A Study of Brain Neuronal and Functional Complexities Estimated Using Multiscale Entropy in Healthy Young Adults
Abstract
:1. Introduction
- Edge level—edge complexity is estimated by the MSE of each edge, calculated from its dFC time series data (eMSE) that is obtained using BOLD PCC.
- Node level—node complexity is estimated by the MSE of each node, calculated from its BOLD time series data (nMSE) or calculated as the mean eMSE of all the edges connected to the node (edge-based nMSE).
- Network level—network complexity is estimated by the mean nMSE or mean edge-based nMSE of all the nodes in the RSN.
- Whole-brain, consisting of only the cortex and subcortical gray matter, level—whole-brain complexity is estimated by the mean nMSE of all the nodes or mean eMSE of all the edges in the brain.
2. Results
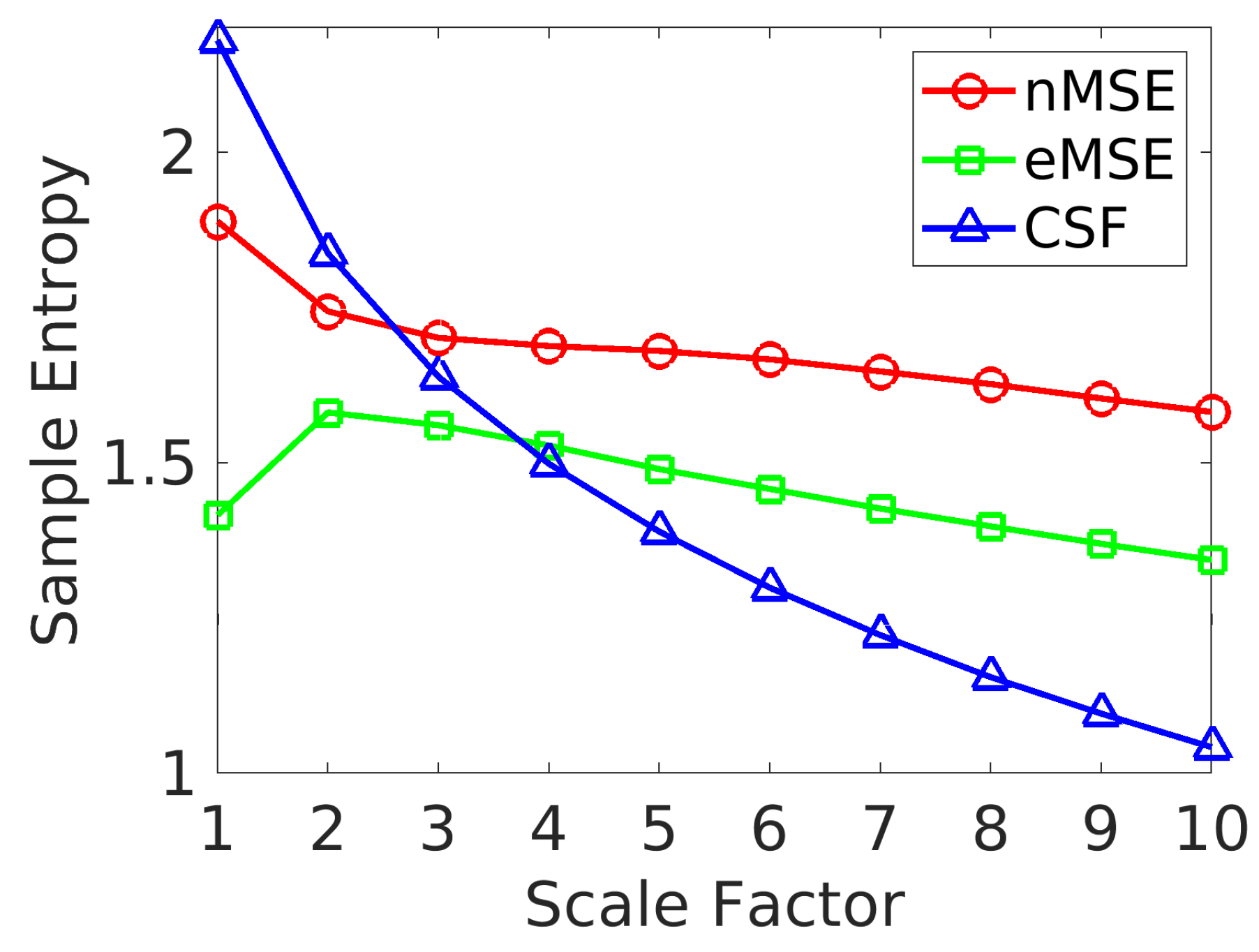
2.1. Relative Error of SampEn
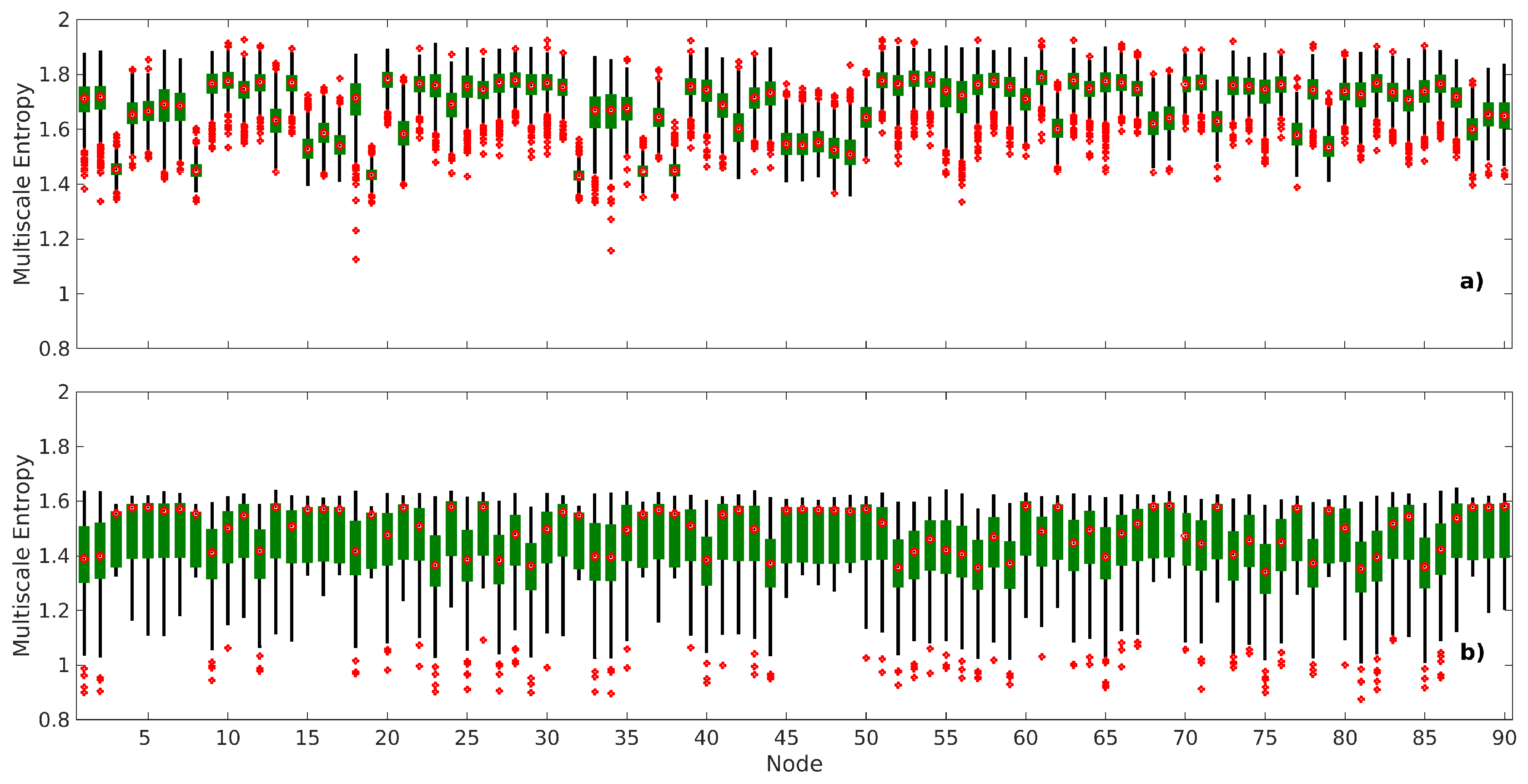
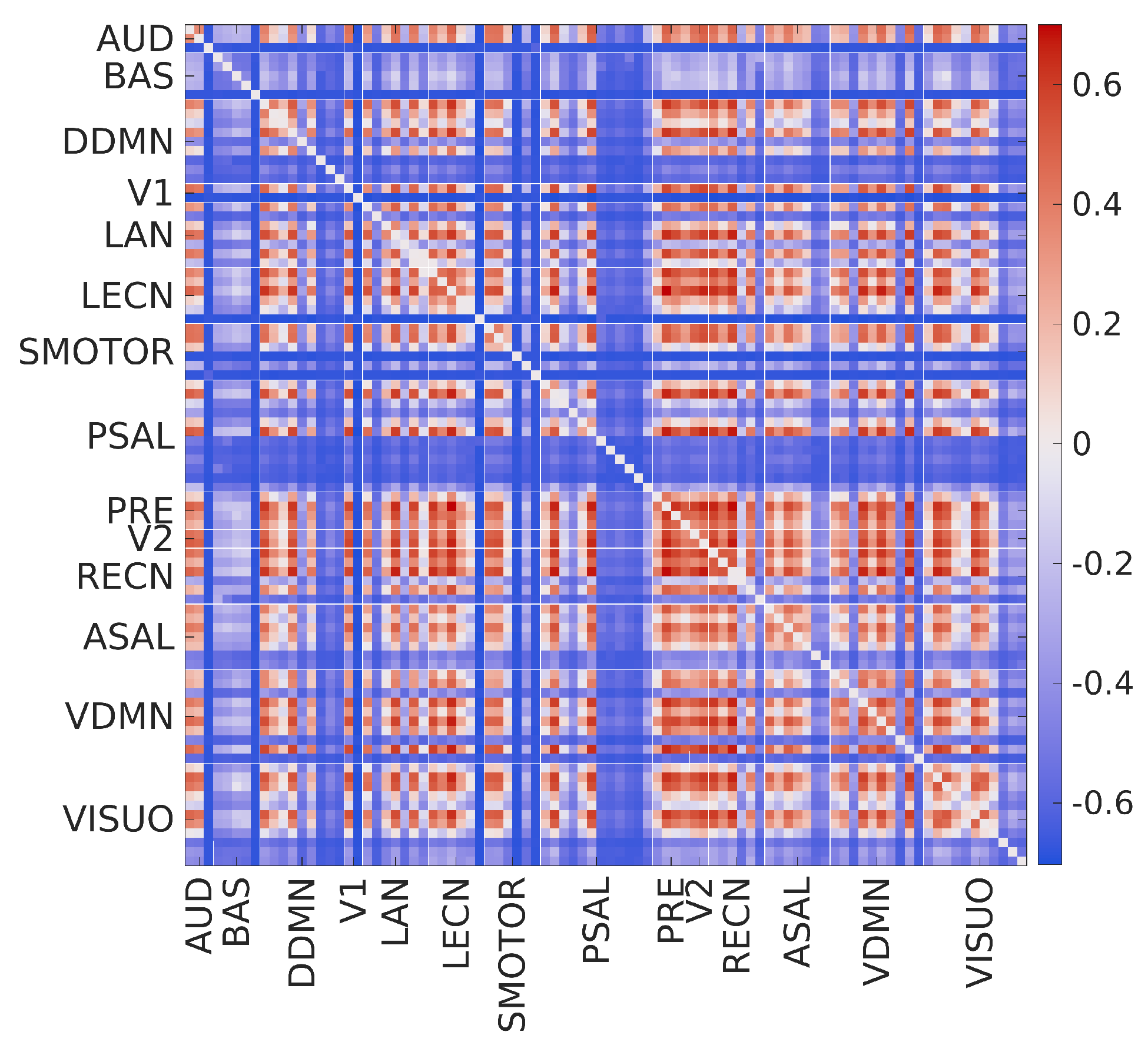
2.2. Brain Complexity Across Different Levels
- The matrix for scale 5 shows that the group mean eMSE values are most similar to the grand mean eMSE values.
- There is a large variation in complexity among all edges at fine scales, which decreases at coarse scales.
- The complexity of intra-network edges is lower than inter-network edges in many RSNs across scales. An example of this is the ventral default mode network (VDMN).
- There is a large variation in complexity of all edges in many nodes in various RSNs at fine scales. An example of this is the dorsal default mode network (DDMN), and these variations decrease at coarse scales.
- About 70% of edges show a decrease in SampEn as the scale increases. The remaining 30% show a diverging pattern with the SampEn increasing, which shows that the complexity is not correlated to the variance of coarse-grained signals [35].
2.3. Cognitive Behavioral Prediction Correlation Scores and Selected Features
3. Discussion
Limitations and Future Direction
4. Materials and Methods
4.1. fMRI Data
- Missing rfMRI sessions—174 subjects were missing in one of the resting-state sessions.
- High average framewise displacement [53]—16 subjects had an average framewise displacement greater than four standard deviations of the group, which introduces head motion artifacts.
- Missing time series data—six subjects had less than 1200 points in one of the sessions.
- Misalignment—one subject was misaligned in standard space and was missing some brain regions.
