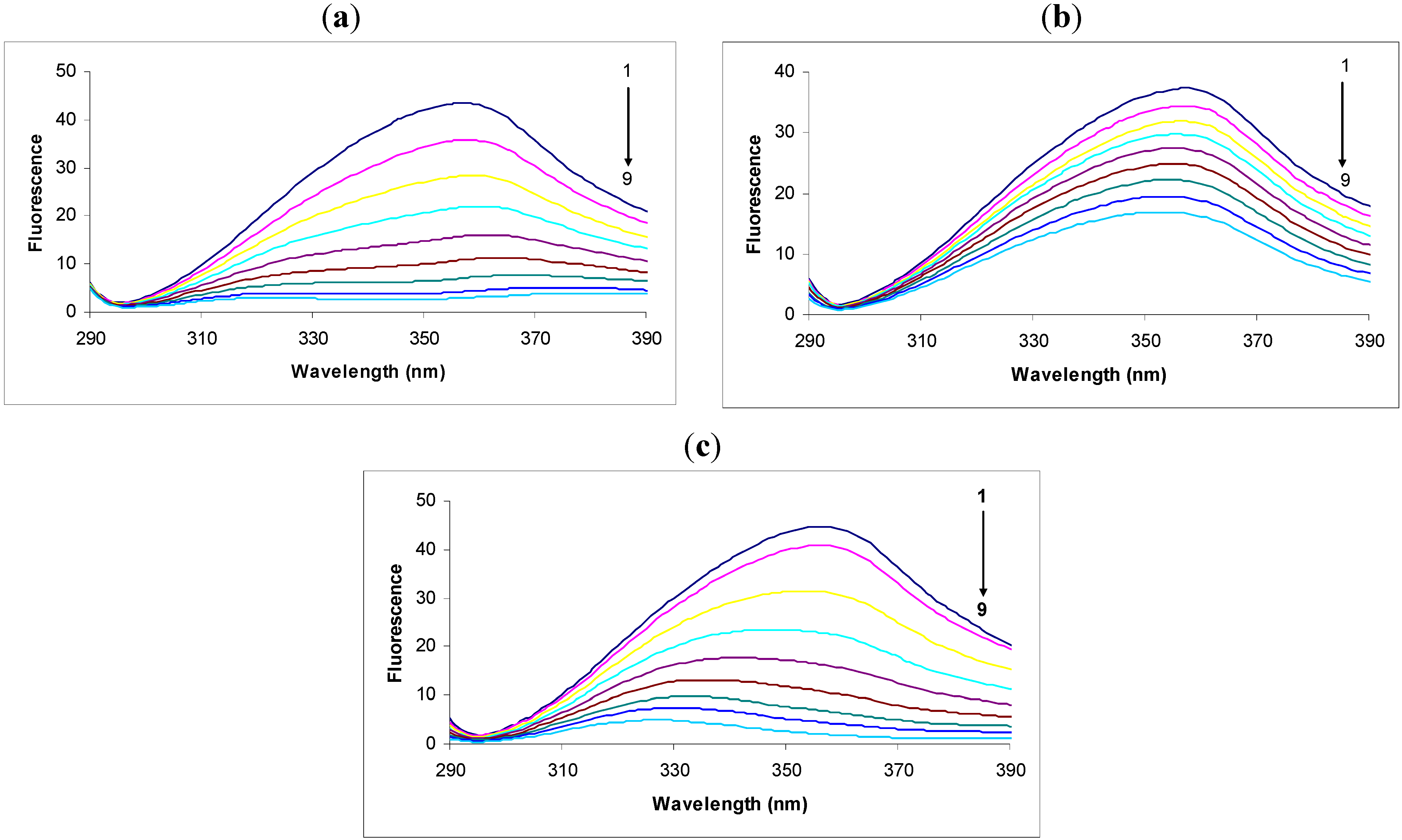
2.1. Steady State Fluorescence Studies
Experiments have shown that fluorescence-based analytical methods are very specific and sensitive, therefore these methods can be extremely useful in the analysis and monitoring of certain environmental pollutants. Dinitrophenols and some nitro- and trinitrophenols are able to quench the fluorescence of compounds, therefore in this work we used their interaction with molecules in the excited state to understand the electron and energy transfer from excited molecules to dinitrophenol.
In the fluorescence spectra of tryptophan we observed that the intensity decreased remarkably in response to the addition of 2,4-DNP (
Figure 1a). This intensity decrease was accompanied by a large shift and apparent splitting of the maximum emission wavelength from 357 nm to 380 nm and 323 nm in the presence of 30.00 × 10
−6 M concentrations of 2,4-DNP. The large displacement of the maximum emission wavelength could be due to the hydrophilic or hydrophobic character of the compounds [
22,
23], thus the theoretical logP was calculated using the specialized Quantitative Structure-Activity Relationship (QSAR) module of the software HyperChem 7.5. Tryptophan has a logP equal to 0.06, 2,4-DNP 1.67 and 2,4-DNP:Tryp complex 0.65. There is a decrease in polarity around tryptophan residues.
