1. Introduction
Recent advances in high computing capability have led to improvements in healthcare systems. With the promise of this advance, healthcare systems are used to improve the quality and effectiveness of healthcare, and to reduce costs in healthcare services. While patients’ health records are stored in different hospitals and databases, it is a challenging process to share and reuse these records. Reusing patients’ health data in any health organization for a health service can accelerate the diagnostic process while reducing material costs in healthcare. For this purpose, Semantic Web technologies can be integrated into healthcare services for modeling and reasoning of healthcare information. In Semantic Web, information is given with a well-defined meaning. It leads to a better collaboration between computers and humans [
1]. Hence, data is represented in a machine-understandable format to support a greater knowledge representation. Therefore, ontologies are used to share and to reuse information between different systems.
An ontology is an explicit specification of a conceptualization [
2]. It describes the entities in a specific domain by defining all the relevant concepts and the relationships between these concepts. Semantic Web technologies use the Resource Description Framework (RDF) [
3] and Web Ontology Language (OWL) [
4] as information description languages to describe metadata, to define ontologies, and to achieve reasoning over the defined ontologies. Storing ontology definitions in specific Uniform Resource Identifier (URI) makes ontologies universal, sharable, and reusable. The ontology description is essential to share the common understanding of the structure of information among people or software agents, to enable reuse of the domain knowledge, to make domain assumptions explicit, to separate the domain knowledge from the operational knowledge, and to analyze the domain knowledge [
5].
In an open world assumption, states contain both positive and negative fluents, and if a fluent does not appear, its value is unknown [
6]. Thus, the belief state corresponds exactly to the set of possible worlds that satisfy this description. Therefore, the information is only meaningful in the environment where it exists and the reliability of the information changes through the environment. In a knowledge base with an open world assumption, access to the information and its dependencies to other sources can affect the meaning and the usage of the information. Ontology developers define ontologies by working with domain experts. Different domain experts mean different personal experiences. Naturally, this will lead to different ontology designs for the same domain. Even though some ontology knowledge bases can connect these different definitions, if the domain definition is not described with its metadata level knowledge, there would be several discrete ontological definitions for the same domain. Consequently, connectivity of ontologies is a breakpoint when it comes to selecting the right ontology for the related domain.
Storing domain information in ontologies eliminates the integration problem of different information sources. The collaboration and connectivity that are provided by Semantic Web technologies and ontologies enable access to the information independent from any data structure. As a result of the unique definition provided by the ontology, the information can be adapted into different structures according to the domain context and the system where this information will be used. This adaptation gives data flexibility that can be used in any information system without any rearrangement of the data. However, access to the information is provided with a common query language and the response time of these queries change according to the internal structure of the information storage environment.
Healthcare systems around the world are changing their information systems in order to share and reuse patient information not only in a department where the information is being created, but also between the departments of an organization and also among different organizations. Furthermore, the healthcare domain is one of the rare areas that has a huge amount of domain knowledge. The increasing availability and growth rate of healthcare information as big data lead to an improvement in overall quality of patient care that is also beneficial for providing a better personalized healthcare service. In order to support the interoperability with Semantic Web technologies, defining and storing healthcare data in ontologies is expected to offer an efficient and effective solution for healthcare information systems. Infectious Disease Ontology (IDO) [
7,
8], Saliva Ontology (SALO) [
9] and Blood Ontology (BLO) [
10] are example ontologies for the healthcare domain that are described by formal ontology languages.
The large scale of information in healthcare needs to be managed in order to make decisions rapidly and to respond to emergency health situations immediately where time is important. For example, a person who has a traffic accident should be taken to the nearest hospital. The doctor must access the patient’s critical health information urgently, for example, to see if he or she has an allergic reaction to any substance or drug. Storing, inserting, retrieving, updating, and querying the big health ontologies efficiently in terms of accessing the right information are as important as developing the ontologies. Therefore, the storage method, especially the ontology store, is critical for the performance of the healthcare information system. Different triple stores store and use ontologies differently. Loading a new ontology, query decomposition, and query result time may differ with how the ontology is represented with the inner structure of the related triple store.