- Low Mini-Mental Status Exam (MMSE) score [54]—33 subjects had an MMSE score of 26 or lower, which can be an indicator of cognitive impairment.
- Missing fluid intelligence score—nine subjects were missing the fluid intelligence score.
4.2. Node and Edge Complexities
4.3. Optimal Parameter Selection for Calculating SampEn of BOLD Signals
4.4. Cognitive Behavioral Prediction
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASAL | Anterior salience network |
| AUD | Auditory network |
| BAS | Basal ganglia network |
| BOLD | Blood oxygen-level dependent |
| CSF | Cerebrospinal fluid |
| DDMN | Dorsal default mode network |
| dFC | Dynamic functional connectivity |
| eMSE | Edge multiscale entropy |
| HCP | Human Connectome Project |
| LAN | Language network |
| LECN | Left executive control network |
| MSE | Multiscale entropy |
| nMSE | Node multiscale entropy |
| PCC | Phase coherence connectivity |
| PRE | Precuneus network |
| PSAL | Posterior salience network |
| rfMRI | Resting-state functional magnetic resonance imaging |
| RECN | Right executive control network |
| RSN | Resting-state networks |
| SampEn | Sample entropy |
| SMOTOR | Sensorimotor network |
| SVR | Support vector regression |
| V1 | Primary visual network |
| V2 | Higher visual network |
| VDMN | Ventral default mode network |
| VISUO | Visuospatial network |
Appendix A. Predicting Fluid Intelligence
| Network | Node Number | Brain Region | Scale Factor | |
|---|---|---|---|---|
| nMSE | Edge-Based nMSE | |||
| BAS | 4 | Left Thalamus, Caudate | 7, 9 | |
| 5 | Right Thalamus, Caudate, Putamen | 1,7 | ||
| DDMN | 12 | Posterior Cingulate Cortex, Precuneus | 1 | |
| LAN | 20 | Inferior Frontal Gyrus | 1, 2 | 1, 2 |
| 22 | Left Middle Temporal Gyrus, Angular Gyrus | 1 | ||
| 23 | Left Middle Temporal Gyrus, Superior Temporal Gyrus, Supramarginal Gyrus, Angular Gyrus | 1 | ||
| 24 | Right Inferior Frontal Gyrus | 5, 10 | ||
| 25 | Right Supramarginal Gyrus, Superior Temporal Gyrus, Middle Temporal Gyrus | 1 | 1 | |
| LECN | 27 | Left Middle Frontal Gyrus, Superior Frontal Gyrus | 1, 2 | 1, 2 |
| 28 | Left Inferior Frontal Gyrus, Orbitofrontal Gyrus | 1, 2 | 1, 2, 3 | |
| 29 | Left Superior Parietal Gyrus, Inferior Parietal Gyrus, Precuneus, Angular Gyrus | 1 | 1, 2 | |
| 30 | Left Inferior Temporal Gyrus, Middle Temporal Gyrus | 2 | 1, 2 | |
| PSAL | 39 | Left Middle Frontal Gyrus | 1, 2 | |
