Characterization of Hydrocortisone Biometabolites and 18S rRNA Gene in Chlamydomonas reinhardtii Cultures
Abstract
:Introduction
Results and Discussion
Identification of the algal strain
Partial sequence of the 18S rRNA
5′attgatcaagaacgaaagttgggggctcgaagacgattagataccgtcgtagtctcaaccataaacgatgccgactagggatt ggcagatgttcttttgatgactctgccagcaccttatgagaaatcaaagtttttgggttccggggggagtatggtcgcaaggctgaa acttaaaggaattgacggaagggcaccaccaggcgtggagcctgcggcttaatttgactcaacacggggaaacttaccaggtc cagacacgggaaggattgacagattgagagctctttcttgattctgtgggtggtggtgcatggccgttcttagttggtgggttgcctt gtcaggttgattccggtaacgaacgagacctcagcctgctaaatagtcagcatcgcacctgcggtgcgccgacttcttagaggg actattggcgtttagccaatggaagtatgaggcgataacaggtctgtgatgcccttagatgttctgggccgcacgcgcgctacact gacgcgaccaacgagcctatccttggccgagaggcccgggtaatc 3′
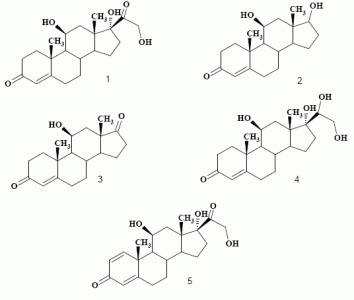
Biotransformation of hydrocortisone (1)

Product characterization
Conclusions
Experimental
General
Chemicals
Collection, preservation and identification of the alga
Incubation conditions
Time course study and the effect of substrate concentration
Products isolation and analysis
18S ribosomal RNA sequencing
Acknowledgements
References
- El-Hady, A. A.; El-Rehim, H. A. A. Production of prednisolone by Pseudomonas oleovorans cells incorporated into PVP/PEO radiation crosslinked hydrogels. J. Biomed. Biotechnol. 2004, 4, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei Yazdi, M.; Arabi, H.; Faramarzi, M. A.; Ghasemi, Y.; Amini, M.; Shokravi, Sh.; Aziz Mohseni, F. Biotransformation of hydrocortisone by a natural isolate of Nostoc muscorum. Phytochemistry 2004, 65, 2205–2209. [Google Scholar] [CrossRef]
- Ghasemi, Y.; Tabatabaei Yazdi, M.; Dehshahri, A.; Niknahad, H.; Shokravi, Sh.; Amini, M.; Ghasemian, A.; Faramarzi, M. A. Algal transformation of hydrocortisone by the cyanobacterium Nostoc ellipsosporum. Chem. Nat. Compd. 2006, 42, 702–705. [Google Scholar] [CrossRef]
- Ghasemi, Y.; Rasoul-Amini, S.; Morowvat, M.H.; Mosavi Azam, S. B.; Shokravi, S.; Mohagheghzadeh, A.; Ghoshoon, M. B.; Raee, M. J. Bioconversion of hydrocortisone by unicellular microalga Oocystis pusilla. Biotechnology 2008, 7, 293–298. [Google Scholar] [CrossRef]
- Faramarzi, M.A.; Adrangi, S.; Yazdi, M.T. Microalgal biotransformaton of steroids. J. Phycol. 2008, 44, 27–37. [Google Scholar] [CrossRef]
- Adham, N. Z.; El-Hady, A. A.; Naim, N. Biochemical studies on the microbial ∆1-dehydrogenation of cortisol by Pseudomonas fluorescens. Process Biochem. 2003, 38, 897–902. [Google Scholar] [CrossRef]
- Bold, H. C.; Wynne, M. J. Introduction to the algae; Prentice-Hall Inc: New Jersey, 1985; pp. 141–144. [Google Scholar]
- Todd, S. J.; Cain, R. B.; Schmidt, S. Biotransformation of naphthalene and diaryl ethers by green microalgae. Biodegradation 2002, 13, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Rittman, B. E. Microbial removal of hazardous organic compounds. Environ. Sci. Technol. 1982, 16, 170A–183A. [Google Scholar] [CrossRef]
- Jacobson, S. N.; Alexander, M. Enhancement of the microbial dehalogenation of a model chlorinated compound. Appl. Environ. Microbiol. 1981, 42, 1062–1066. [Google Scholar] [PubMed]
- Liebe, B.; Fock, H.P. Growth and adaptation of the green alga Chlamydomonas reinhardtii on diesel exhaust particle extracts. J. Gen. Microbiol. 1992, 16, 973–978. [Google Scholar] [CrossRef]
- Lei, A. P.; Wong, Y. S.; Tam, N. F. Y. Removal of pyrene by different microalgal species. Water Sci. Technol. 2002, 46, 195–201. [Google Scholar] [PubMed]
