysis

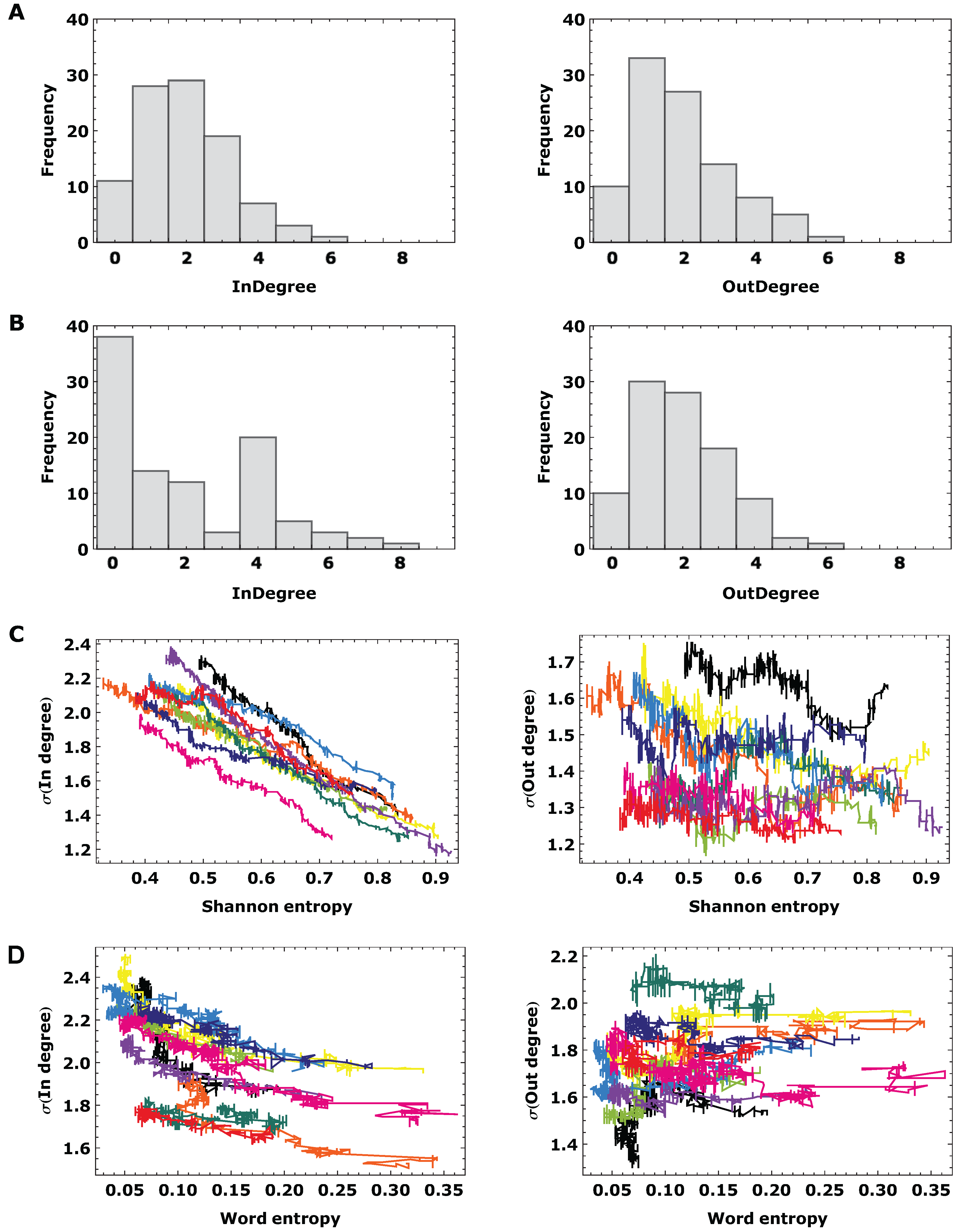
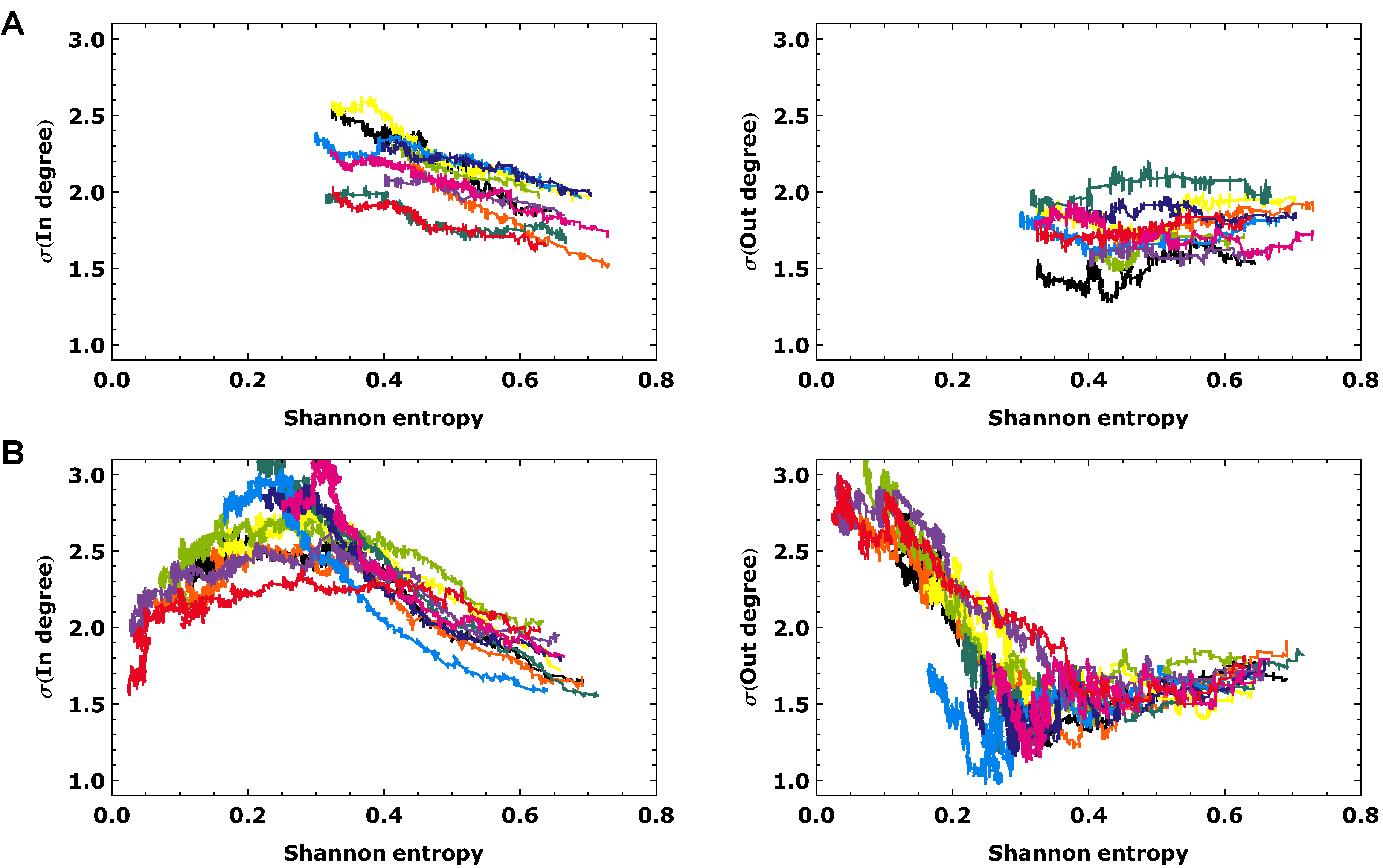
4. Conclusions
- Indirectly, these binary dynamics measure whether fluctuations in the neighborhood of a node will propagate to the node under consideration, or whether they will rather be dampened out. In fact, when one discusses a simple system of linear ODEs on the graph and monitors transient lengths, one sees a clear correlation with our two entropies.