| 40 | Left Supramarginal Gyrus, Inferior Parietal Gyrus | 1, 2 | 1, 2 | |
| 44 | Right Supramarginal Gyrus, Inferior Parietal Gyrus | 1 | 1 | |
| PRE | 52 | Precuneus | 1 | 1 |
| 53 | Left Angular Gyrus | 1, 2 | 1, 2, 3 | |
| 54 | Right Angular Gyrus | 1 | 1, 2 | |
| RECN | 57 | Right Middle Frontal Gyrus, Right Superior Frontal Gyrus | 1 | 1, 2 |
| 58 | Right Middle Frontal Gyrus | 1, 2, 10 | 1, 2, 3 | |
| 59 | Right Inferior Parietal Gyrus, Supramarginal Gyrus, Angular Gyrus | 1 | 1, 2 | |
| 60 | Right Superior Frontal Gyrus | 9, 10 | ||
| 61 | Left Crus 1, Crus II, Lobule VI | 1, 2 | ||
| 62 | Right Caudate | 7, 9 | 1 | |
| ASAL | 63 | Left Middle Frontal Gyrus | 1 | |
| 66 | Right Middle Frontal Gyrus | 1, 2 | 1, 2 | |
| 67 | Right Insula | 1 | ||
| VDMN | 73 | Left Middle Occipital Gyrus | 1, 2 | 1, 2 |
| 75 | Precuneus | 1 | ||
| 78 | Right Angular Gyrus, Middle Occipital Gyrus | 1 | 1, 2 | |
| VISUO | 85 | Right Inferior Parietal Lobule | 1 | 1 |
| 86 | Right Frontal Operculum. Inferior Frontal Gyrus | 1 | 1 | |
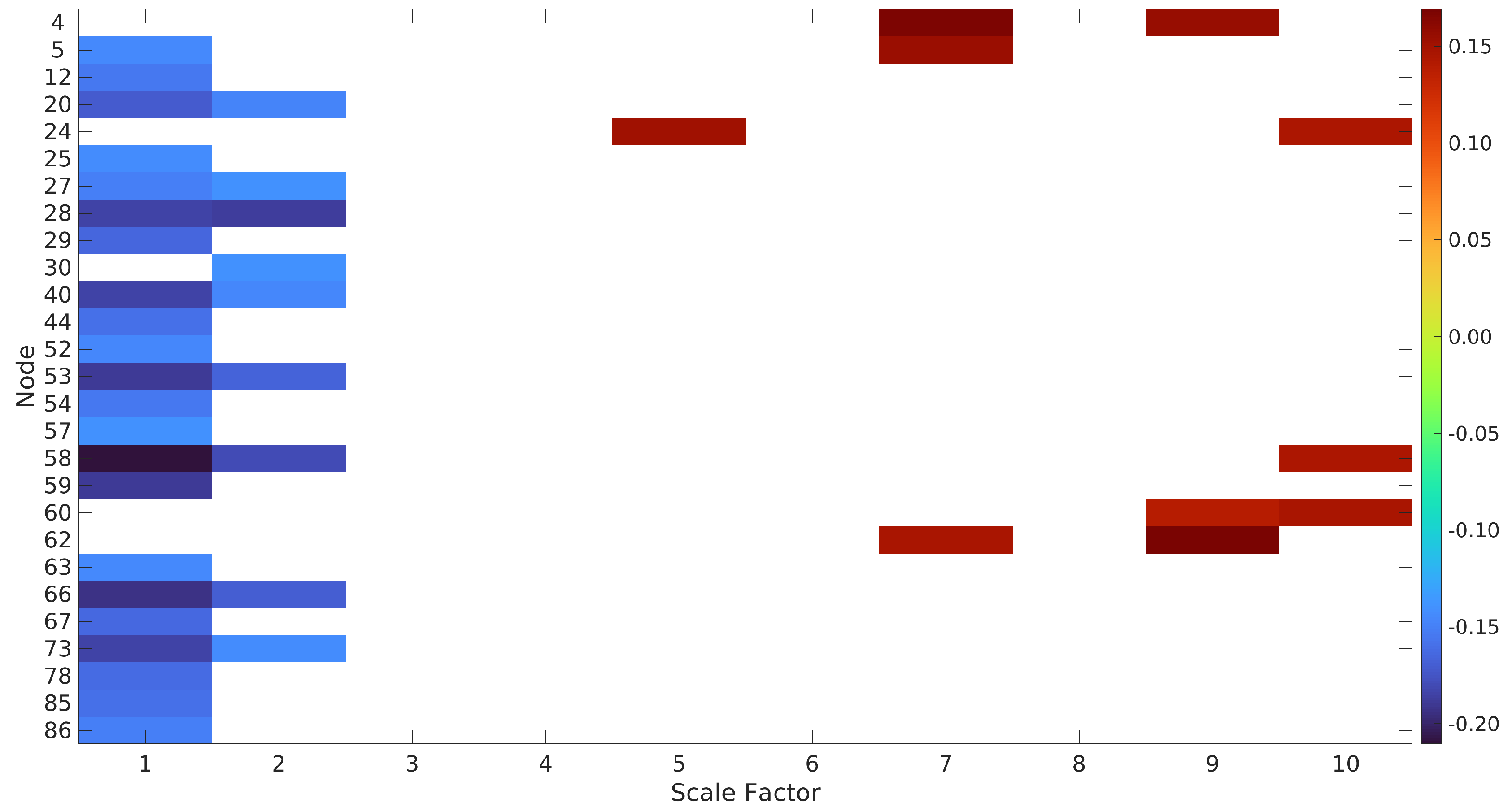
References
- Smith, S.M.; Vidaurre, D.; Beckmann, C.F.; Glasser, M.F.; Jenkinson, M.; Miller, K.L.; Nichols, T.E.; Robinson, E.C.; Salimi-Khorshidi, G.; Woolrich, M.W.; et al. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 2013, 17, 666–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirer, W.R.; Ryali, S.; Rykhlevskaia, E.; Menon, V.; Greicius, M.D. Decoding Subject-Driven Cognitive States with Whole-Brain Connectivity Patterns. Cereb. Cortex 2011, 22, 158–165. [Google Scholar] [CrossRef]
- Lee, M.; Smyser, C.; Shimony, J. Resting-State fMRI: A Review of Methods and Clinical Applications. Am. J. Neuroradiol. 2013, 34, 1866–1872. [Google Scholar] [CrossRef] [PubMed]
- Preti, M.G.; Bolton, T.A.; Van De Ville, D. The dynamic functional connectome: State-of-the-art and perspectives. NeuroImage 2017, 160, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Spinelli, E.G.; Cividini, C.; Agosta, F. Resting State Dynamic Functional Connectivity in Neurodegenerative Conditions: A Review of Magnetic Resonance Imaging Findings. Front. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.A.; Damaraju, E.; Plis, S.M.; Erhardt, E.B.; Eichele, T.; Calhoun, V.D. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 2014, 24, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Deco, G.; Cabral, J.; Woolrich, M.W.; Stevner, A.B.; van Hartevelt, T.J.; Kringelbach, M.L. Single or multiple frequency generators in on-going brain activity: A mechanistic whole-brain model of empirical MEG data. NeuroImage 2017, 152, 538–550. [Google Scholar] [CrossRef]
