Synthesis and Theoretical Study of Molecularly Imprinted Nanospheres for Recognition of Tocopherols
Abstract
:1. Introduction

2. Results and Discussion
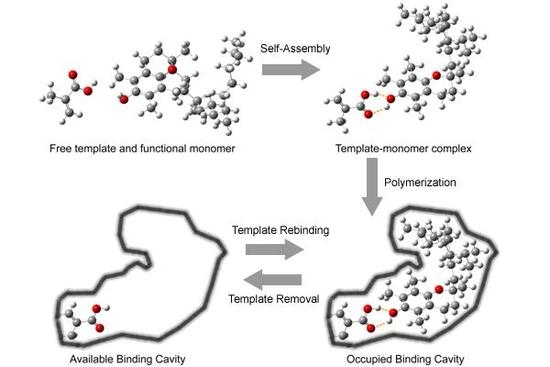
2.1. Preparation of tocopherol-imprinted polymers

2.2. Recognition properties of tocopherol-imprinted polymers





2.3. Molecular modeling of template-monomer complex
| E (a.u.) | Δ E (a.u.) a | Δ E (kJ mol-1) b | |
|---|---|---|---|
| TP | –1285.682 | ||
| TPA | –1438.350 | ||
| MAA | –306.475 | ||
| TP–MAA(1) | –1592.180 | –0.023 | –61.042 |
| TP–MAA(2) | –1592.167 | –0.010 | –27.159 |
| TP–MAA(3) | –1592.172 | –0.015 | –40.222 |
| TPA–MAA(1) | –1744.844 | –0.020 | –51.514 |
| TPA–MAA(2) | –1744.840 | –0.015 | –39.900 |
| TPA–MAA(3) | –1744.839 | –0.015 | –38.108 |


| EHOMO | ELUMO | EHOMO–LUMO | |
|---|---|---|---|
| TP | –7.442 | –1.143 | 6.299 |
| TPA | –5.371 | 0.346 | 5.717 |
| MAA | –5.492 | 0.236 | 5.728 |
| TP–MAA(1) | –5.446 | –1.032 | 4.414 |
| TP–MAA(2) | –5.184 | –1.359 | 3.826 |
| TP–MAA(3) | –5.749 | –0.839 | 4.911 |
| TPA–MAA(1) | –5.664 | –0.604 | 5.059 |
| TPA–MAA(2) | –5.646 | –0.930 | 4.716 |
| TPA–MAA(3) | –5.959 | –0.778 | 5.181 |
3. Conclusions
4. Experimental
4.1. Chemicals
4.2. Preparation of molecularly imprinted polymers
4.3. Preparation of molecularly imprinted nanospheres
4.4. Scanning Electron Microscopy (SEM)
4.5. Binding analysis
4.6. Molecular modeling analysis

 represents the interaction energy,
represents the interaction energy,  represents the energy of template-monomer complex,
represents the energy of template-monomer complex,  represents the energy of template molecule, and
represents the energy of template molecule, and  is the energy of functional monomer molecules.
is the energy of functional monomer molecules.Acknowledgements
References
- Lonn, E. Effects of long-term vitamin E supplementation on cardiovascular events and cancer: A randomized controlled trial. J. Am. Med. Assoc. 2005, 293, 1338–1347. [Google Scholar] [CrossRef]
- Coulter, I.D.; Hardy, M.L.; Morton, S.C.; Hilton, L.G.; Tu, W.; Valentine, D.; Shekelle, P.G. Antioxidants vitamin C and vitamin E for the prevention and treatment of cancer. J. Gen. Intern. Med. 2006, 21, 735–744. [Google Scholar] [CrossRef]
- Burton, G.W.; Doba, T.; Gabe, E.; Hughes, L.; Lee, F.L.; Prasad, L.; Ingold, K.U. Autoxidation of biological molecules. 4. Maximizing the antioxidant activity of phenols. J. Am. Chem. Soc. 1985, 107, 7053–7065. [Google Scholar] [CrossRef]
- Mustacich, D.J.; Bruno, R.S.; Traber, M.G.; Vitamin, E. In Vitamins and Hormones; Litwack, G., Ed.; Academic Press: San Diego, CA, USA, 2007; Vol. 76, pp. 1–21. [Google Scholar]
- Neužil, J.; Witting, P.K.; Stocker, R. α-Tocopheryl hydroquinone is an efficient multifunctional inhibitor of radical-initiated oxidation of low density lipoprotein lipids. Proc. Natl. Acad. Sci. USA 1997, 94, 7885–7890. [Google Scholar] [CrossRef]
- Nagao, T.; Kobayashi, T.; Hirota, Y.; Kitano, M.; Kishimoto, N.; Fujita, T.; Watanabe, Y.; Shimada, Y. Improvement of a process for purification of tocopherols and sterols from soybean oil deodorizer distillate. J. Mol. Catal. B - Enzym. 2005, 37, 56–62. [Google Scholar] [CrossRef]
- Martins, P.F.; Batistella, C.B.; Maciel-Filho, R.; Wolf-Maciel, M.R. Comparison of two different strategies for tocopherols enrichment using a molecular distillation process. Ind. Eng. Chem. Res. 2006, 45, 753–758. [Google Scholar] [CrossRef]
- Nagesha, G.K.; Subramanian, R.; Udaya Sankar, K. Processing of tocopherol and FA systems using a nonporous denser polymeric membrane. J. Am. Oil Chem. Soc. 2003, 80, 397–402. [Google Scholar] [CrossRef]
