Functional Analysis of Polyphenol Oxidases by Antisense/Sense Technology
Abstract
:Introduction
Polyphenol oxidase (PPO) and PPO genes
| Plant | Genomic/ cDNA | Tissue source | Number of genes | Transcript size (Kb) | Protein size (kDa) | Reference |
|---|---|---|---|---|---|---|
| Tomato | Genomic | NA 1 | 7 | 2.0 | 66-71 (57-62) 2 | [9c] |
| Potato | cDNA | Leaf/young tuber | 6 | 2.0 | 67-68 (57) | [9b, 9g] |
| Tobacco | cDNA | Stigma/style | 1 (≥2) 3 | 2.3 | 68 (58) | [9i] |
| Apple | Genomic/cDNA | Leaf/fruit peel/ young fruit | 2 (≥4) | 2.0-2.2 | 65-66 (55-57) | [9e, 65, 10b] |
| Japanese pear | Genomic | Leaf | 1 (≥2) | NA | 66 (56) | [16] |
| Broad bean | cDNA | Mature leaf | 3 | 2.2 | 68-70 (58-60) | [9a, 66] |
| Pineapple | cDNA | Blackheart flesh | 2 (≥4) | 2.1 | 69 (58) | [9l] |
| Sweet potato | Genomic | NA | 2 | 2.1 | 66 (56) | [67] |
| Grape | cDNA | Immature berry | 1 | 2.2 | 67 (40) | [68, 9d] |
| Apricot | cDNA | Immature fruit | 1 (≥2) | 2.2 | 67 (56) | [17c] |
| Plum (Nai) | cDNA | Leaf/pulp | 2 | 2.2 | 67-68 (57) | [69] |
| Hybrid poplar | cDNA | Leaf (systemically wounded) | 1 (2) | 2.1 | 64 (57) | [9j] |
| Trembling aspen | cDNA | Wounded leaf | 1 (2) | 2.1 | 65 (59) | [9k] |
| Pokeweed | cDNA | Suspension culture | 2 | 2.1-2.3 | 65 (54) | [9f] |
| Lettuce | cDNA | Young leaf | 1 | 2.1 | 68 (57) | [70] |
| Spinach | cDNA | Primary leaf/ etiolated cotyledon | 2 | 2.5 | 73 (62-64) | [71] |
| Banana | cDNA | Flesh and peel of small fruit | 4 (≥4) | 2.1 | 67 (56) | [12a, 70] |
| Sugarcane | cDNA | Immature stem | 1 (≥2) | 2.2 | 67 (59) | [9h] |