- Glerean, E.; Salmi, J.; Lahnakoski, J.M.; Jääskeläinen, I.P.; Sams, M. Functional Magnetic Resonance Imaging Phase Synchronization as a Measure of Dynamic Functional Connectivity. Brain Connect. 2012, 2, 91–101. [Google Scholar] [CrossRef]
- Cabral, J.; Kringelbach, M.L.; Deco, G. Functional connectivity dynamically evolves on multiple time-scales over a static structural connectome: Models and mechanisms. NeuroImage 2017, 160, 84–96. [Google Scholar] [CrossRef]
- Liu, J.; Liao, X.; Xia, M.; He, Y. Chronnectome fingerprinting: Identifying individuals and predicting higher cognitive functions using dynamic brain connectivity patterns. Hum. Brain Mapp. 2018, 39, 902–915. [Google Scholar] [CrossRef]
- Menon, S.S.; Krishnamurthy, K. A Comparison of Static and Dynamic Functional Connectivities for Identifying Subjects and Biological Sex Using Intrinsic Individual Brain Connectivity. Sci. Rep. 2019, 9, 5729. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Fu, Z.; Calhoun, V.D. Classification and Prediction of Brain Disorders Using Functional Connectivity: Promising but Challenging. Front. Neurosci. 2018, 12, 525. [Google Scholar] [CrossRef] [PubMed]
- Easson, A.K.; McIntosh, A.R. BOLD signal variability and complexity in children and adolescents with and without autism spectrum disorder. Dev. Cogn. Neurosci. 2019, 36, 100630. [Google Scholar] [CrossRef] [PubMed]
- Pool, R. Is it healthy to be chaotic? Science 1989, 243, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Vaillancourt, D.E.; Newell, K.M. Changing complexity in human behavior and physiology through aging and disease. Neurobiol. Aging 2002, 23, 1–11. [Google Scholar] [CrossRef]
- Sokunbi, M.O. Sample entropy reveals high discriminative power between young and elderly adults in short fMRI data sets. Front. Neuroinform. 2014, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Li, Y.; Childress, A.R.; Detre, J.A. Brain Entropy Mapping Using fMRI. PLoS ONE 2014, 9, e89948. [Google Scholar] [CrossRef]