In this study, two triple stores, AllegroGraph [
11] and Oracle 12c [
12], are compared according to their performance for health domain information. AllegroGraph and Oracle 12c are recommended by W3C [
13] as efficient triple stores that contain more than one billion triples. Thus, we chose these two stores, AllegroGraph and Oracle 12c, for performance evaluation of big healthcare data. For this purpose, the following steps are performed:
Exploring the ontologies for the health domain and selecting the suitable ontologies;
Loading the selected ontologies into data stores;
Inserting, updating, and deleting the triples;
Choosing the relevant queries for the health domain;
Performing the selected queries to compare the performance of two data stores based on their response time.
The paper is organized as follows:
Section 2 presents the related work.
Section 3 describes the materials and methods of the evaluation. The evaluation results are given in
Section 4. Finally,
Section 5 contributes and outlines the direction of the future work.
2. Related Work
Semantic Web is the extension of the current web where information is given in a well-defined meaning and leads to a better collaboration between computers and humans [
14]. The Semantic Web studies focus on developing domain specific ontologies as well as semantic recommendation systems. Recently, ontology-based data access technologies (OBDA) became a popular approach to store semantic data on relational databases. In [
15], an open-source system architecture named Ontop is presented to query relational data sources through the semantical representation of the related domain. The architecture of Ontop is used as the query transformation component of the Optique Platform developed in [
16]. The Optique Platform provides a visual query interface for end users to formulate user information needs by unlocking the access to Big Data.
In the literature, there are several RDF data stores to store, query, and manipulate these ontologies. Various works also compare these RDF data stores [
17,
18,
19,
20,
21]. In addition to comparing the RDF stores, RDF loaders are also compared in [
22]. Distinct from our work; [
17,
18,
19] use generated data, [
17,
18] use insufficiently low data for the performance evaluation, [
17,
18,
19,
20,
22] do not use medical data, and [
19,
22] compare both AllegroGraph and Oracle 12c. These studies use different metrics to query RDF data, but none of these studies compare the performance of triple stores using CONSTRUCT queries. In this work, we use real medical data instead of the generated data and execute CONSTRUCT queries.
The data set of The University Ontology Benchmark (UOBM) [
23] is used as a benchmark to compare the RDF data stores; Jena TDB [
24], Virtuoso [
25], AllegroGraph [
11], BigOWLIM [
26], Eclipse RDF4J (Sesame) [
27], and Oracle [
12]. Fourteen different comparison variables are used for the benchmarking. RDF data stores are categorized as simple, memory based, or database based according to their architecture, forward-based, reverse-based, or hybrid chaining inference methods. Ontology loading time, the query response time, the query languages, the accuracy, and the completeness of the returned results of the query are also included as the physical benchmarking variables. Based on query capabilities; the comparison between The Simple Protocol and RDF Query Language (SPARQL) [
28] and the standard RDF query language, and also the scalability of complex queries is discussed. Furthermore, the benchmarking also contains evaluations that are performed according to the integration of the inference engine, clustering capabilities, end user interface support, relational database support, and interoperability of different platforms and operating systems.
RDF triple stores model data using RDF, store RDF triples, and are queried using SPARQL. RDF triple stores, 4Store [
29], BigData [
30], OWLIM-SE [
31], Mulgara [
32], and Virtuoso [
33] are evaluated in [
33]. The data sets that are used in [
33] are not simulated and they are real world data sets that are already used in various applications. Data sets from Cell Cycle Ontology [
34], Allie Ontology [
35], PDBj (Protein Data Bank Japan) [
36], UniProt (Universal Protein Resource) [
37], and DDBJ (DNA Data Bank of Japan) [
38] are loaded to the RDF triple stores. Inside these datasets, DDBJ is the biggest data set with 10 billion triples. The ontology loading time, the memory location, the query response time, query accuracy, and environmental situations are used as the comparison metrics. According to these metrics; Cell Cycle Ontology has the fastest loading time for the 4Store repository, DDBJ ontology has the fastest loading time for Virtuoso. Based on memory allocation; Cell Cycle Ontology is stored with the smallest memory allocation in the BigData repository and OWLIM-SE uses the smallest memory while storing DDBJ ontology. When the query response times are compared, Cell Cycle Ontology gives the fastest response time through all repositories. However, the fastest response time for nine of nineteen queries belongs to the OWLIM-SE. This work is helpful in comparing different data sets with different triple repositories. However, the most used triple repositories, AllegroGraph and Oracle RDF triple stores, are not included in [
33].