| Wheat | Genomic/ cDNA | Immature seed/ QTL/EST | 3 (≥6) | 2.0 | 57-68 (50-58) | [9m, 12b, 72] |
| Red clover | cDNA | Leaf | 3 (3-5) | 1.9 | 68-71 (57-60) | [9n] |
| Alfafa | Genomic | NA | 1 | NA | 69 (58) | [73] |
| Creosate bush | cDNA | Leaf | 1 | 1.8 | 66 (43) | [51] |
| Snapdragon | cDNA | Petal | 1 | 1.7 | 64 (39) | [4] |
| Tea | Genomic | NA | 3 | NA | 65-68 (54-57) | [74] |
- 1
- NA: not available.
- 2
- Predicted protein size (predicted mature protein size after N-terminal cleavage of a transit peptide and/or C-terminal cleavage).
- 3
- The number in parenthesis indicates expected number of genes in the genome estimated from Southern analysis.
Antisense/sense transgenic tomato
Role of PPO in disease resistance

Role of PPO in insect resistance
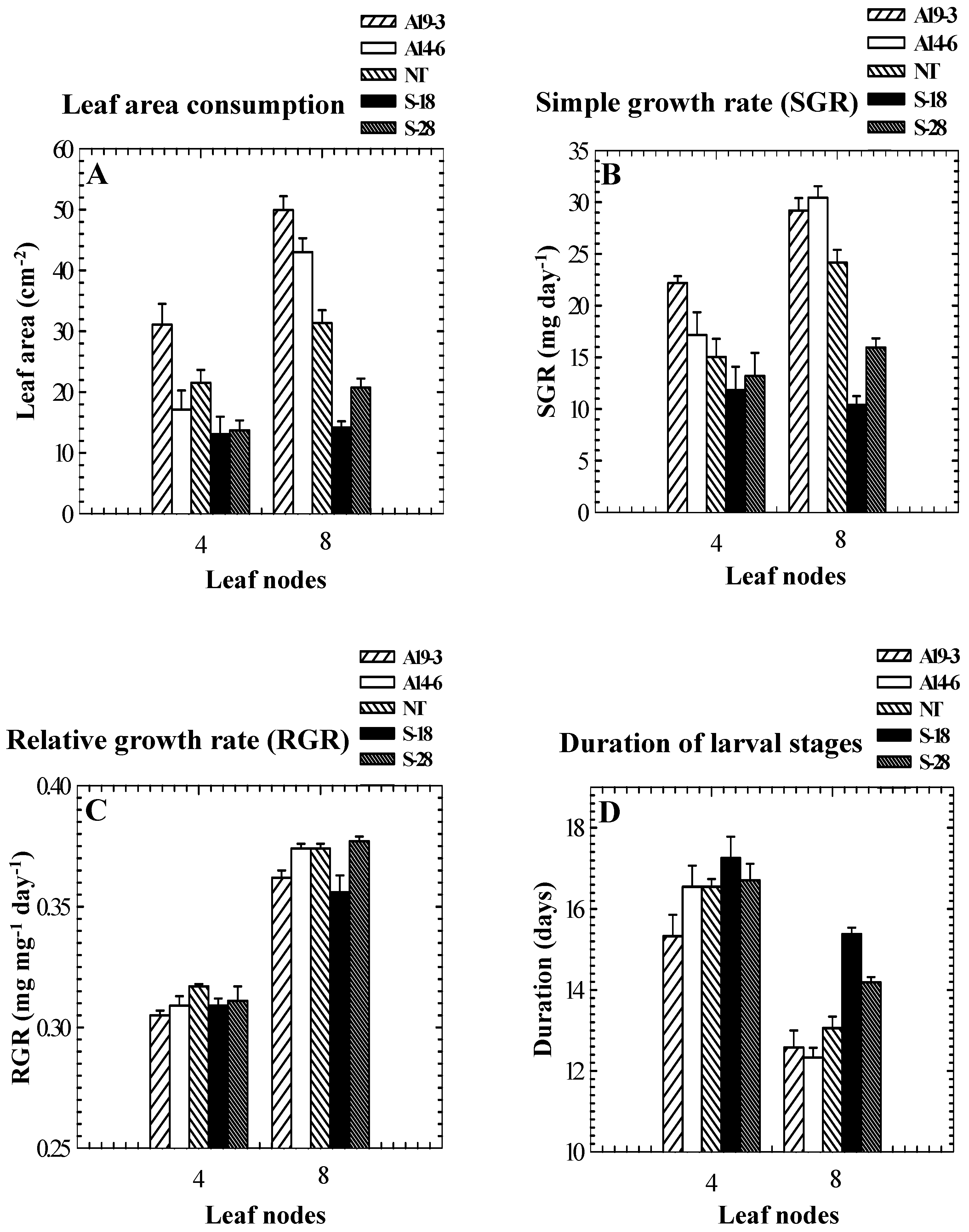
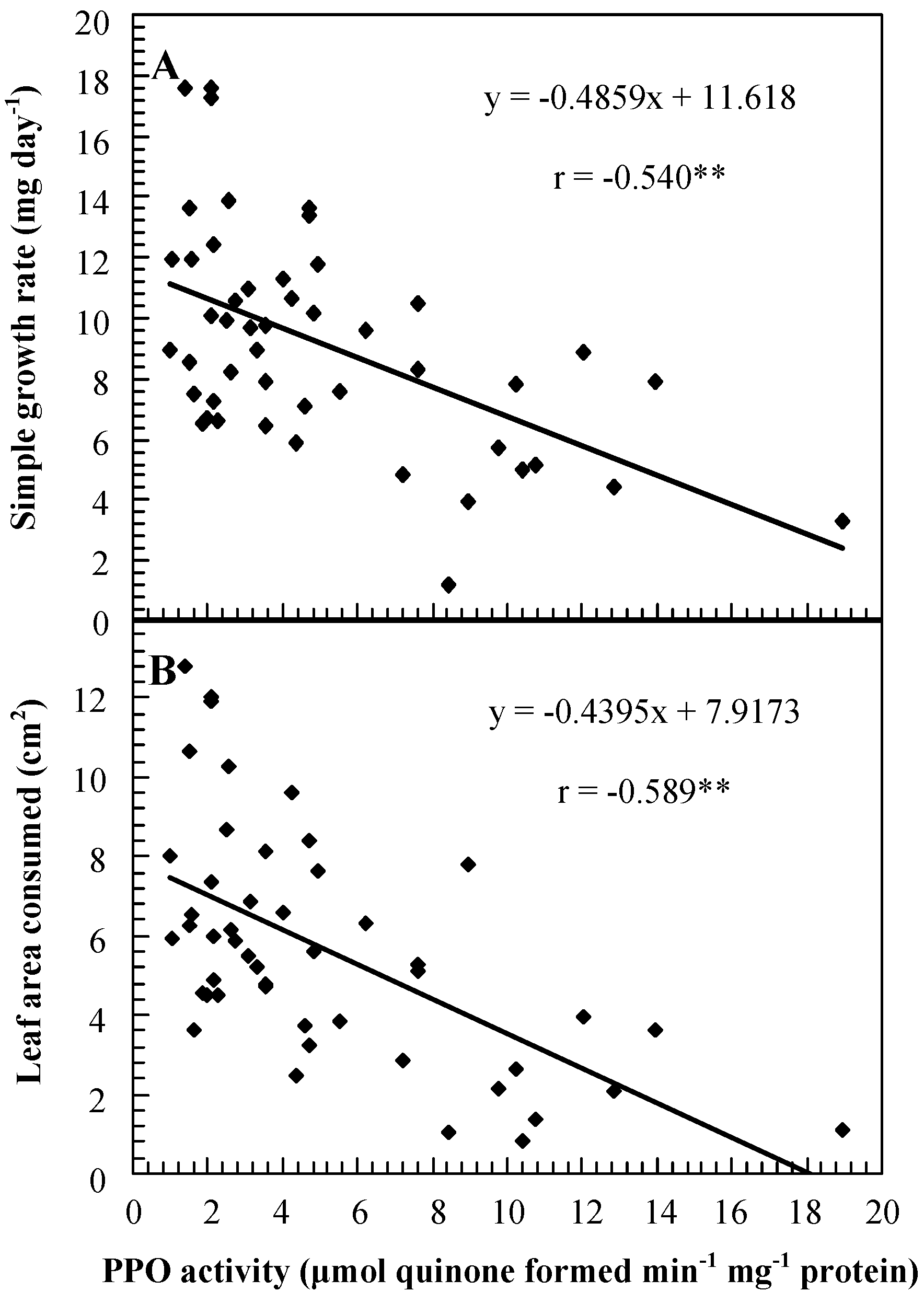
| Genotype | PPO activity (μmol quinone formed min-1 mg-1 protein) 1 | Percent mortality | Weight gain (mg)/larva | Foliage consumption (mg)/larva | Date of feeding assay |
|---|---|---|---|---|---|
| A14-6 2 | ND 3 | 45 a | 43 ± 1.9 a 4 | 123 ± 6.4 a | July 1993 |
| NT | 16.0 | 68 b | 32 ± 4.1 b | 76 ± 5.0 b | |
| A14-6 | ND | 3 | 50 ± 2.6 | 131 ± 12.3 | Dec. 1993 |
| NT | 7.1 | 10 | 44 ± 3.7 | 115 ± 13.7 |
- 1
- Values are from leaf nodes 1-2 and do not reflect the PPO activity of foliage fed to larvae (nodes 3-6).
- 2
- A14-6, A19-3, transgenic lines with suppressed PPO activity; NT, nontransformed control; S-18, S-28, transgenic lines overexpressing PPO activity.
- 3
- ND: not detectable.
- 4
- Data are presented as means ± SE. Data not followed by the same letter in a column are significantly different (p<0.05).