- When formally re-writing the CA update rules as coupled ODEs (with, e.g., a switching of a state corresponding to a large term in the corresponding ODE) one could in some cases allow for a biological interpretation of the update rules, as the right-hand sides of such ODE systems can be viewed as a “list” of biological processes to the dynamics of the system at hand.
Acknowledgements
References
- Jeong, H.; Tombor, B.; Albert, R.; Oltvai, Z.N.; Barabási, A.L. The large-scale organization of metabolic networks. Nature 2000, 407, 651–654. [Google Scholar]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabási, A.L. Hierarchical organization of modularity in metabolic networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef] [PubMed]
- Guimera, R.; Amaral, L. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Itzkovitz, S.; Kashtan, N.; Levitt, R.; Shen-Orr, S.; Ayzenshtat, I.; Sheffer, M.; Alon, U. Superfamilies of evolved and designed networks. Science 2004, 303, 1538–1542. [Google Scholar] [CrossRef] [PubMed]
- Alon, U. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 2007, 8, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Lesne, A. Complex networks: From graph theory to biology. Lett. Math. Phys. 2006, 78, 235–262. [Google Scholar] [CrossRef]
- Strogatz, S. Exploring complex networks. Nature 2001, 410, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Rosvall, M.; Bergstrom, C. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 2008, 105, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Arenas, A.; Díaz-Guilera, A.; Pérez-Vicente, C.J. Synchronization reveals topological scales in complex networks. Phys. Rev. Lett. 2006, 96, 114102. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Alexander, R.P.; Kim, P.M.; Emonet, T.; Gerstein, M.B. Understanding modularity in molecular networks requires dynamics. Sci. Signal. 2009, 2, pe44. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A. Circuit topology and the evolution of robustness in two-gene circadian oscillators. Proc. Natl. Acad. Sci. USA 2005, 102, 11775. [Google Scholar] [CrossRef] [PubMed]
- Schulman, L.; Gaveau, B. Dynamical distance: Coarse grains, pattern recognition, and network analysis. Bull. Sci. Math. 2005, 129, 631–642. [Google Scholar]
- Marr, C.; Hütt, M. Topology regulates pattern formation capacity of binary cellular automata on graphs. Physica A 2005, 354, 641–662. [Google Scholar] [CrossRef]
- Marr, C.; Hütt, M.T. Outer-totalistic cellular automata on graphs. Phys. Lett. A 2009, 373, 546–549. [Google Scholar] [CrossRef]
- Kashtan, N.; Alon, U. Spontaneous evolution of modularity and network motifs. Proc. Natl. Acad. Sci. USA 2005, 102, 13773–13778. [Google Scholar] [CrossRef] [PubMed]
- Kashtan, N.; Noor, E.; Alon, U. Varying environments can speed up evolution. Proc. Natl. Acad. Sci. USA 2007, 104, 13711. [Google Scholar] [CrossRef] [PubMed]
- Kashtan, N.; Parter, M.; Dekel, E.; Mayo, A.E.; Alon, U. Extinctions in heterogeneous environments and the evolution of modularity. Evolution 2009, 63, 1964–1975. [Google Scholar] [CrossRef] [PubMed]
- Kaluza, P.; Mikhailov, A.S. Evolutionary design of functional networks robust against noise. Europhys. Lett. 2007, 79, 48001. [Google Scholar] [CrossRef]
- Kaluza, P.; Ipsen, M.; Vingron, M.; Mikhailov, A. Design and statistical properties of robust functional networks: A model study of biological signal transduction. Phys. Rev. E 2007, 75, 15101. [Google Scholar] [CrossRef]