In our work, AllegroGraph and Oracle 12c RDF triple repositories are examined to evaluate their query capabilities and query response time by using GENE Ontology [
39] while GENE, Gene Expression Ontology (GEXO), Regulation of Transcription Ontology (RETO), and Regulation of Gene Expression Ontology (REXO) are evaluated for loading times. These ontologies are the most known and used health ontologies in the healthcare domain. GENE Ontology reveals information regarding the role of an organism’s gene products. GENE Ontology has been cited in over 5000 reviewed articles and has been used in research studies for informing or validating hypotheses [
40]. Researchers who develop health domain specific ontologies load their ontologies to BioPortal (
https://bioportal.bioontology.org/) which is a well-known repository of biomedical ontologies. BioPortal is an open access portal. GEXO integrates fragments of GENE Ontology and the Molecular Interaction ontology (MI). RETO is an ontology for the domain of gene transcription regulation and also integrates fragments of GENE Ontology and MI ontology. REXO is an ontology for the domain of gene expression regulation. The ontology integrates fragments of GENE Ontology and MI ontology. GEXO, RETO, and REXO ontologies are based on GENE Ontology.
AllegroGraph is a high-performance graph-based database that is used as a backend to develop the Semantic Web applications. It has Java, Python, Lisp, and Prolog interfaces and gives support for languages such as SPARQL, RDFS++, Prolog, and TwinQL. AllegroGraph provides efficient memory usage by the help of disk-based storage. AllegroGraph supports OWL 2 reasoning and Prolog sentences. Moreover, AllegroGraph defines additional features such as social networking analysis, free-text indexing, named graphs for weights, dynamic and automatic indexing, and efficient range queries.
Oracle 12c stores ontologies in relational databases with RDF and RDFS support. Starting with Oracle version 10gR2, RDF statements can be stored in databases with their natural triple structure. The support for OWL language begins with Oracle version 11g and continues with Oracle version 12c. Oracle provides full support to store, query, and manipulate the ontology data as triple. Oracle 12c also supports inferencing by defining rules for ontologies in addition to the Oracle-supplied rule bases such as RDF and RDFS. As is known, creating indexes improve the performance of certain semantic queries. For this purpose, Oracle has built-in functions to create semantic indexes. Although it is a relational database, it also supports SPARQL queries in addition to SQL queries with its semantic subprograms.
3. Materials and Methods
Resource Description Framework (RDF,
https://www.w3.org/RDF/) is designed by the World Wide Web Consortium (W3C,
https://www.w3.org/) as a metadata model. RDF is a standard model that is used for conceptual description and modeling of data in web resources. In RDF, each source is defined with a
triple that consists of a subject, predicate, and an object. Traditional data warehouses are based on relational databases and are used to work with relational data. However, when storing RDF data, the triple data structure should be maintained for better representation and knowledge representation. Various RDF stores were developed to store RDF data for this necessity. In this work, AllegroGraph and Oracle 12c triple stores are examined.
Web Ontology Language (OWL,
https://www.w3.org/OWL/) is also a W3C specification that is designed to represent knowledge and the relationship between things in a semantically rich manner. OWL represents the web content that is supported by XML (
https://www.w3.org/XML/), RDF, and RDF Schema (RDF-S,
https://www.w3.org/TR/rdf-schema/) by presenting a formal semantic and additional description constructs. Furthermore, Semantic Web knowledge is expressed with ontologies. Thus, the content of knowledge is not only interpreted by humans, but also by machines. Therefore, applications can process this knowledge as humans do.