Role of PPO during water stress
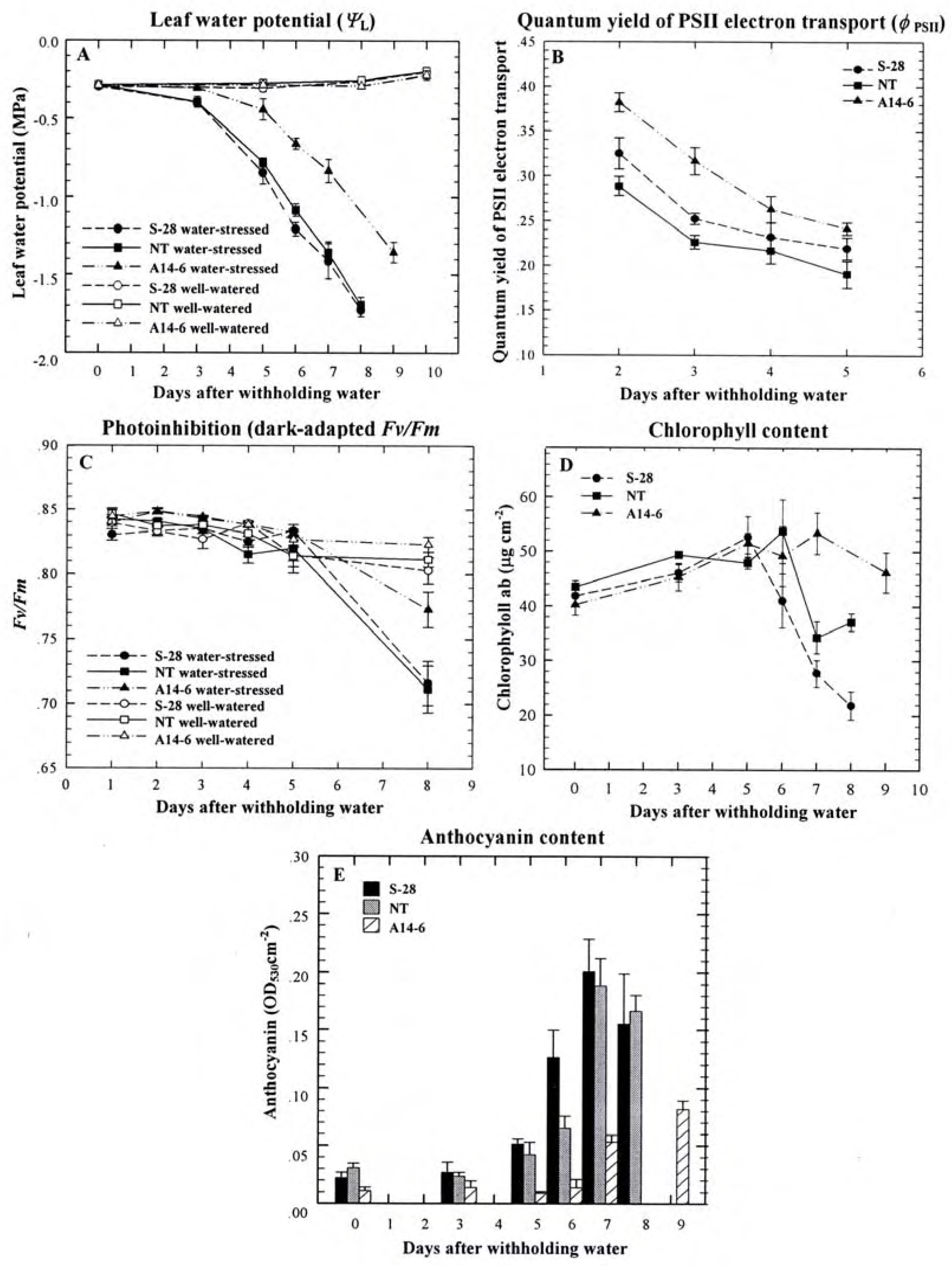
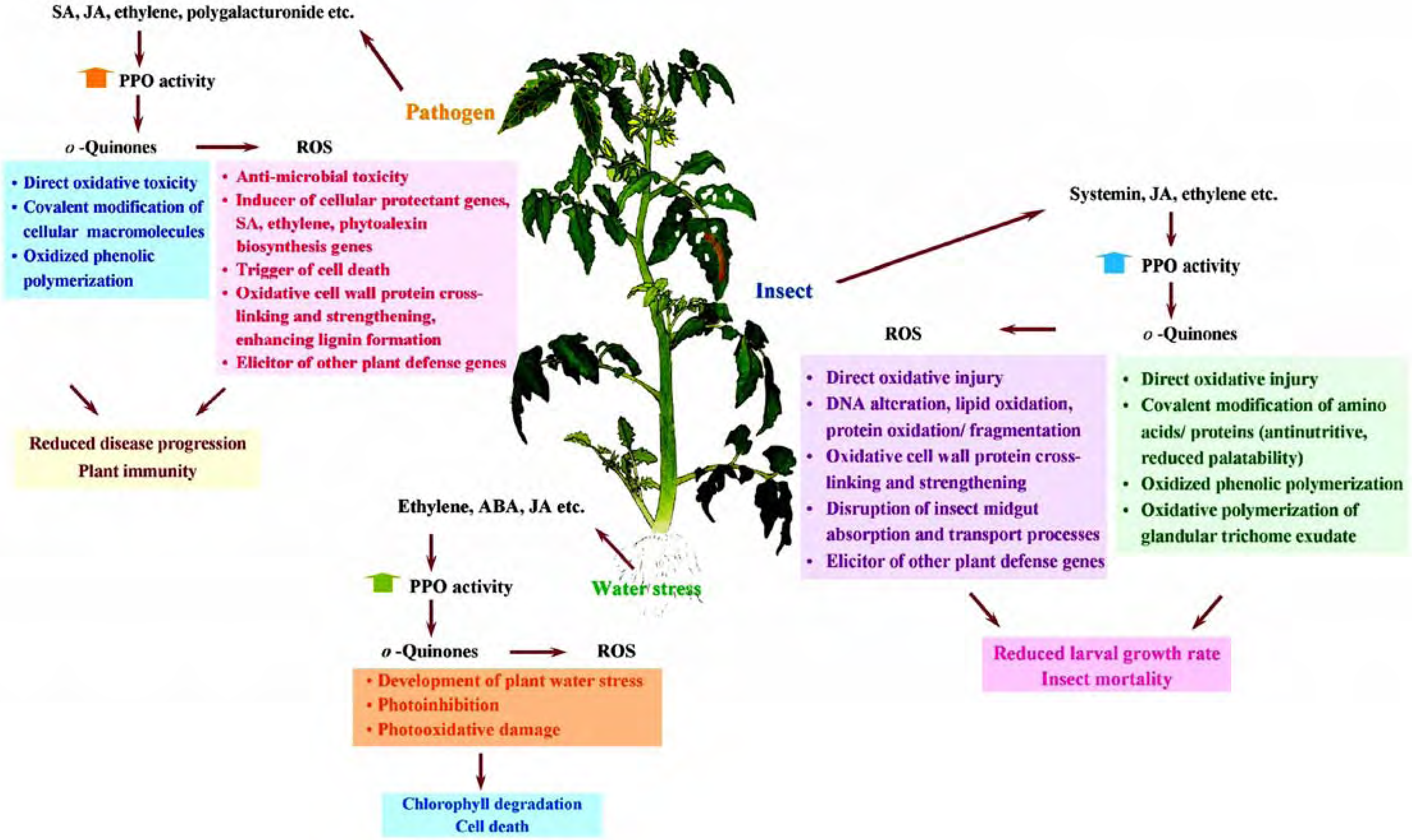
Other possible roles
Application of transgenic plants with modified PPO activity
Conclusion and future prospects
Acknowledgments
References and Notes
- Mayer, A. M.; Harel, E. Phenoloxidases and Their Significance in Fruit and Vegetables. In Food Enzymology; Fox P., F., Ed.; Elsevier: New York, 1991; pp. 373–398. [Google Scholar] Friedman, M. Chemistry, Biochemistry, and Dietary Role of Potato Polyphenols. J. Agric. Food Chem. 1997, 45, 1523–1540. [Google Scholar]
- Kojima, M.; Takeuchi, W. Detection and Characterization of p-Coumaric Acid Hydroxylase in Mungbean, Vigna mungo, Seedlings. J. Biochem. 1989, 105, 265–270. [Google Scholar]
- Vaughn, K. C.; Lax, A. R.; Duke, S. O. Polyphenol Oxidase: The Chloroplast Oxidase With No Established Function. Physiol. Plant. 1988, 72, 659–665. [Google Scholar] Trebst, A.; Depka, B. Polyphenol Oxidase and Photosynthesis Research. Photosynth. Res. 1995, 46, 41–44. [Google Scholar]
- Nakayama, T.; Yonekura-Sakakibara, K.; Sato, T.; Kikuchi, S.; Fukui, Y.; Fukuchi-Mizutani, M.; Ueda, T.; Nakao, M.; Tanaka, Y.; Kusumi, T.; Nishino, T. Aureusidin Synthase: A Polyphenol Oxidase Homolog Responsible for Flower Coloration. Science 2000, 290, 1163–1166. [Google Scholar] [CrossRef]
- Felton, G. W.; Donato, K. K.; Del Vecchio, R. J.; Duffey, S. S. Activation of Plant Foliar Oxidases by Insect Feeding Reduces Nutritive Quality of Foliage for Noctuid Herbivores. J. Chem. Ecol. 1989, 15, 2667–2694. [Google Scholar] Stout, M. J.; Workman, K. V.; Bostock, R. M.; Duffey, S. S. Stimulation and Attenuation of Induced Resistance by Elicitors and Inhibitors of Chemical Induction in Tomato (Lycopersicon esculentum) Foliage. Entomol. Exper. Appli. 1998, 86, 267–279. [Google Scholar] Li, L.; Steffens, J. C. Overexpression of Polyphenol Oxidase in Transgenic Tomato Plants Results in Enhanced Bacterial Disease Resistance. Planta 2002, 215, 239–247. [Google Scholar] Thipyapong, P.; Hunt, M. D.; Steffens, J. C. Antisense Downregulation of Polyphenol Oxidase Results in Enhanced Disease Susceptibility. Planta 2004, 220, 105–117. [Google Scholar] Wang, J.; Constabel, C. P. Polyphenol Oxidase Overexpression in Transgenic Populus Enhances Resistance to Herbivory by Forest Tent Caterpillar (Malacosoma disstria). Planta 2004, 220, 87–96. [Google Scholar] Barbehenn, R. V.; Jones, C. P.; Yip, L.; Tran, L.; Constabel, C. P. Does the Induction of Polyphenol Oxidase Defend Trees against Caterpillars? Assessing Plant Defenses One at a Time with Transgenic Poplar. Oecologia 2007, 220. in press. [Google Scholar]
- Mayer, A. M.; Harel, E. Polyphenol Oxidases in Plants. Phytochemistry 1979, 18, 193–215. [Google Scholar] Constabel, C. P.; Bergey, D. R.; Ryan, C. A. Systemin Activates Synthesis of Wound-inducible Tomato Leaf Polyphenol Oxidase via the Octadecanoid Defense Signaling Pathway. Proc. Natl. Acad. Sci. USA 1995, 92, 407–411. [Google Scholar] Thipyapong, P.; Steffens, J. C. Tomato Polyphenol Oxidase: Differential Response of the Polyphenol Oxidase F Promoter to Injuries and Wound Signals. Plant Physiol. 1997, 115, 409–418. [Google Scholar] Maki, H.; Morohashi, Y. Development of Polyphenol Oxidase Activity in the Micropylar Endosperm of Tomato Seeds. J. Plant Physiol. 2006, 163, 1–10. [Google Scholar]
- Steffens, J. C.; Harel, E.; Hunt, M. D. (1994). Polyphenol Oxidase. In Genetic Engineering of Plant Secondary Metabolism; Ellis, B. E., et al., Eds.; Plenum Press: New York, 1994; pp. 275–312. [Google Scholar]
- Gandía-Herrero, F.; Escribano, J.; García-Carmona, F. Characterization of the Monophenolase Activity of Tyrosinase on Betaxanthins: The Tyramine-betaxanthin/dopamine-betaxanthin Pair. Planta 2005, 222, 307–318. [Google Scholar] [CrossRef]