- Lake, D.E.; Richman, J.S.; Griffin, M.P.; Moorman, J.R. Sample entropy analysis of neonatal heart rate variability. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2002, 283, R789–R797. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.C.; Tsai, S.J.; Lin, C.P.; Peng, C.K. A strategy to reduce bias of entropy estimates in resting-state fMRI signals. Front. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale Entropy Analysis of Complex Physiologic Time Series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.C.; Huang, C.C.; Yeh, H.L.; Liu, M.E.; Hong, C.J.; Tu, P.C.; Chen, J.F.; Huang, N.E.; Peng, C.K.; Lin, C.P.; et al. Complexity of spontaneous BOLD activity in default mode network is correlated with cognitive function in normal male elderly: a multiscale entropy analysis. Neurobiol. Aging 2013, 34, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.X.; Yan, L.; Wang, D.J. Multiple time scale complexity analysis of resting state FMRI. Brain Imaging Behav. 2014. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Wang, B.; Zhou, M.; Xue, J.; Shapour, H.; Cao, R.; Cui, X.; Wu, J.; Xiang, J. Dynamic Complexity of Spontaneous BOLD Activity in Alzheimer’s Disease and Mild Cognitive Impairment Using Multiscale Entropy Analysis. Front. Neurosci. 2018, 12, 677. [Google Scholar] [CrossRef] [PubMed]
- Grieder, M.; Wang, D.J.J.; Dierks, T.; Wahlund, L.O.; Jann, K. Default Mode Network Complexity and Cognitive Decmidrule in Mild Alzheimer’s Disease. Front. Neurosci. 2018, 12, 770. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Han, S.; Zhao, J.; Chen, H. Resting-State Brain Activity Complexity in Early-Onset Schizophrenia Characterized by a Multi-scale Entropy Method. In Intelligence Science and Big Data Engineering; Springer: Cham, Switzerland, 2017; pp. 580–588. [Google Scholar] [CrossRef]
- Jia, Y.; Gu, H. Identifying nonlinear dynamics of brain functional networks of patients with schizophrenia by sample entropy. Nonlinear Dyn. 2019, 96, 2327–2340. [Google Scholar] [CrossRef]
- Jia, Y.; Gu, H.; Luo, Q. Sample entropy reveals an age-related reduction in the complexity of dynamic brain. Sci. Rep. 2017, 7, 7990. [Google Scholar] [CrossRef]
- Essen, D.C.V.; Smith, S.M.; Barch, D.M.; Behrens, T.E.J.; Yacoub, E.; Ugurbil, K. The WU-Minn Human Connectome Project: An overview. NeuroImage 2013, 80, 62–79. [Google Scholar] [CrossRef] [Green Version]
- Gray, J.R.; Burgess, G.C.; Schaefer, A.; Yarkoni, T.; Larsen, R.J.; Braver, T.S. Affective personality differences in neural processing efficiency confirmed using fMRI. Cogn. Affect. Behav. Neurosci. 2005, 5, 182–190. [Google Scholar] [CrossRef]