Description Logic based tools, such as Fact [
41] and Racer [
42], are used to inference small-size ontologies. However, these tools are not successful in querying large amounts of data. Therefore, when a real world semantic knowledge base is considered, the intended results cannot be achieved. For this purpose, triple stores such as AllegroGraph, Oracle 12c, Virtuoso, and 4store are used to store, update, and query ontologies that consist of large amounts of semantic data. Moreover, if the knowledge base grows with time, the query time will also increase. In addition to the query performance, results of queries have to be complete and accurate.
In this study, two triple stores are examined to store, to update, and to query large ontologies. For this purpose, ontologies developed for the healthcare domain are used to measure the performance of semantic knowledge bases. Furthermore, the required time to access information is also measured and the accessed information is controlled to check whether the query result is accurate or not. The information access time depending on the complexity of queries is also measured. Hence, we first installed the related tools, then updated the healthcare ontologies, and specified criteria for the comparison. Based on these criteria, we created queries and queried ontologies. Later, we compared the query results and evaluated them. The performance measurements are obtained in a virtual machine with specifications of 2.40 GHz Intel(R) CPU, 16 GB of RAM (Intel CPU, Kingston RAM, Asus Motherboard, assembled in Izmir, Turkey). The following subsections explain AllegroGraph and Oracle 12c, data sets that are used, and queries that are executed over these data sets, respectively.
3.1. AllegroGraph and Oracle 12c
AllegroGraph [
11] is a RDF triple store and stores RDF triples. It is used by various open source, academic research and commercial projects [
43,
44,
45]. AllegroGraph supports Java, Python, Ruby, Perl, C#, Lisp, and Prolog interfaces, and also SPARQL, RDFS++, and Prolog reasoning. AllegroGraph is used to store several different data sets.
Oracle is a well-known relational database management system. Oracle 10gR2 was released in 2005 and is the first version of the Oracle database that supports RDF and RDF-S, and allows the storage of triples in the database. It provides functionalities to users such as storing their ontologies in relational databases and querying these ontologies that are used as information bases for Semantic Web applications. Oracle 11g (released in 2007) supports the OWL ontology definition language in addition to RDF and RDF-S support. In this study, Oracle 12c [
12], which was released in June 2013, is used. The semantic properties of the Oracle 12c database and their relations are shown in
Figure 1 [
46]. As seen in
Figure 1, Oracle 12c can contain both semantic and relational data at the same time.
DB-Engines (
http://db-engines.com/en/) collects and presents information on database management systems, and also ranks them in different lists according to the scope of usage such as RDF store, relational DBMS, key-value store, document store, etc. According to DB-Engines, Oracle is the most popular system for relational DBMS and in overall database management systems. Oracle is the first relational database management system that supports RDF storage. Therefore, using Oracle as a RDF store simplifies the integration of existing traditional data with triples. In the RDF store ranking of DB-Engines, AllegroGraph is listed in fifth place. AllegroGraph supports SPARQL which is the standard query language of W3C. AllegroGraph also supports Prolog and languages such as JavaScript to query triples. Furthermore, it provides mechanisms for reasoning by supporting RDF, RDFS, and OWL-DL predicates. In order to execute semantic web rules, AllegroGraph is integrated with Racer, which is a semantic web reasoning system.
The efficient way to load semantic data is bulk loading. In this study, bulk loading is used to insert semantic data from ontologies into Oracle 12c triple store. However, semantic data can also be added triple by triple by using the INSERT statement in SPARQL. In Oracle 12c, semantic data can be queried efficiently. Additionally, relational and semantic data can be queried by means of ontologies in order to find relations between relational and semantic data. After loading semantic data, the power of querying semantic data can be improved by using rules and inference engines. Inference provides logical results with regards to data and rules. However, in this study, the inferred triples are not evaluated in order to not make the evaluation more complex.
3.2. Data Sets
In this study, we used the following ontologies: Regulation of Gene Expression Ontology (REXO) [
47], Regulation of Transcription Ontology (RETO) [
48], Gene Expression Ontology (GEXO) [
49], Cell Cycle Ontology (CCO) [
34], and GENE Ontology [
39]. The number of classes, triples, individuals, property, and depths of these ontologies are shown in
Table 1.