- King, J.W.; Favati, F.; Taylor, S.L. Production of tocopherol concentrates by supercritical fluid extraction and chromatography. Separ. Sci. Technol. 1996, 31, 1843–1857. [Google Scholar] [CrossRef]
- Masqué, N.; Marcé, R.M.; Borrull, F. Molecularly imprinted polymers: new tailor-made materials for selective solid-phase extraction. TrAC Trend. Anal. Chem. 2001, 20, 477–486. [Google Scholar]
- Ye, L.; Mosbach, K. Molecular imprinting: synthetic materials as substitutes for biological antibodies and receptors. Chem. Mater. 2008, 20, 859–868. [Google Scholar] [CrossRef]
- Bergmann, N.M.; Peppas, N.A. Molecularly imprinted polymers with specific recognition for macromolecules and proteins. Prog. Polym. Sci. 2008, 33, 271–288. [Google Scholar]
- Ghasemzadeh, N.; Nyberg, F.; Hjertén, S. Highly selective artificial gel antibodies for detection and quantification of biomarkers in clinical samples. I. Spectrophotometric approach to design the calibration curve for the quantification. J. Sep. Sci. 2008, 31, 3945–3953. [Google Scholar] [CrossRef]
- Piacham, T.; Josell, Å.; Arwin, H.; Prachayasittikul, V.; Ye, L. Molecularly imprinted polymer thin films on quartz crystal microbalance using a surface bound photo-radical initiator. Anal. Chim. Acta 2005, 536, 191–196. [Google Scholar] [CrossRef]
- Turiel, E.; Tadeo, J.L.; Cormack, P.A.G.; Martin-Esteban, A. HPLC imprinted-stationary phase prepared by precipitation polymerisation for the determination of thiabendazole in fruit. Analyst 2005, 130, 1601–1607. [Google Scholar] [CrossRef]
- Haginaka, J. Molecularly imprinted polymers for solid-phase extraction. Anal. Bioanal. Chem. 2004, 379, 332–334. [Google Scholar] [CrossRef]
- Mosbach, K.; Yu, Y.; Andersch, J.; Ye, L. Generation of new enzyme inhibitors using imprinted binding sites: The anti-idiotypic approach, a step toward the next generation of molecular imprinting. J. Am. Chem. Soc. 2001, 123, 12420–12421. [Google Scholar]
- Hillberg, A.L.; Brain, K.R.; Allender, C.J. Molecular imprinted polymer sensors: Implications for therapeutics. Adv. Drug Deliver. Rev. 2005, 57, 1875–1889. [Google Scholar]
- Vlatakis, G.; Andersson, L.I.; Muller, R.; Mosbach, K. Drug assay using antibody mimics made by molecular imprinting. Nature 1993, 361, 645–647. [Google Scholar]
- Hart, B.R.; Shea, K.J. Synthetic peptide receptors: Molecularly imprinted polymers for the recognition of peptides using peptide-metal interactions. J. Am. Chem. Soc. 2001, 123, 2072–2073. [Google Scholar] [CrossRef]
- Boonpangrak, S.; Prachayasittikul, V.; Bülow, L.; Ye, L. Molecularly imprinted polymer microspheres prepared by precipitation polymerization using a sacrificial covalent bond. J. Appl. Polym. Sci. 2006, 99, 1390–1398. [Google Scholar] [CrossRef]
- Boonpangrak, S.; Whitcombe, M.J.; Prachayasittikul, V.; Mosbach, K.; Ye, L. Preparation of molecularly imprinted polymers using nitroxide-mediated living radical polymerization. Biosens. Bioelectron. 2006, 22, 349–354. [Google Scholar] [CrossRef]
- Allender, C.J.; Richardson, C.; Woodhouse, B.; Heard, C.M.; Brain, K.R. Pharmaceutical applications for molecularly imprinted polymers. Int. J. Pharm. 2000, 195, 39–43. [Google Scholar] [CrossRef]
- Sellergren, B.; Allender, C.J. Molecularly imprinted polymers: a bridge to advanced drug delivery. Adv. Drug Deliver. Rev. 2005, 57, 1733–1741. [Google Scholar] [CrossRef]
- Piacham, T.; Isarankura Na Ayudhya, C.; Prachayasittikul, V.; Bülow, L.; Ye, L. A polymer supported manganese catalyst useful as a superoxide dismutase mimic. Chem. Commun. 2003, 1254–1255. [Google Scholar]
- Sellergren, B.; Karmalkar, R.N.; Shea, K.J. Enantioselective ester hydrolysis catalyzed by imprinted polymers. J. Org. Chem. 2000, 65, 4009–4027. [Google Scholar] [CrossRef]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–27. [Google Scholar] [CrossRef]