- Cary, J. W.; Lax, A. R.; Flurkey, W. H. Cloning and Characterization of cDNAs Coding for Vicia faba Polyphenol Oxidase. Plant Mol. Biol. 1992, 20, 245–253. [Google Scholar] Hunt, M. D.; Eannetta, N. T.; Yu, H.; Newman, S. M.; Steffens, J. C. cDNA Cloning and Expression of Potato Polyphenol Oxidase. Plant Mol. Biol. 1993, 21, 59–68. [Google Scholar] Newman, S. M.; Eannetta, N. T.; Yu, H.; Prince, J. P.; Carmen de Vicente, M.; Tanksley, S. D.; Steffens, J. C. Organisation of the Tomato Polyphenol Oxidase Gene Family. Plant Mol. Biol. 1993, 21, 1035–1051. [Google Scholar] Dry, I. B.; Robinson, S. P. Molecular Cloning and Characterization of Grape Berry Polyphenol Oxidase. Plant Mol. Biol. 1994, 26, 495–502. [Google Scholar] Boss, P. K.; Gardner, R. C.; Janssen, B.-J.; Ross, G. S. An Apple Polyphenol Oxidase cDNA is Up-regulated in Wounded Tissues. Plant Mol. Biol. 1995, 27, 429–433. [Google Scholar] Joy IV, R. W.; Sugiyama, M.; Fukuda, H.; Komamine, A. Cloning and Characterization of Polyphenol Oxidase cDNAs of Phytolacca americana. Plant Physiol 1995, 107, 1083–1089. [Google Scholar] Thygesen, P. W.; Dry, I. B.; Robinson, S. P. Polyphenol Oxidase in Potato: A Multigene Family that Exhibits Differential Expression Patterns. Plant Physiol 1995, 109, 525–531. [Google Scholar] Bucheli, C. S.; Dry, I. B.; Robinson, S. P. Isolation of a Full-length cDNA Encoding Polyphenol Oxidase from Sugarcane, a C4 Grass. Plant Mol. Biol. 1996, 31, 1233–1238. [Google Scholar] Goldman, M. H. S.; Seurinck, J.; Marins, M.; Goldman, G. H.; Mariani, C. A Tobacco Flower-specific Gene Encodes a Polyphenol Oxidase. Plant Mol. Biol. 1998, 36, 479–485. [Google Scholar] Constabel, C. P.; Yip, L.; Patton, J. J.; Christopher, M. E. Polyphenol Oxidase from Hybrid Poplar: Cloning and Expression in Response to Wounding and Herbivory. Plant Physiol. 2000, 124, 285–295. [Google Scholar] Haruta, M.; Pedersen, J. A.; Constabel, C. P. Polyphenol Oxidase and Herbivore Defense in Trembling Aspen (Populus tremuloides): cDNA Cloning, Expression, and Potential Substrates. Plant Physiol. 2001, 112, 552–558. [Google Scholar] Stewart, R. J.; Sawyer, B. J. B.; Bucheli, C. S.; Robinson, S. P. Polyphenol Oxidase is Induced by Chilling and Wounding in Pineapple. Aust. J. Plant Physiol. 2001, 28, 181–191. [Google Scholar] Demeke, T.; Morris, C. F. Molecular Characterization of Wheat Polyphenol Oxidase (PPO). Theor. Appl. Genet. 2002, 104, 813–818. [Google Scholar] Sullivan, M. L.; Hatfield, R. D.; Thoma, S. L.; Samac, D. A. Cloning and Characterization of Red Clover Polyphenol Oxidase cDNAs and Expression of Active Protein in Escherichia coli and Transgenic Alfalfa. Plant Physiol. 2004, 136, 3234–3244. [Google Scholar]
- Thipyapong, P.; Joel, D. M.; Steffens, J. C. Differential Expression and Turnover of the Tomato Polyphenol Oxidase Gene Family during Vegetative and Reproductive Development. Plant Physiol. 1997, 113, 707–718. [Google Scholar] Kim, J. Y.; Seo, Y. S.; Kim, J. E.; Sung, S.-K.; Song, K. J.; An, G.; Kim, W. T. Two Polyphenol Oxidases are Differentially Expressed during Vegetative and Reproductive Development and in Response to Wounding in the Fuji Apple. Plant Sci. 2001, 161, 1145–1152. [Google Scholar]
- Ono, E.; Hatayama, M.; Isono, Y.; Sato, T.; Watanabe, R.; Yonekura-Sakakibara, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Kusumi, T.; Nishino, T.; Nakayama, T. Localization of a Flavonoid Biosynthetic Polyphenol Oxidase in Vacuoles. Plant J. 2006, 45, 133–143. [Google Scholar] [CrossRef]
- Gooding, P. S.; Bird, C.; Robinson, S. P. Molecular Cloning and Characterisation of Banana Fruit Polyphenol Oxidase. Planta 2001, 213, 748–757. [Google Scholar] Anderson, J. V.; Morris, C. F.; Robinson, S. P. Characterization and Expression of Polyphenol Oxidase in Developing Grain of Wheat (Triticum aestivum L.). In Plant Biology 2003; Honolulu, Hawaii, USA, 2003. [Google Scholar]
- Thipyapong, P.; Melkonian, J.; Wolfe, D. W.; Steffens, J. C. Suppression of Polyphenol Oxidases Increases Stress Tolerance in Tomato. Plant Sci. 2004, 167, 693–703. [Google Scholar] [CrossRef]