- Bilker, W.B.; Hansen, J.A.; Brensinger, C.M.; Richard, J.; Gur, R.E.; Gur, R.C. Development of Abbreviated Nine-Item Forms of the Raven’s Standard Progressive Matrices Test. Assessment 2012, 19, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.R.; Chabris, C.F.; Braver, T.S. Neural mechanisms of general fluid intelligence. Nat. Neurosci. 2003, 6, 316–322. [Google Scholar] [CrossRef] [PubMed]
- McDonough, I.M.; Nashiro, K. Network complexity as a measure of information processing across resting-state networks: evidence from the Human Connectome Project. Front. Hum. Neurosci. 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Costa, Goldberger, and Peng Reply. Phys. Rev. Lett. 2004, 92, 089804. [Google Scholar] [CrossRef]
- Wang, D.J.J.; Jann, K.; Fan, C.; Qiao, Y.; Zang, Y.F.; Lu, H.; Yang, Y. Neurophysiological Basis of Multi-Scale Entropy of Brain Complexity and Its Relationship with Functional Connectivity. Front. Neurosci. 2018, 12. [Google Scholar] [CrossRef]
- Gignac, G.E.; Bates, T.C. Brain volume and intelligence: The moderating role of intelligence measurement quality. Intelligence 2017, 64, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Gignac, G.E.; Szodorai, E.T. Effect size guidelines for individual differences researchers. Personal. Individ. Differ. 2016, 102, 74–78. [Google Scholar] [CrossRef]
- Finn, E.S.; Shen, X.; Scheinost, D.; Rosenberg, M.D.; Huang, J.; Chun, M.M.; Papademetris, X.; Constable, R.T. Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat. Neurosci. 2015, 18, 1664–1671. [Google Scholar] [CrossRef]
- Noble, S.; Spann, M.N.; Tokoglu, F.; Shen, X.; Constable, R.T.; Scheinost, D. Influences on the Test–Retest Reliability of Functional Connectivity MRI and its Relationship with Behavioral Utility. Cereb. Cortex 2017, 27, 5415–5429. [Google Scholar] [CrossRef]
- Saxe, G.N.; Calderone, D.; Morales, L.J. Brain entropy and human intelligence: A resting-state fMRI study. PLoS ONE 2018. [Google Scholar] [CrossRef]
- Vakorin, V.A.; Lippe, S.; McIntosh, A.R. Variability of Brain Signals Processed Locally Transforms into Higher Connectivity with Brain Development. J. Neurosci. 2011, 31, 6405–6413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIntosh, A.R.; Vakorin, V.; Kovacevic, N.; Wang, H.; Diaconescu, A.; Protzner, A.B. Spatiotemporal Dependency of Age-Related Changes in Brain Signal Variability. Cereb. Cortex 2014, 24, 1806–1817. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.H.; Cole, M.W. Higher Intelligence Is Associated with Less Task-Related Brain Network Reconfiguration. J. Neurosci. 2016, 36, 8551–8561. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, M.A.; Anderson, J.S.; Spreng, R.N. Fluid and flexible minds: Intelligence reflects synchrony in the brain’s intrinsic network architecture. Netw. Neurosci. 2017, 1, 192–207. [Google Scholar] [CrossRef] [PubMed]