As seen in
Table 1, most of the healthcare ontologies have approximately 2 million triples. Despite the number of triples in the ontologies, it is difficult to find a data set with an adequate number of individuals for evaluation. The existing ontologies in the literature are inadequate in terms of the number of individuals. Therefore, we used GENE, GEXO, RETO, and REXO ontologies to compare the loading times of AllegroGraph and Oracle 12c. The CCO ontology could not be loaded in our evaluation because AllegroGraph’s free version only allows loading up to 5 million triples. As the GENE ontology has a certain number of individuals while the rest of the ontologies do not have any individuals, we used GENE ontology to compare query response times while the other ontologies do not have any individuals. Queries were developed in SPARQL 1.1 language [
50] and handled on GENE ontology. Finally, the query response times were measured for the comparison.
3.3. Queries
In this subsection, the executed queries are explained. In this study, we created five queries for the evaluation. These five queries have been chosen at semantically increasing difficulty. In addition, the structure of these queries were based on the basic structural property of the cases used to test RDF stores that are presented in [
33]. However, in this study, these five queries are constructed specifically for GENE ontology, while cases presented in [
33] are constructed for Cell Cycle Ontology [
34].
The first query is a simple composite triple query. This query is used to measure the response time of triple stores for simple query usages.
Query 1: This is a simple multiple sentenced query as shown in
Figure 2.
Genes that are molecular_function, have synonym as “MOO Activity”, and have type as GO_0016701 are queried. Query 1 is a simple “GENE” query. By creating the first query, we aimed to measure the response of triple stores to simple queries that include multiple inner queries.
Query 2: The second query, shown in
Figure 3, is constructed by merging two simple queries with the “UNION” operand. The “UNION” operator is used to measure the response times to merge query spaces into a single result set for each RDF store. Query 2 returns the same result with Query 1. Both queries are used to capture how triple stores will respond to the same queries that are developed in different formats.
Query 3: In the third query, the difference between RDF triple data stores is examined. For this purpose, the usage of the REGEX function is handled inside the “FILTER” option. The REGEX function is a Boolean string function that returns results according to a given string. In this query,
genes that are molecular_function, type of GO_0016701, and have an exact synonym starting with the letters “myo” are being queried. Query 3, shown in
Figure 4, is used to measure the capabilities for string functions and indexing of Oracle 12c and AllegroGraph triple stores.