- Hind, G.; Marshak, D. R.; Coughlan, S. J. Spinach Thylakoid Polyphenol Oxidase: Cloning, Characterization, and Relation to a Putative Protein Kinas. Biochemistry 1995, 34, 8157–8164. [Google Scholar] [CrossRef]
- Haruta, M.; Murata, M.; Kadokura, H.; Homma, S. Immunological and Molecular Comparison of Polyphenol Oxidase in Rosaceae Fruit Trees. Phytochemistry 1999, 50, 1021–1025. [Google Scholar] [CrossRef]
- Nishimura, M.; Fukuda, C.; Murata, M.; Homma, S. Cloning and Some Properties of Japanese Pear (Pyrus pyrifolia) Polyphenol Oxidase, and Changes in Browning Potential during Fruit Maturation. J. Sci. Food Agric. 2003, 83, 1156–1162. [Google Scholar] [CrossRef]
- Shahar, T.; Hennig, N.; Gutfinger, T.; Hareven, D.; Lifschitz, E. The Tomato 66.3-kD Polyphenoloxidase Gene: Molecular Identification and Developmental Expression. Plant Cell 1992, 4, 135–147. [Google Scholar] Thipyapong, P.; Hunt, M. D.; Steffens, J. C. Systemic Wound Induction of Potato (Solanum tuberosum) Polyphenol Oxidase. Plant Cell 1995, 40, 673–676. [Google Scholar] Chevalier, T.; de Rigal, D.; Mbéguié-A-Mbéguié, D.; Gauillard, F.; Richard-Forget, F.; Fils-Lycaon, B. R. Molecular Cloning and Characterization of Apricot Fruit Polyphenol Oxidase. Plant Physiol. 1999, 119, 1261–1269. [Google Scholar]
- Gleave, A. P. A Versatile Binary Vector System With a T-DNA Organisational Structure Conducive to Efficient Integration of Cloned DNA into the Plant Genome. Plant Mol. Biol. 1992, 20, 1203–1207. [Google Scholar] [CrossRef]
- Thipyapong, P. Polyphenol Oxidase Gene Family: Differential Expression during Vegetative and Reproductive Development, and in Response to Injuries, and Defensive Functional Analysis. Ph.D. Thesis, Cornell University, Ithaca, New York, 1997. [Google Scholar]
- Levine, A.; Pennell, R. I.; Alvarez, M. E.; Palmer, R.; Lamb, C. Calcium-mediated Apoptosis in a Plant Hypersensitive Disease Resistance Response. Current Biol. 1996, 6, 427–437. [Google Scholar] Chamnongpol, S.; Willekens, H.; Moeder, W.; Langebartels, C.; Sandermann, H., Jr.; Montagu, M. V.; Inzé, D.; Camp, W. V. Defense Activation and Enhanced Pathogen Tolerance Induced by H2O2 in Transgenic Tobacco. Proc. Natl. Acad. Sci. USA 1998, 95, 5818–5823. [Google Scholar] Grant, J. J.; Loake, G. J. Role of Reactive Oxygen Intermediates and Cognate Redox Signaling in Disease Resistance. Plant Physiol. 2000, 124, 21–29. [Google Scholar] Garcia-Olmedo, F.; Rodriguez-Palenzuela, P.; Molina, A.; Alamillo, J. M.; Lopez-Solanilla, E.; Berrocal-Lobo, M.; Poza-Carrion, C. Antibiotic Activities of Peptides, Hydrogen Peroxide and Peroxynitrite in Plant Defence. FEBS Letters 2001, 498, 219–222. [Google Scholar]
- Campos, A. D.; Ferreira, A. G.; Vozarí Hampe, M. M.; Antunes, I. F.; Brancão, N.; da Silveira, E. P.; Osório, V. A.; Augustin, E. Peroxidase and Polyphenol Oxidase Activity in Bean Anthracnose Resistance. Pesq. agropec. bras., Brasilia. 2004, 39, 637–643. [Google Scholar]
- Tyagi, M.; Kayastha, A. M.; Sinha, B. The Role of Peroxidase and Polyphenol Oxidase Isozymes in Wheat Resistance to Alternaria triticina. Biol. Plant. 2000, 43, 559–562. [Google Scholar] Raj, S. N.; Sarosh, B. R.; Shetty, H. S. Induction and Accumulation of Polyphenol Oxidase Activities as Implicated in Development of Resistance Against Pearl Millet Downy Mildew Disease. Funct. Plant Biol. 2006, 33, 563–571. [Google Scholar]
- Krishnamoorthy, V.; Kumar, N.; Angappan, K.; Soorianathasundaram, K. Evaluation of New Banana Hybrids Against Black Leaf Streak Disease. Infomusa 2004, 13, 25–27. [Google Scholar]
- Banga, S. S.; Kaur, K.; Ahuja, K. L.; Banga, S. K. Introgression and Biochemical Manifestation of the Gene(s) for White Rust Resistance in Indian Mustard (Brassicajuncea (L.) Coss.) [online]. 2004. available from: http://www.cropscience.org.au/icsc2004/poster/3/7/3/296_banga.htm.