- Puoci, F.; Cirillo, G.; Curcio, M.; Iemma, F.; Spizzirri, U.G.; Picci, N. Molecularly imprinted solid phase extraction for the selective HPLC determination of α-tocopherol in bay leaves. Anal. Chim. Acta 2007, 593, 164–170. [Google Scholar] [CrossRef]
- Faizal, C.K.M.; Kikuchi, Y.; Kobayashi, T. Molecular imprinting targeted for α-tocopherol by calix[4]resorcarenes derivative in membrane scaffold prepared by phase inversion. J. Membrane Sci. 2009, 334, 110–116. [Google Scholar] [CrossRef]
- Faizal, C.K.M.; Kobayashi, T. Tocopherol-targeted membrane adsorbents prepared by hybrid molecular imprinting. Polym. Eng. Sci. 2008, 48, 1085–1093. [Google Scholar] [CrossRef]
- Puoci, F.; Cirillo, G.; Curcio, M.; Iemma, F.; Parisi, O.I.; Castiglione, M.; Picci, N. Molecularly imprinted polymers for α-tocopherol delivery. Drug Deliv. 2008, 15, 253–258. [Google Scholar] [CrossRef]
- Andersson, L.I. Application of molecular imprinting to the development of aqueous buffer and organic solvent based radioligand binding assays for (S)-propranolol. Anal. Chem. 1996, 68, 111–117. [Google Scholar] [CrossRef]
- Haginaka, J. Monodispersed, molecularly imprinted polymers as affinity-based chromatography media. J. Chromatogr. B 2008, 866, 3–13. [Google Scholar] [CrossRef]
- Yoshimatsu, K.; Reimhult, K.; Krozer, A.; Mosbach, K.; Sode, K.; Ye, L. Uniform molecularly imprinted microspheres and nanoparticles prepared by precipitation polymerization: The control of particle size suitable for different analytical applications. Anal. Chim. Acta 2007, 584, 112–121. [Google Scholar] [CrossRef]
- Dorothea, V.; Katharina, L.; Iris, K.; Herwig, B.; Günter, E.M.T. Molecularly imprinted polymer nanospheres as synthetic affinity receptors obtained by miniemulsion polymerisation. Macromol. Chem. Phys. 2002, 203, 1965–1973. [Google Scholar] [CrossRef]
- Cacho, C.; Turiel, E.; Martin-Esteban, A.; Pérez-Conde, C.; Cámara, C. Characterisation and quality assessment of binding sites on a propazine-imprinted polymer prepared by precipitation polymerisation. J. Chromatogr. B 2004, 802, 347–353. [Google Scholar]
- Sajonz, P.; Kele, M.; Zhong, G.; Sellergren, B.; Guiochon, G. Study of the thermodynamics and mass transfer kinetics of two enantiomers on a polymeric imprinted stationary phase. J. Chromatogr. A 1998, 810, 1–17. [Google Scholar]
- Pérez-Moral, N.; Mayes, A.G. Novel MIP formats. Bioseparation 2001, 10, 287–299. [Google Scholar] [CrossRef]
- Pérez-Moral, N.; Mayes, A.G. Comparative study of imprinted polymer particles prepared by different polymerisation methods. Anal. Chim. Acta 2004, 504, 15–21. [Google Scholar] [CrossRef]
- Oxelbark, J.; Legido-Quigley, C.; Aureliano, C.S.A.; Titirici, M.-M.; Schillinger, E.; Sellergren, B.; Courtois, J.; Irgum, K.; Dambies, L.; Cormack, P.A.G.; Sherrington, D.C.; De Lorenzi, E. Chromatographic comparison of bupivacaine imprinted polymers prepared in crushed monolith, microsphere, silica-based composite and capillary monolith formats. J. Chromatogr. A 2007, 1160, 215–226. [Google Scholar]
- Mayes, A.G. Polymerisation techniques for the formation of imprinted beads. In Molecularly Imprinted Polymers: Man-made Mimics of Antibodies and Their Applications in Analytical Chemistry; Sellergren, B., Ed.; Elsevier: Amsterdam, The Netherlands, 2001; pp. 305–324. [Google Scholar]
- Nicholls, I.A.; Adbo, K.; Andersson, H.S.; Andersson, P.O.; Ankarloo, J.; Hedin-Dahlström, J.; Jokela, P.; Karlsson, J.G.; Olofsson, L.; Rosengren, J.; Shoravi, S.; Svenson, J.; Wikman, S. Can we rationally design molecularly imprinted polymers? Anal. Chim. Acta 2001, 435, 9–18. [Google Scholar] [CrossRef]