- Wang, X. D.; Liu, X. J.; Yang, S.; Li, A. L.; Yang, Y. L. Removal and toxicological response of triazophos by Chlamydomonas reinhardtii. Bull. Environ. Contam. Toxicol. 2007, 78, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Hirooka, T.; Nagase, H.; Hirata, K.; Miyamoto, K. Degradation of 2,4-dinitrophenol by a mixed culture of photoautotrophic microorganisms. Biochem. Eng. J. 2006, 29, 157–162. [Google Scholar] [CrossRef]
- Ghasemi, Y.; Faramarzi, M. A.; Arjmand-Inalou, M.; Mohagheghzadeh, A.; Shokravi, S.; Morowvat, M. H. Side-chain cleavage and C-20 ketone reduction of hydrocortisone by a natural isolate of Chroococcus dispersus. Ann. Microbiol. 2007, 57, 577–581. [Google Scholar] [CrossRef]
- Tabatabaei Yazdi, M.; Ghasemi, Y.; Ghasemian, A.; Shokravi, Sh.; Niknahad, H.; Amini, M.; Dehshahri, A.; Faramarzi, M. A. Bioconversion of hydrocortisone by cyanobacterium Fischerella ambigua PTCC 1635. World J. Microbiol. Biotechnol. 2005, 21, 811–814. [Google Scholar] [CrossRef]
- Wadhwa, L.; Smith, K. E. Progesterone side-chain cleavage by Bacillus sphaericus. FEMS Microbiol. Lett. 2000, 192, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Rehm, H. J.; Reed, G. Biotechnology; Verlag Chemic GmbH: Weinheim, 1984; Vol. 6a, pp. 31–78. [Google Scholar]
- Ghasemi, Y.; Tabatabaei Yazdi, M.; Shafiee, A.; Amini, M.; Shokravi, Sh.; and Zarrini, G. Parsiguine, A novel antimicrobial substance from Fischerella ambigua. Pharm. Biol. 2004, 42, 318–322. [Google Scholar] [CrossRef]
- Allen, M.M. Simple conditions for unicellular blue-green algae on plates. J. Appl. Phycol. 1968, 4, 1–4. [Google Scholar] [CrossRef]
- John, D.M.; Whitton, B. A.; Brook, A. J. The Freshwater Algal Flora of the British Isles, an Identification Guide to Freshwater and Terrestrial Algae; Cambridge University Press: Cambridge, 2003; pp. 39–43. [Google Scholar]
- Prescott, G. W. Algae of the western great lake areas; W. M. C. Brown Company Publisher: Dubuque, Iowa, 1962. [Google Scholar]
- Soltani, N.; Khavari-Nejad, R.A.; Tabatabaei Yazdi, M.; Shokravi, Sh.; Fernández-Valiente, E. Variation of Nitrogenase activity, Photosynthesis, and pigmentation of cyanobacterium Fischerella sp. FS18 under different irradiance and pH. World J. Microbiol. Biotechnol. 2006, 22, 571–576. [Google Scholar] [CrossRef]
- Andersen, R.A. Algal culturing techniques; Elsevier Academic Press: New York, 2005; pp. 239–287. [Google Scholar]
- Nubel, U.; Garcia, P. F.; Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Applied Environ. Microbiol. 1997, 63, 3327–3332. [Google Scholar]
- Blast: http://www.ncbi.nlm.nih.gov/blast/Blast.cgi.
- GeneDoc: http://www.nrbsc.org/gfx/genedoc/index.html.
- Sample Availability: Contact the authors.
© 2008 by the authors. Licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ghasemi, Y.; Rasoul-Amini, S.; Morowvat, M.H.; Raee, M.J.; Ghoshoon, M.B.; Nouri, F.; Negintaji, N.; Parvizi, R.; Mosavi-Azam, S.B. Characterization of Hydrocortisone Biometabolites and 18S rRNA Gene in Chlamydomonas reinhardtii Cultures. Molecules 2008, 13, 2416-2425. https://doi.org/10.3390/molecules13102416
Ghasemi Y, Rasoul-Amini S, Morowvat MH, Raee MJ, Ghoshoon MB, Nouri F, Negintaji N, Parvizi R, Mosavi-Azam SB. Characterization of Hydrocortisone Biometabolites and 18S rRNA Gene in Chlamydomonas reinhardtii Cultures. Molecules. 2008; 13(10):2416-2425. https://doi.org/10.3390/molecules13102416
Chicago/Turabian StyleGhasemi, Younes, Sara Rasoul-Amini, Mohammad Hossein Morowvat, Mohammad Javad Raee, Mohammad Bagher Ghoshoon, Fatemeh Nouri, Narges Negintaji, Rezvan Parvizi, and Seyed Bagher Mosavi-Azam. 2008. "Characterization of Hydrocortisone Biometabolites and 18S rRNA Gene in Chlamydomonas reinhardtii Cultures" Molecules 13, no. 10: 2416-2425. https://doi.org/10.3390/molecules13102416