- Tomiyama, K.; Stahmann, M. A. Alteration of Oxidative Enzymes in Potato Tuber Tissue by Infection with Phytophthora infestans. Plant Physiol. 1964, 39, 483–490. [Google Scholar] [CrossRef]
- Mohammadi, M.; Kazemi, H. Changes in Peroxidase and Polyphenol Oxidase Activities in Susceptible and Resistant Wheat Heads Inoculated with Fusarium graminearum and Induced Resistance. Plant Sci. 2002, 162, 491–498. [Google Scholar] Honty, K.; Hevesi, M.; Tóth, M.; Stefanovíts-Bányai, É. Some Biochemical Changes in Pear Fruit Tissue Induced by Erwinia aymlovora. Acta Biol. Szeged 2005, 49, 127–129. [Google Scholar]
- Stahmann, M. A.; Clare, B. G.; Woodbury, W. Increase Disease Resistance and Enzyme Activity Induced by Ethylene and Ethylene Production by Black Rot Infected Sweet Potato Tissue. Plant Physiol. 1966, 41, 1505–1512. [Google Scholar] [CrossRef]
- Liang, Y. C.; Sun, W. C.; Si, J.; Römheld, V. Effects of Foliar- and Root-applied Silicon on the Enhancement of Induced Resistance to Powdery Mildew in Cucumis sativus. Plant Pathol. 2005, 54, 678–685. [Google Scholar] [CrossRef]
- Mathur, N.; Vyas, A. Biochemical Changes in Ziziphus xylopyrus by VA Mycorrhizae. Bot. Bull. Acad. Sin. 1996, 37, 209–212. [Google Scholar] Panwar, J.; Vyas, A. AM Fungi: A Biological Approach Towards Conservation of Endangered Plants in Thar Desert, India. Curr. Sci. 2002, 82, 576–578. [Google Scholar]
- Kavitha, P. G.; Varadarajan, R.; Jonathan, E. I. Induced Systemic Resistance of Pseudomonas fluorescens Against Root-knot Nematode, Meloidogyne incognita in Tomato. Resistant Pest Management Newsletter n.d., [ http://whalonlab.msu.edu/rpmnews/vol.15_no.2/globe/Kavitha_etal.htm], 15.
- Gailite, A.; Samsone, I.; Ievinsh, G. Ethylene is Involved in Trichoderma-induced Resistance of Bean Plants Against Pseudomonas syringae. Acta Universitatis Latviensis, Biology 2005, 691, 59–70. [Google Scholar]
- Wegener, C. B.; Olsen, O. Heterologous Pectate Lyase Isoenzymes are Not Different in Their Effects on Soft Rot Resistance in Transgenic Potatoes. Physiol. Mol. Plant Pathol. 2004, 65, 59–66. [Google Scholar] [CrossRef]
- Punja, Z. K. Genetic Engineering of Plants to Enhance Resistance to Fungal Pathogens-A Review of Progress and Future Prospects. Can. J. Plant Pathol. 2001, 23, 216–235. [Google Scholar]
- Castañera, P.; Steffens, J. C.; Tingey, W. M. Biological Performance of Colorado Potato Beetle Larvae on Potato Genotypes with Differing Levels of Polyphenol Oxidase. J. Chem. Ecol. 1996, 22, 91–101. [Google Scholar] Ramiro, D. A.; Guerreiro-Filho, O.; Mazzafera, P. Phenol Contents, Oxidase Activities, and the Resistance of Coffee to the Leaf Miner Leucoptera coffeella. J. Chem. Ecol. 2006, 32, 1997–1988. [Google Scholar]
- Duffey, S. S.; Stout, M. J. Antinutritive and Toxic Components of Plant Defense Against Insects. Arch. Insect Biochem. Physiol. 1996, 32, 3–37. [Google Scholar] [CrossRef]
- Thaler, J. S.; Karban, R.; Ullman, D. E.; Boege, K.; Bostock, R. M. Cross-talk Between Jasmonate and Salicylate Plant Defense Pathways: Effects on Several Plant Parasites. Oecologia 2002, 131, 227–235. [Google Scholar] Cooper, W. C.; Jia, L.; Goggin, F. L. Acquired and R-gene-mediated Resistance Against the Potato Aphid in Tomato. J. Chem. Ecol. 2004, 30, 2527–2542. [Google Scholar]
- Ren, F.; Lu, Y.-T. Overexpression of Tobacco Hydroxyproline-rich Glycopeptide Systemin Precursor A Gene in Transgenic Tobacco Enhances Resistance Against Helicoverpa armigera Larvae. Plant Sci. 2006, 171, 286–292. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.; McCaig, B. C.; Wingerd, B. A.; Wang, J.; Whalon, M. E.; Pichersky, E.; Howe, G. A. The Tomato Homolog of Coronatine-insensitive1 is Required for the Maternal Control of Seed Maturation, Jasmonate-signaled Defense Responses, and Glandular Trichome Developmen. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef]
- Felton, G. W.; Donato, K. K.; Broadway, R. M.; Duffey, S. S. Impact of Oxidized Plant Phenolics on the Nutritional Quality of Dietary Protein to a Noctuid Herbivore, Spodoptera exigua. J. Insect Physiol. 1992, 38, 277–285. [Google Scholar] [CrossRef]
- Ananthakrishnan, T. N. Applied Chemical Ecology: Induced and Transgenic Defences in Insect-plant Interactions. Current Sci. 2003, 84, 492–493. [Google Scholar]
- Hoover, K.; Stout, M. J.; Alaniz, S. A.; Hammock, B. D.; Duffey, S. S. Influence of Induced Plant Defenses in Cotton and Tomato on the Efficacy of Baculoviruses on Noctuid Larvae. J. Chem. Ecol. 1998, 24, 253–271. [Google Scholar]
- Tolbert, N. E. Activation of Polyphenol Oxidase of Chloroplasts. Plant Physiol. 1973, 51, 234–244. [Google Scholar] Vaughn, K. C.; Duke, S. O. Function of Polyphenol Oxidase in Higher Plants. Physiol. Plant. 1984, 60, 106–112. [Google Scholar]
- Badger, M. R.; von Caemmerer, S.; Ruuska, S.; Nakano, H. Electron Flow to Oxygen in Higher Plants and Algae: Rates and Control of Direct Photoreduction (Mehler Reaction) and Rubisco Oxygenase. Phil. Trans. R. Soc. Lond. B. 2000, 355, 1433–1446. [Google Scholar] Haupt-Herting, S.; Fock, H. P. Oxygen Exchange in Relation to Carbon Assimilation in Water-stressed Leaves during Photosynthesis. Annals Bot. 2002, 89, 851–859. [Google Scholar]