- Isarankura-Na-Ayudhya, C.; Nantasenamat, C.; Buraparuangsang, P.; Piacham, T.; Ye, L.; Bülow, L.; Prachayasittikul, V. Computational insights on sulfonamide imprinted polymers. Molecules 2008, 13, 3077–3091. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Naenna, T.; Prachayasittikul, V. Quantitative structure-imprinting factor relationship of molecularly imprinted polymers. Biosens. Bioelectron. 2007, 22, 3309–3317. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Naenna, T.; Isarankura Na Ayudhya, C.; Prachayasittikul, V. Quantitative prediction of imprinting factor of molecularly imprinted polymers by artificial neural network. J. Comput. Aid. Mol. Des. 2005, 19, 509–524. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Tantimongcolwat, T.; Naenna, T.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Prediction of selectivity index of pentachlorophenol-imprinted polymers. EXCLI J. 2006, 5, 150–163. [Google Scholar]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Naenna, T.; Prachayasittikul, V. Prediction of bond dissociation enthalpy of antioxidant phenols by support vector machine. J. Mol. Graph. Model. 2008, 27, 188–196. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Tansila, N.; Naenna, T.; Prachayasittikul, V. Prediction of GFP spectral properties using artificial neural network. J. Comput. Chem. 2007, 28, 1275–1289. [Google Scholar] [CrossRef]
- Thippakorn, C.; Suksrichavalit, T.; Nantasenamat, C.; Tantimongcolwat, T.; Isarankura-Na-Ayudhya, C.; Naenna, T.; Prachayasittikul, V. Modeling the LPS neutralization activity of anti-endotoxins. Molecules 2009, 14, 1869–1888. [Google Scholar] [CrossRef]
- Worachartcheewan, A.; Nantasenamat, C.; Naenna, T.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Modeling the activity of furin inhibitors using artificial neural network. Eur. J. Med. Chem. 2009, 44, 1664–1673. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Naenna, T.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Recognition of DNA Splice Junction via Machine Learning Approaches. EXCLI J. 2005, 4, 114–129. [Google Scholar]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Naenna, T.; Prachayasittikul, V. A practical overview of quantitative structure-activity relationship. EXCLI J. 2009, 8, 74–88. [Google Scholar]
- Karelson, M.; Lobanov, V.S.; Katritzky, A.R. Quantum-Chemical Descriptors in QSAR/QSPR Studies. Chem. Rev. 1996, 96, 1027–1044. [Google Scholar] [CrossRef]
- Ye, L.; Cormack, P.A.G.; Mosbach, K. Molecularly imprinted monodisperse microspheres for competitive radioassay. Anal. Commun. 1999, 36, 35–38. [Google Scholar] [CrossRef]
- Sample Availability: Contact the authors.
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Piacham, T.; Nantasenamat, C.; Suksrichavalit, T.; Puttipanyalears, C.; Pissawong, T.; Maneewas, S.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Synthesis and Theoretical Study of Molecularly Imprinted Nanospheres for Recognition of Tocopherols. Molecules 2009, 14, 2985-3002. https://doi.org/10.3390/molecules14082985
Piacham T, Nantasenamat C, Suksrichavalit T, Puttipanyalears C, Pissawong T, Maneewas S, Isarankura-Na-Ayudhya C, Prachayasittikul V. Synthesis and Theoretical Study of Molecularly Imprinted Nanospheres for Recognition of Tocopherols. Molecules. 2009; 14(8):2985-3002. https://doi.org/10.3390/molecules14082985
Chicago/Turabian StylePiacham, Theeraphon, Chanin Nantasenamat, Thummaruk Suksrichavalit, Charoenchai Puttipanyalears, Tippawan Pissawong, Supanee Maneewas, Chartchalerm Isarankura-Na-Ayudhya, and Virapong Prachayasittikul. 2009. "Synthesis and Theoretical Study of Molecularly Imprinted Nanospheres for Recognition of Tocopherols" Molecules 14, no. 8: 2985-3002. https://doi.org/10.3390/molecules14082985