Query 4: In the fourth query, shown in
Figure 5, the “FILTER” option which was used with the REGEX function in Query 3 is replaced with a complete individual value. In Query 4, only the individuals that have the string value matching with the given value are returned as a result set. In order to make the query more complex, more than one value of the “FILTER” option is added and connected with the “AND” operation. Thus, the response of each triple data store into more than one “FILTER” option can be scored.
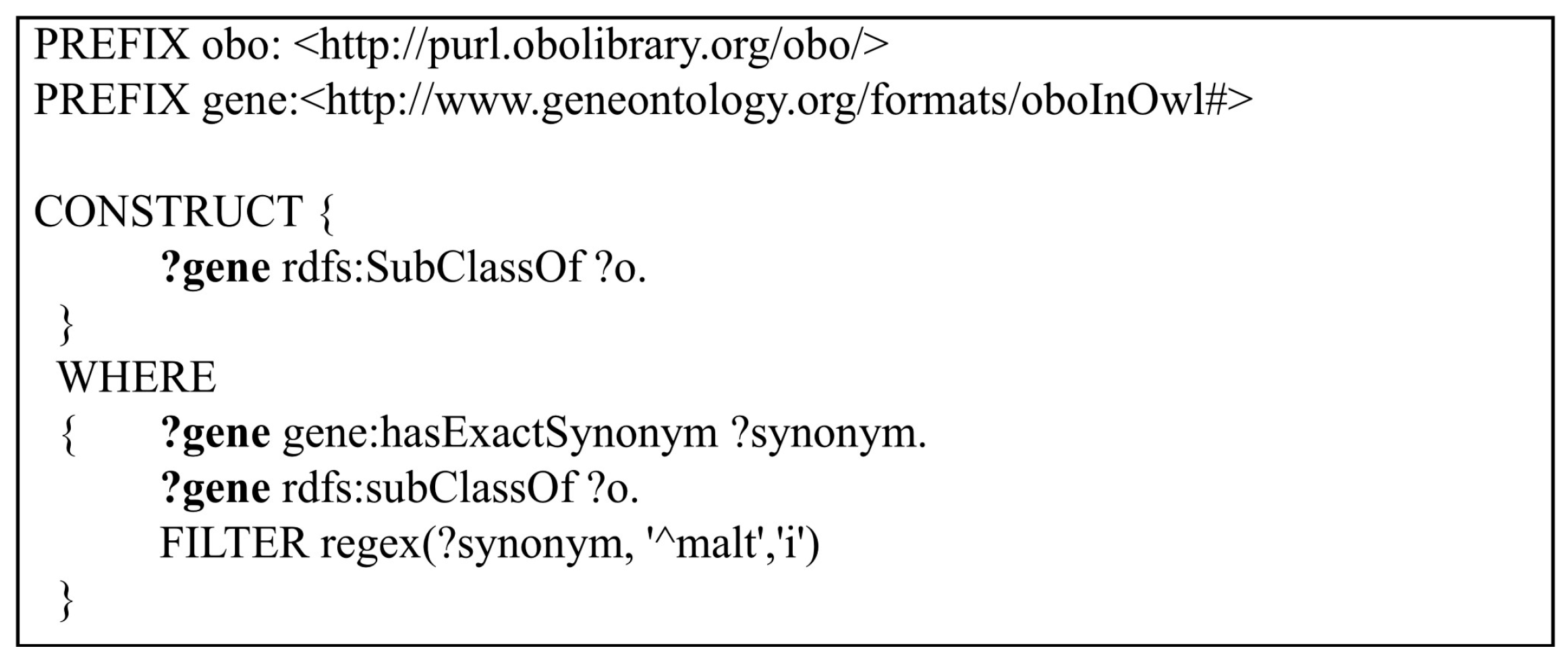
Query 5: In the fifth query, “CONSTRUCT” queries are validated instead of “SELECT” queries. CONSTRUCT queries are used to create a graph response that satisfies the result set restricted with the WHERE clause. CONSTRUCT queries are hard to defragment inside the layered structure of the triple stores which may cause infinite loops and return unrelated results. Thus, in Query 5, CONSTRUCT queries are used to reveal how fast each triple store creates a semantic graph of a given query from its semantic model. Despite using a simple “CONSTRUCT” query to return a graph result, a complex query that has more than one “FILTER” option is constructed. Query 5 is shown in
Figure 6. In this query, the differences between AllegroGraph and Oracle 12c while creating a graph with any given filter option are examined. In Query 5,
the genes which have synonyms starting with the letters “malt” are being queried. The graph that is constructed by the matched genes and their super classes are returned as a result.
The difference between the ontologies’ loading time and querying time of the GENE ontology is compared with the queries. The evaluation results are presented in the next section.
4. Results and Discussion
The queries given in
Section 3 are first experimented in the AllegroGraph SPARQL endpoint. The entire loading and query times are evaluated in the Jena API [
51] that is written in the Java Programming Language. Jena uses description logic to create a connected query space and provides an interface to use and to manipulate ontologies inside Java. Before bulk loading the semantic data, the loading ontology must be complete and accurate which means that the ontology should not contain any conflicting information. Any inconsistency in the ontology causes a loading error and needs to be corrected before loading the ontology into the triple store. Despite the open world assumption of the ontology, ontological structures are described with a closed world assumption where the inconsistency in an ontology can result in the failure of the ontological model. AllegroGraph and Oracle 12c are both using the open world assumption while representing and saving the ontological structures. Additionally, they both support Jena. As a result, converting ontological structures into Jena may lead to the loss of semantics. However, this is a reasonable risk in order to compare the query performances of these RDF triple stores.