Figure 1.
Fluorescence spectra of tryptophan (1.0 μg/mL) in the absence (black, spectrum 1) or presence of: (a) 2.4-DNP (6.65 × 10−6; 10 × 10−6; 13.33 × 10−6; 16.66 × 10−6; 20.00 × 10−6; 23.33 × 10−6; 26.66 × 10−6; 30.00 × 10−6 M, respectively, spectra 2-9); (b) 2.6-DNP (6.65 × 10−6; 10 × 10−6; 13.33 × 10−6; 16.66 × 10−6; 20.00 × 10−6; 23.33 × 10−6; 26.66 × 10−6 respectively 30.00 × 10−6 M, spectra 2-9); (c) 2.4-DNOC (6.65 × 10−6; 10 × 10−6; 13.33 × 10−6; 16.66 × 10−6; 20.00 × 10−6; 23.33 × 10−6; 26.66 × 10−6 respectively 30.00 × 10−6 M, spectra 2-9).
Figure 1.
Fluorescence spectra of tryptophan (1.0 μg/mL) in the absence (black, spectrum 1) or presence of: (a) 2.4-DNP (6.65 × 10−6; 10 × 10−6; 13.33 × 10−6; 16.66 × 10−6; 20.00 × 10−6; 23.33 × 10−6; 26.66 × 10−6; 30.00 × 10−6 M, respectively, spectra 2-9); (b) 2.6-DNP (6.65 × 10−6; 10 × 10−6; 13.33 × 10−6; 16.66 × 10−6; 20.00 × 10−6; 23.33 × 10−6; 26.66 × 10−6 respectively 30.00 × 10−6 M, spectra 2-9); (c) 2.4-DNOC (6.65 × 10−6; 10 × 10−6; 13.33 × 10−6; 16.66 × 10−6; 20.00 × 10−6; 23.33 × 10−6; 26.66 × 10−6 respectively 30.00 × 10−6 M, spectra 2-9).
We observed that the intensity of the fluorescence quenching spectra also decreased in response to the addition of 2,6-DNP (
Figure 1b). This decrease in intensity was accompanied by a small shift of the maximum emission wavelength from 357 nm to 353 nm in the presence of 30.00 × 10
−6 M 2,6-DNP. According to the theoretical calculations we performed, 2,6-DNP has the same value of logP as its isomer, but the maximum emission wavelength shift was much smaller, and hence this factor does not have a decisive effect on the maximum emission wavelength shift. Another important factor is probably the heat of formation of the complex with 2,6-DNP, which is higher in this case than in the complex with 2,4-DNP. Therefore, the likelihood of 2,6-DNP:tryptophan complex formation is lower for the same energy consumption.
Studying the spectra of fluorescence quenching in response to the addition of DNOC we observed that the intensity decreased greatly (
Figure 1c). Thus, the presence of the compound at a concentration of 30.00 × 10
−6 M produced a large shift of the maximum emission wavelength from 357 nm to 327 nm. The shifted emission maxima is almost similar with that of 2,4-DNP, both of them having -NO
2 groups in the same position, and an inductive effect over these overlapped groups was noticed, probably do to an conjugation effect (—E effect). Also, the logP value calculated for DNOC was 2.14 and for the complex it was −1.34, so the complex is soluble in water.
Fluorescence quenching can occur through two different mechanisms, static and dynamic, respectively [
24]. We analysed the fluorescence quenching data according to the Stern-Volmer Equation (1):
where F
0 is the initial intensity of the fluorescent molecule fluorescence signal, F is the fluorescence intensity after adding fluorescence quench molecular solution, K
SV is the Stern-Volmer constant and [Q] is the concentration extinguisher fluorescence [
21,
24].