- Humeau-Heurtier, A. The Multiscale Entropy Algorithm and Its Variants: A Review. Entropy 2015, 17, 3110–3123. [Google Scholar] [CrossRef] [Green Version]
- Faes, L.; Porta, A.; Javorka, M.; Nollo, G. Efficient Computation of Multiscale Entropy over Short Biomedical Time Series Based on Linear State-Space Models. Complexity 2017, 2017, 1768264. [Google Scholar] [CrossRef]
- Faes, L.; Marinazzo, D.; Stramaglia, S. Multiscale Information Decomposition: Exact Computation for Multivariate Gaussian Processes. Entropy 2017, 19, 408. [Google Scholar] [CrossRef]
- Faes, L.; Pereira, M.A.; Silva, M.E.; Pernice, R.; Busacca, A.; Javorka, M.; Rocha, A.P. Multiscale information storage of linear long-range correlated stochastic processes. Phys. Rev. E 2019, 99, 032115. [Google Scholar] [CrossRef] [Green Version]
- Kosciessa, J.Q.; Kloosterman, N.A.; Garrett, D.D. Standard multiscale entropy reflects spectral power at mismatched temporal scales: What’s signal irregularity got to do with it? bioRxiv 2019. [Google Scholar] [CrossRef]
- Glasser, M.F.; Sotiropoulos, S.N.; Wilson, J.A.; Coalson, T.S.; Fischl, B.; Andersson, J.L.; Xu, J.; Jbabdi, S.; Webster, M.; Polimeni, J.R.; et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 2013, 80, 127. [Google Scholar] [CrossRef]
- Salimi-Khorshidi, G.; Douaud, G.; Beckmann, C.F.; Glasser, M.F.; Griffanti, L.; Smith, S.M. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. NeuroImage 2014, 90, 46. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.S.; Mitra, A.; Laumann, T.O.; Seitzman, B.A.; Raichle, M.; Corbetta, M.; Snyder, A.Z. Data quality influences observed links between functional connectivity and behavior. Cereb. Cortex 2017, 27, 4492–4502. [Google Scholar] [CrossRef] [PubMed]
- Dubois, J.; Galdi, P.; Paul, L.K.; Adolphs, R. A distributed brain network predicts general intelligence from resting-state human neuroimaging data. Philos. Trans. R. Soc. Biol. Sci. 2018, 373, 20170284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feinberg, D.A.; Moeller, S.; Smith, S.M.; Auerbach, E.; Ramanna, S.; Glasser, M.F.; Miller, K.L.; Ugurbil, K.; Yacoub, E. Multiplexed Echo Planar Imaging for Sub-Second Whole Brain FMRI and Fast Diffusion Imaging. PLoS ONE 2010, 5, e15710. [Google Scholar] [CrossRef] [PubMed]