- Asada, K.; Takahashi, M. Production and Scavenging of Active Oxygen in Chloroplasts. In Photoinhibition; Kyle, D. J., Osmond, C. B., Arntzen, C. J., Eds.; Elsevier: Amsterdam, 1987; pp. 227–287. [Google Scholar]
- Demmig, B.; Bjorkman, O. Comparison of the Effect of Excessive Light on Chlorophyll Fluorescence (77 K) and Photon Yield of O2 Evolution in Leaves of Higher Plants. Planta 1987, 171, 171–184. [Google Scholar] [CrossRef]
- English-Loeb, G.; Stout, M. J.; Duffey, S. S. Drought Stress in Tomatoes: Changes in Plant Chemistry and Potential Nonlinear Consequences for Insect Herbivores. OIKOS 1997, 79, 456–468. [Google Scholar] [CrossRef]
- Shivishankar, S. Polyphenol Oxidase Isozymes in Coconut Genotypes under Water Stress. Plant Physiol. Biochem. 1988, 15, 87–91. [Google Scholar]
- Niknam, V.; Razavi, N.; Ebrahimzadeh, H.; Sharifizadeh, B. Effect of NaCl on Biomass, Protein and Proline Contents, and Antioxidant Enzymes in Seedlings and Calli of Two Trigonella Species. Biol. Plant. 2006, 50, 591–596. [Google Scholar] [CrossRef]
- Balakumar, T.; Gayathri, B.; Anbudurai, P. R. Oxidative Stress Injury in Tomato Plants Induced by Supplemental UV-B Radiation. Biol. Plant. 1997, 39, 215–221. [Google Scholar] [CrossRef]
- Sherman, T. D.; Vaughn, K. C.; Duke, S. O. A Limited Survey of the Phylogenetic Distribution of Polyphenol Oxidase. Phytochemistry 1991, 30, 2499–2506. [Google Scholar] [CrossRef]
- Cho, M.-H.; Moinuddin, S. G. A.; Helms, G. L.; Hishiyama, S.; Eichinger, D.; Davin, L. B.; Lewis, N. G. (+)-Larreatricin Hydroxylase, an Enantio-specific Polyphenol Oxidase from the Creosote Bush (Larrea tridentata). Proc. Natl. Acad. Sci. USA 2003, 100, 10641–10646. [Google Scholar] [CrossRef]
- Vaughn, K. C.; Duke, S. O. Tissue Localization of Polyphenol Oxidase in Sorghum. Protoplasma 1981, 108, 319–327. [Google Scholar] Vaughn, K. C.; Duke, S. O. Tentoxin Effects on Sorghum: The Role of Polyphenol Oxidase. Protoplasma 1982, 110, 48–53. [Google Scholar]
- Franke, R.; Humphreys, J. M.; Hemm, M. R.; Denault, J. W.; Ruegger, M. O.; Cusumano, J. C.; Chapple, C. The Arabidopsis REF8 Gene Encodes the 3-Hydroxylase of Phenylpropanoid Metabolism. Plant J. 2002, 30, 33–45. [Google Scholar] [CrossRef]
- Sommer, A.; Néeman, E.; Steffens, J. C.; Mayer, A. M.; Harel, E. Import, Targeting and Processing of a Plant Polyphenol Oxidase. Plant Physiol. 1994, 105, 1301–1311. [Google Scholar]
- Sullivan, M. L.; Hatfield, R. D. Polyphenol Oxidase and o-Diphenols Inhibit Postharvest Proteolysis in Red Clover and Alfalfa. Crop Sci. 2006, 46, 662–670. [Google Scholar] [CrossRef]
- Ebrahimzadeh, H.; Abrishamchi, P. Changes in IAA, Phenolic Compounds, Peroxidase, IAA Oxidase, and Polyphenol Oxidase in Relation to Flower Formation in Crocus sativus. Russian J. Plant Physiol. 2001, 48, 190–195. [Google Scholar] Lavid, N.; Schwartz, A.; Lewinsohn, E.; Tel-Or, E. Phenols and Phenol Oxidases are Involved in Cadmium Accumulation in the Water Plants Nymphoides peltata (Menyanthaceae) and Nymphaeae (Nymphaeaceae). Planta 2001, 214, 189–195. [Google Scholar]
- Spagna, G.; Barbagallo, R. N.; Chisari, M.; Branca, F. Characterization of a Tomato Polyphenol Oxidase and Its Role in Browning and Lycopene Content. J. Agric. Food Chem. 2005, 53, 2032–2038. [Google Scholar] [CrossRef]
- Bachem, C. W. B.; Speckmann, G. J.; van der Linde, P. C. G.; Verheggen, F. T. M.; Hunt, M. D.; Steffens, J. C.; Zabeau, M. Antisense Expression of Polyphenol Oxidase Genes Inhibits Enzymatic Browning in Potato Tubers. Biotechnology 1994, 12, 1101–1105. [Google Scholar] Coetzer, C.; Corsini, D.; Love, S.; Pavek, J.; Tumer, N. Control of Enzymatic Browning in Potato (Solanum tuberosum L.) by Sense and Antisense RNA from Tomato Polyphenol Oxidase. J. Agri. Food Chem. 2001, 49, 652–657. [Google Scholar]
- O’Neill, G. New Greens Beat the Browning Blues. ECOS. 1994. [ http://www.publish.csiro.au/?act=view_file&file_id=EC82p12.pdf], 82. (b)CSIRO. Field Trial of GM Grapevines - Evaluation of Berry Colour, Sugar Composition, Flower and Fruit Development and Pollen Flow Study [online]. 2003. available from: http://www.ogtr.gov.au/rtf/ir/dir031finalrarmp.rtf. [Google Scholar]
- Queensland Department of Primary Industries. Field Trial of Pineapple Plants Modified for Blackheart Reduction and to Delay Flowering [online]. 2003. available from: http://www.ogtr.gov.au/rtf/ir/dir028finalrarmp2.rtf. [Google Scholar]
- Murata, M.; Haruta, M.; Murai, N.; Tanikawa, N.; Nishimura, M.; Homma, S.; Itoh, Y. Transgenic Apple (Malus × domestica) Shoot Showing Low Browning Potential. J. Agric. Food Chem. 2000, 48, 5243–5248. [Google Scholar] Cao, Y. H.; Zhang, Z.; Yao, Q. H.; Peng, R. H.; Xiong, A. S.; Li, X. Suppression of Apple Polyphenol Oxidase by Double-stranded RNA (RNAi). Acta Biol. Exper. Sinica 2004, 37, 487–493. (in Chinese). [Google Scholar] Okanagan Biotechnology Inc. Products & Services: Apple Varieties/ The Non-browning Apple [online]. 2005. available from: http://www.okanaganbiotechnology.com/apple-non-browning.php.