Loading and using ontologies by Jena starts with the matching of ontological structures with Jena descriptions and ends with saving this Jena description space in a local data store. Before querying the ontology model, connection with the triple data store server, initialization of the Jena ontology model, and binding the connection to the model has to be completed. The query response time can be measured after completing these phases. Then, the query starts with the execution of the queries into Jena Models. The query time ends with the return of the query results from the triple data store into the Jena result set.
As AllegroGraph supports Jena API, Jena is used to calculate the differences between loading times of GENE, GEXO, RETO, and REXO ontologies into triple data stores. The loading times for AllegroGraph and Oracle 12c are given in
Table 2. The entire loading times are converted to seconds.
After creating the Jena ontology model and loading the ontology into this model, construction of the queries is done. In
Table 3, the query times of each query are given. The entire querying times are measured in terms of Java world time and converted into milliseconds.
AllegroGraph and Oracle 12c can be used to store similar data, but they have different loading and querying times of the same data set. In
Table 2, it is clear that the two data stores have large loading time differences. The reason behind this difference is the infrastructure and creation purpose of these data stores. Oracle 12c stores structural data according to its traditional database structures. However, AllegroGraph is a pure graph-based RDF triple store. Thus, AllegroGraph has a better performance in loading ontological structures which are essentially graph-based descriptions. As the triple number increases, the loading time also increases. However, the number of triples do not affect Oracle 12c as it affects AllegroGraph. Besides, the accuracy of the query results is the same for both of the triple stores.
Figure 7 and
Figure 8 show the graphical representation of the loading and querying times.
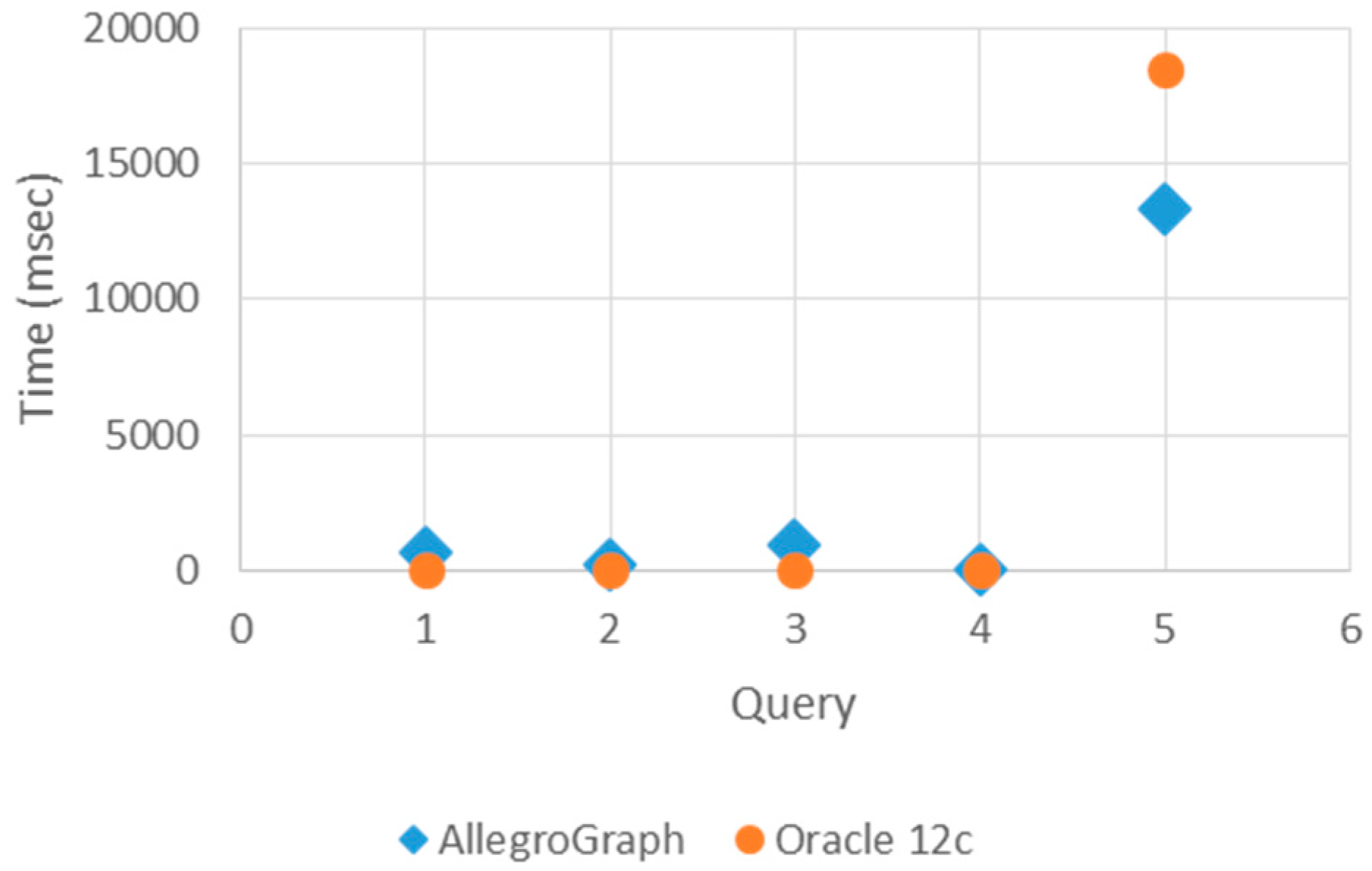
In
Figure 8, query times show that Oracle 12c responds to simple queries that are basic and indexed, such as Query 1–3, more rapidly than AllegroGraph. Oracle 12c responds to the basic and indexed queries more quickly. However, it responds slower to more complex queries such as Query 4 and Query 5. The AllegroGraph response time does not depend on the complexity of the query. Consequently, the number of triples in query space do not affect AllegroGraph’s response time.