- Kaluza, P.; Vingron, M.; Mikhailov, A. Self-correcting networks: Function, robustness, and motif distributions in biological signal processing. Chaos 2008, 18, 026113. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Shibata, T.; Kuramoto, Y.; Mikhailov, A.S. Evolutionary design of oscillatory genetic networks. Eur. Phys. J. B 2010, 76, 167–178. [Google Scholar] [CrossRef]
- von Neumann, J. The general and logical theory of automata. In Design of Computers, Theory of Automata and Numerical Analysis; J. von Neumann, Collected Works; Taub, A.H., Ed.; Pergamon Press: London, UK, 1963; Volume 5, pp. 288–326. [Google Scholar]
- Gardner, M. Mathematical games: The fantastic combinations of John Conway’s new solitaire game “Life”. Sci. Am. 1970, 10, 120. [Google Scholar] [CrossRef]
- Dresden, M.; Wong, D. Life games and statistical models. Proc. Natl. Acad. Sci. USA 1975, 72, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Wolfram, S. Statistical mechanics of cellular automata. Rev. Mod. Phys. 1983, 55, 601. [Google Scholar] [CrossRef]
- Wolfram, S. Universality and complexity in cellular automata. Physica D 1984, 10, 1. [Google Scholar] [CrossRef]
- Deutsch, A.; Dormann, S. Cellular Automaton Modeling and Biological Pattern Formation; Birkhäuser: Boston, MA, USA, 2005. [Google Scholar]
- Schulman, L.; Seiden, P. Percolation and galaxies. Science 1986, 233, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Seiden, P.; Celado, F. A model for simulating cognate recognition and response in the immune system. J. Theor. Biol. 1992, 158, 329–357. [Google Scholar] [CrossRef]
- Marr, C.; Hütt, M. Similar impact of topological and dynamic noise on complex patterns. Phys. Lett. A 2006, 349, 302–305. [Google Scholar] [CrossRef]
- Müller-Linow, M.; Marr, C.; Hütt, M.T. Topology regulates the distribution pattern of excitations in excitable dynamics on graphs. Phys. Rev. E 2006, 74, 1–7. [Google Scholar] [CrossRef]
- Müller-Linow, M.; Hilgetag, C.C.; Hütt, M.T. Organization of excitable dynamics in hierarchical biological networks. PLoS Comput. Biol. 2008, 4, e1000190. [Google Scholar] [CrossRef] [PubMed]
- Marr, C.; Müller-Linow, M.; Hütt, M.T. Regularizing capacity of metabolic networks. Phys. Rev. E 2007, 75, 1–6. [Google Scholar] [CrossRef]
- Amaral, L.; Goldberger, A. Emergence of complex dynamics in a simple model of signaling networks. Proc. Natl. Acad. Sci. USA 2004, 101, 15551–15555. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.A.; Mathur, A.; Diermeier, D.; Amaral, L.A.N. Efficient system-wide coordination in noisy environments. Proc. Natl. Acad. Sci. USA 2004, 101, 12085. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440. [Google Scholar] [CrossRef] [PubMed]
- Graham, I.; Matthai, C.C. Investigation of the forest-fire model on a small-world network. Phys. Rev. E 2003, 68, 036109. [Google Scholar] [CrossRef]
- Shen-Orr, S.; Milo, R.; Mangan, S.; Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002, 31, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Riehl, W.J.; Krapivsky, P.L.; Redner, S.; Segrè, D. Signatures of arithmetic simplicity in metabolic network architecture. PLoS Comput. Biol. 2010, 6, e1000725. [Google Scholar] [CrossRef] [PubMed]
- Maslov, S.; Krishna, S.; Pang, T.; Sneppen, K. Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc. Natl. Acad. Sci. USA 2009, 106, 9743. [Google Scholar] [PubMed]
- Samal, A.; Singh, S.; Giri, V.; Krishna, S.; Raghuram, N.; Jain, S. Low degree metabolites explain essential reactions and enhance modularity in biological networks. BMC Bioinformatics 2006, 7, 118–118. [Google Scholar] [CrossRef] [PubMed]