- Bijsterbosch, J.; Smith, S.M.; Beckmann, C.F. Introduction to Resting State FMRI Functional Connectivity; Oxford University Press: Oxford, UK, 2017; p. 34. [Google Scholar]
- Cabral, J.; Vidaurre, D.; Marques, P.; Magalhães, R.; Silva Moreira, P.; Miguel Soares, J.; Deco, G.; Sousa, N.; Kringelbach, M.L. Cognitive performance in healthy older adults relates to spontaneous switching between states of functional connectivity during rest. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Dosenbach, N.U.F.; Nardos, B.; Cohen, A.L.; Fair, D.A.; Power, J.D.; Church, J.A.; Nelson, S.M.; Wig, G.S.; Vogel, A.C.; Lessov-Schlaggar, C.N.; et al. Prediction of Individual Brain Maturity Using fMRI. Science 2010, 329, 1358–1361. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Su, M.; Li, L.; Shu, H.; Gong, G. Individualized Prediction of Reading Comprehension Ability Using Gray Matter Volume. Cereb. Cortex 2018, 28, 1656–1672. [Google Scholar] [CrossRef]
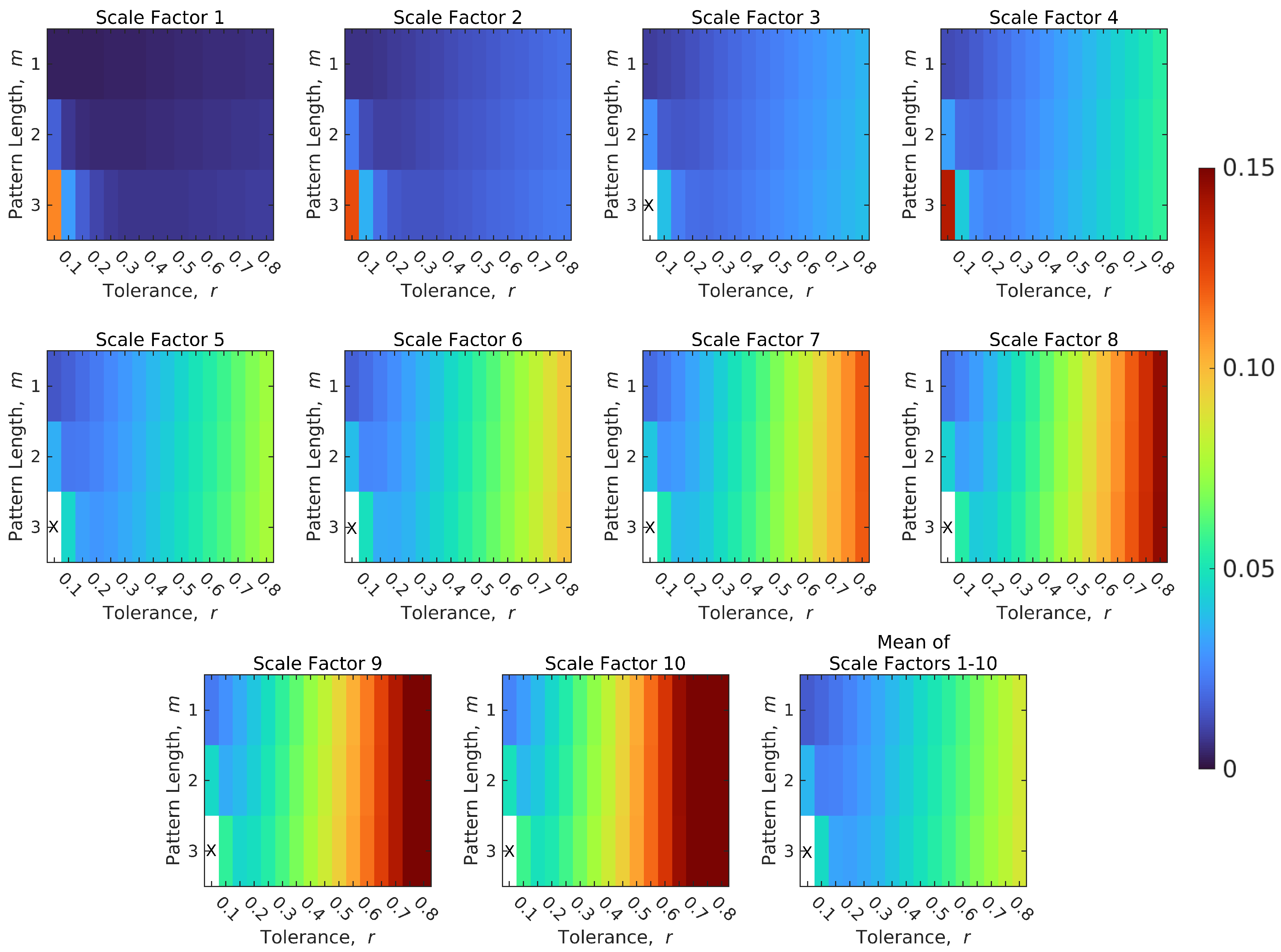






| Complexity Level | Pattern Length, m | Tolerance, r | Number of Selected Features | |||
|---|---|---|---|---|---|---|
| 25 | 50 | 100 | 900 | |||
| nMSE | 1 | 0.05 | 0.168 | 0.174 | 0.134 | 0.178 |
| 2 | 0.10 | 0.231 | 0.179 | 0.200 | 0.150 | |
| 3 | 0.20 | 0.200 | 0.249 | 0.133 | 0.158 | |
| Edge-based nMSE | 1 | 0.05 | 0.079 | 0.239 | 0.319 | 0.237 |
| 2 | 0.10 | 0.159 | 0.152 | 0.246 | 0.181 | |
| 3 | 0.20 | 0.178 | 0.202 | 0.195 | 0.207 | |
| eMSE | 1 | 0.05 | 0.180 | 0.127 | 0.171 | 0.165 |
| 2 | 0.10 | 0.160 | 0.238 | 0.210 | 0.126 | |
| 3 | 0.20 | 0.237 | 0.240 | 0.234 | 0.165 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menon, S.S.; Krishnamurthy, K. A Study of Brain Neuronal and Functional Complexities Estimated Using Multiscale Entropy in Healthy Young Adults. Entropy 2019, 21, 995. https://doi.org/10.3390/e21100995
Menon SS, Krishnamurthy K. A Study of Brain Neuronal and Functional Complexities Estimated Using Multiscale Entropy in Healthy Young Adults. Entropy. 2019; 21(10):995. https://doi.org/10.3390/e21100995
Chicago/Turabian StyleMenon, Sreevalsan S., and K. Krishnamurthy. 2019. "A Study of Brain Neuronal and Functional Complexities Estimated Using Multiscale Entropy in Healthy Young Adults" Entropy 21, no. 10: 995. https://doi.org/10.3390/e21100995