Consequently, AllegroGraph is a more efficient triple store to load healthcare ontologies as opposed to Oracle 12c. However, for long-living software system applications, a frequent software maintenance is an undesired situation. The ontology for describing the information base of the system must be loaded to the triple store once, and then the information of the system grows by inserting or deleting the instances without bulk load. As every individual has a considerable amount of health records along his or her life, healthcare information is a good example of big data application areas. Furthermore, as genetic factors are important for diagnosis, the personal health records for a person could also be queried also after his or her death for the family health portrait. In addition, time is one of the most important factors in diagnosis and treatment. Therefore, accessing the required health data in a timely manner is crucial. Considering all of these facts, the query response times of triple stores are the key factor for choosing the appropriate triple store for the healthcare domain. As a result, Oracle 12c is faster than AllegroGraph according to the query times. Hence, when query response time has significance, Oracle 12c as a triple store will be more appropriate for living healthcare information systems.
5. Conclusions
AllegroGraph and Oracle 12c are RDF triple stores that are used to store big data sets. Both of the triple stores are widely used to store different types of data. AllegroGraph can store both graph-based data and RDF ontologies. Oracle 12c, with the help of its Semantic Web extension, is used to store structured data with regards to RDF and OWL ontologies. In this study, the ontology loading and querying time differences between AllegroGraph and Oracle 12c RDF triple stores are compared and the reasons behind these differences are discussed.
AllegroGraph and Oracle 12c both respond accurately to the queries on the same data set. Oracle 12c’s loading time is slower than AllegroGraph. On the contrary, AllegroGraph is loading and matching RDF/OWL data faster than Oracle 12c due to its design purpose. Oracle has a slow loading time on big data sets due to its layered infrastructure. However, Oracle 12c queries faster than AllegroGraph when the data is already loaded. Despite its fast response time in basic queries, Oracle 12c becomes slower when it responds to queries that are based on description logic or graph-based ontological structures. When the query space grows large enough or becomes more complete, AllegroGraph also becomes slower as expected.
Besides all the comparisons given, AllegroGraph and Oracle 12c store big healthcare data sets accurately and query them efficiently. As a conclusion, AllegroGraph is the more suitable candidate to store very fast changing data sets that need to be loaded repeatedly into triple store. Oracle 12c is more suitable to store big data sets that are static. Also, when query time is more important than the loading time, Oracle 12c is a more feasible triple store than AllegroGraph.