- Almaas, E. Optimal flux patterns in cellular metabolic networks. Chaos 2007, 17, 026107. [Google Scholar] [CrossRef] [PubMed]
- Almaas, E.; Kovács, B.; Vicsek, T.; Oltvai, Z.N.; Barabási, A.L. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 2004, 427, 839–843. [Google Scholar] [PubMed]
- Basler, G.; Grimbs, S.; Ebenhöh, O.; Selbig, J.; Nikoloski, Z. Evolutionary significance of metabolic network properties. J. R. Soc. Interface 2011. [Google Scholar] [CrossRef] [PubMed]
- Basler, G.; Ebenhöh, O.; Selbig, J.; Nikoloski, Z. Mass-balanced randomization of metabolic networks. Bioinformatics 2011, 27, 1397–1403. [Google Scholar] [CrossRef] [PubMed]
- Price, N.D.; Reed, J.L.; Palsson, B.O. Genome-scale models of microbial cells: Evaluating the consequences of constraints. Nat. Rev. Microbiol. 2004, 2, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.J.; Prakash, P.; Edwards, J.S. Advances in flux balance analysis. Curr. Opin. Biotechnol. 2003, 14, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Feist, A.M.; Palsson, B.O. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 2008, 26, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zeng, A.P. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics 2003, 19, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Erdős, P.; Rényi, A. On random graphs. Publ. Math. 1959, 6, 290. [Google Scholar]
- Trusina, A.; Maslov, S.; Minnhagen, P.; Sneppen, K. Hierarchy measures in complex networks. Phys. Rev. Lett. 2004, 92, 178702. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, S.; Gelatt, C.D., Jr.; Vecchi, M.P. Optimization by simulated annealing. Science 1983, 220, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509. [Google Scholar] [PubMed]
- Turing, A. The chemical basis of morphogenesis. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 1952, 237, 37–72. [Google Scholar] [CrossRef]
- Nakao, H.; Mikhailov, A.S. Turing patterns in network-organized activator-inhibitor systems. Nature 2010, 6, 544–550. [Google Scholar] [CrossRef]
- Drossel, B. Random Boolean networks. In Reviews of Nonlinear Dynamics and Complexity; Schuster, H.G., Ed.; Wiley: Weinheim, Germany, 2008. [Google Scholar]
- Peixoto, T.; Drossel, B. Boolean networks with reliable dynamics. Phys. Rev. E 2009, 80, 056102. [Google Scholar] [CrossRef]
- Krawitz, P.; Shmulevich, I. Basin entropy in Boolean network ensembles. Phys. Rev. Lett. 2007, 98, 158701. [Google Scholar] [CrossRef] [PubMed]
- Davidich, M.; Bornholdt, S. Boolean network model predicts cell cycle sequence of fission yeast. PLoS One 2008, 3, e1672. [Google Scholar] [CrossRef] [PubMed]
- Braunewell, S.; Bornholdt, S. Superstability of the yeast cell-cycle dynamics: Ensuring causality in the presence of biochemical stochasticity. J. Theor. Biol. 2007, 245, 638–643. [Google Scholar] [CrossRef] [PubMed]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Marr, C.; Hütt, M.-T. Cellular Automata on Graphs: Topological Properties of ER Graphs Evolved towards Low-Entropy Dynamics. Entropy 2012, 14, 993-1010. https://doi.org/10.3390/e14060993
Marr C, Hütt M-T. Cellular Automata on Graphs: Topological Properties of ER Graphs Evolved towards Low-Entropy Dynamics. Entropy. 2012; 14(6):993-1010. https://doi.org/10.3390/e14060993
Chicago/Turabian StyleMarr, Carsten, and Marc-Thorsten Hütt. 2012. "Cellular Automata on Graphs: Topological Properties of ER Graphs Evolved towards Low-Entropy Dynamics" Entropy 14, no. 6: 993-1010. https://doi.org/10.3390/e14060993