- Vickers, J. E.; Grof, C. P. L.; Bonnett, G. D.; Jackson, P. A.; Knight, D. P.; Roberts, S. E.; Robinson, S. P. Overexpression of Polyphenol Oxidase in Transgenic Sugarcane Results in Darker Juice and Raw Sugar. Crop Sci. 2005, 45, 354–362. [Google Scholar] [CrossRef]
- Robinson, S. P.; Loveys, B. R.; Chacko, E. K. Polyphenol Oxidase Enzymes in the Sap and Skin of Mango Fruit. Aust. J. Plant Physiol. 1993, 20, 99–107. [Google Scholar]
- Okanagan Biotechnology Inc. Products & Services: Cherry Varieties/ PPO Inhibited Cherry [online]. 2005. available from: http://www.okanaganbiotechnology.com/cherry-ppo-inhibited.php.
- Haruta, M.; Murata, M.; Hiraide, A.; Kadokura, H.; Yamasaki, M.; Sakuta, M.; Shimizu, S.; Homma, S. Cloning Genomic DNA Encoding Apple Polyphenol Oxidase and Comparison of the Gene Product in Escherichia coli and in Apple. Biosci. Biotechnol. Biochem. 1998, 62, 358–362. [Google Scholar] [CrossRef]
- Robinson, S. P.; Dry, I. B. Broad Bean Leaf Polyphenol Oxidase is a 60-kilodalton Protein Susceptible to Proteolytic Cleavage. Plant Physiol. 1992, 99, 317–323. [Google Scholar] [CrossRef]
- Greving, J.; Gerdemann, C.; Spener, F.; Krebs, B. Sequencing and Cloning of Genomic DNA Encoding Ipomoea batatas Catechol Oxidase, NCBI#AJ309175 [online]. 2001. [01/04/09], available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=29691901, http://www.ebi.uniprot.org/uniprot-srv/results/gridView.do;jsessionid=C3912C67B...
- Rathjen, A. H.; Robinson, S. P. Aberrant Processing of Polyphenol Oxidase in a Variegated Grapevine Mutant. Plant Physiol. 1992, 99, 1619–1625. [Google Scholar] [CrossRef]
- (a)Pan, D.; Chen, G.; Lai, Z.; Lv, L. Molecular Cloning of Full Length cDNA Encoding Polyphenol Oxidase in Pulp of Nai (Prunus salicina Lindl. var. cordata Y. He & J. Y. Zhang), NCBI#AAW58109, AAW59443, AY865623, AY870335 [online]. 2005; [05/08/03], available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=71738563, http:// www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=71738562. [Google Scholar] (b)Pan, D.; Chen, G.; Lai, Z.; Lv, L.; Peng, S.; Lai, Z.; Guo, Z. Isolation of Full Length Nai (Prunus salicina Lindl.) Leaf Polyphenol Oxidase, NCBI AAW65103 [online]. 2005. available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=58047496. [Google Scholar]
- Robinson, S. P. Polyphenol Oxidase Genes from Banana, Lettuce, Tobacco and Pineapple. US Pat. 2005. available from: http://www.freshpatents.com/Polyphenol-oxidase-genes-from-banana,-lettuce,-tobacco-and-pineapple-dt20050901ptan20050191739.php.
- Hind, G.; Marshak, D. R.; Coughlan, S. J. Spinach Thylakoid Polyphenol Oxidase: Cloning, Characterization, and Relation to a Putative Protein Kinase. Biochemistry 1995, 34, 8157–8164. [Google Scholar] Sokolenko, A.; Fulgosi, H.; Gal, A.; Altschmied, L.; Ohad, I.; Herrmann, R. G. The 64 kDa Polypeptide of Spinach may Not be the LHCII Kinase, But a Lumen-located Polyphenol Oxidase. FEBS Letters 1995, 371, 176–180. [Google Scholar]
- Jukanti, A. K.; Bruckner, P. L.; Fischer, A. M. Evaluation of Wheat Polyphenol Oxidase Genes. Cereal Chem. 2004, 81, 481–485. [Google Scholar] Raman, R.; Raman, H.; Johnstone, K.; Lisle, C.; Smith, A.; Matin, P.; Allen, H. Genetic and in Silico Comparative Mapping of the Polyphenol Oxidase Gene in Bread Wheat (Triticum aestivum L.). Funct. Integr. Genomics 2005, 5, 185–200. [Google Scholar]
- Sullivan, M. L.; Evans, M.; Thoma, S. L.; Samac, D. A.; Hatfield, R. D. Alfalfa Polyphenol Oxidase is Expressed in Flowers and Seed Pods, NCBI#AAP33165 [online]. 2003. [03/05/15], available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=30790423. [Google Scholar]
- (a)Raizada, J.; Kumar, S.; Ahuja, P. S. Cloning and Characterization of Polyphenol Oxidase Gene in Camellia sinensis (L.) O. kuntze, NCBI#AAT75166 [online]. 2004. [04/08/20], available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=50346969. [Google Scholar] (b)Li, B.; Chen, Z. Z.; Li, Q. H. Cloning Polyphenol Oxidase in Tea Variety Camellia ptilophylla, NCBI#ABF19601 [online]. 2006. [06/05/06], available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=94450151. [Google Scholar] (c)Li, B.; Chen, Z. Z.; Li, Q. H. Cloning Polyphenol Oxidase in Tea Variety Camellia sinensis var. assamica cv. Yinghong 9, NCBI#ABF19602 [online]. 2006. [06/05/06], available from: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=94450153. [Google Scholar]
- Sample Availability: Available from the authors.
© 2007 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Thipyapong, P.; Stout, M.J.; Attajarusit, J. Functional Analysis of Polyphenol Oxidases by Antisense/Sense Technology. Molecules 2007, 12, 1569-1595. https://doi.org/10.3390/12081569
Thipyapong P, Stout MJ, Attajarusit J. Functional Analysis of Polyphenol Oxidases by Antisense/Sense Technology. Molecules. 2007; 12(8):1569-1595. https://doi.org/10.3390/12081569
Chicago/Turabian StyleThipyapong, Piyada, Michael J. Stout, and Jutharat Attajarusit. 2007. "Functional Analysis of Polyphenol Oxidases by Antisense/Sense Technology" Molecules 12, no. 8: 1569-1595. https://doi.org/10.3390/12081569
APA StyleThipyapong, P., Stout, M. J., & Attajarusit, J. (2007). Functional Analysis of Polyphenol Oxidases by Antisense/Sense Technology. Molecules, 12(8), 1569-1595. https://doi.org/10.3390/12081569