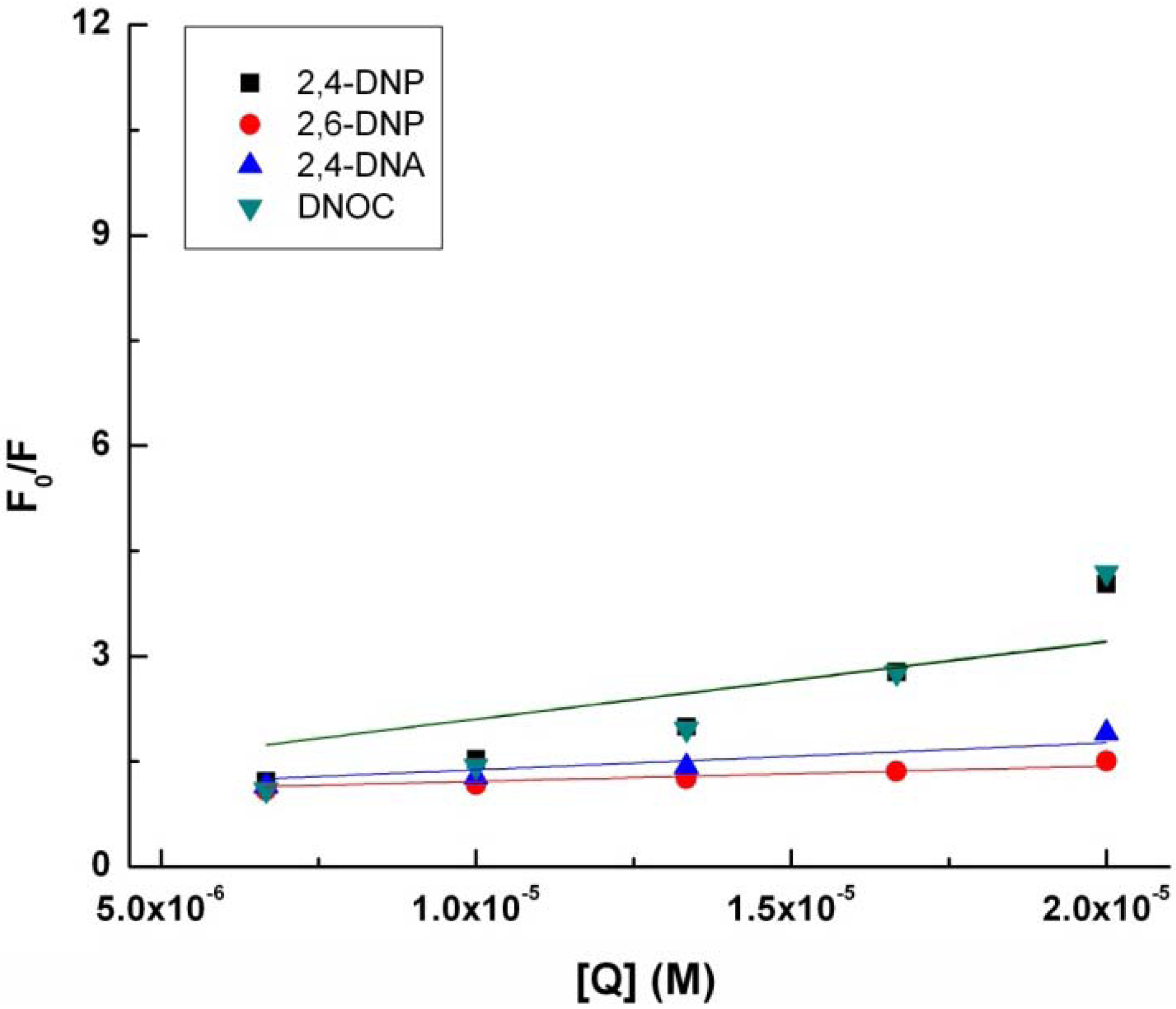
With quenching the tryptophan fluorescence in the presence of 2,4-DNP, 2,6-DNP, 2,4-DNA and DNOC, K
SV, the Stern-Volmer constant increases in the following order: 2,6-DNP, 2,4-DNA, 2,4-DNP and DNOC (
Table 1,
Figure 2). The difference between the Stern-Volmer constants of 2,4-DNP and DNOC is negligible. In contrast, 2,4-DNA and 2,6-DNP had smaller tryptophan fluorescence quenching constants than 2,4-DNP. From the above results we observe that phenolic hydroxyl blocking or changing the NO
2 group positions lead to reduced tryptophan fluorescence quenching by dinitrophenol derivatives due to the different resulting electronic effects and conjugation.
Table 1.
Fluorescence quenching characteristics obtained using linear regression Stern-Volmer Equation (1).
Table 1.
Fluorescence quenching characteristics obtained using linear regression Stern-Volmer Equation (1).
| Compound | Ksv (M−1) | r2 |
|---|
| 2,4-DNP | 110116 ± 19365 | 0.941 |
| 2,6-DNP | 21760 ± 1731 | 0.998 |
| 2,4-DNA | 38270 ± 4723 | 0.992 |
| DNOC | 111048 ± 22833 | 0.919 |
Figure 2.
Modified Stern-Volmer plot for the binding of 2.4-DNP, 2.6-DNP, DNA, and DNOC to tryptophan.
Figure 2.
Modified Stern-Volmer plot for the binding of 2.4-DNP, 2.6-DNP, DNA, and DNOC to tryptophan.
Deviations from linearity may exist, they can be positive or negative. If, as in this case, there are positive deviations from the straight line, concave with respect to the Y axis, these may be due to the simultaneous presence of dynamic and static extinction when an excited fluorophore is extinguished due to collision with a fluorescence extinguisher [
21,
24,
25]. Therefore, in these cases, the Stern-Volmer linear regression can be obtained using the Stern-Volmer expresion according to Equation (2):
where F
0 is the initial intensity of the fluorescent molecule fluorescence signal, F is the fluorescence intensity after adding fluorescence quench molecular solution, K
SV is the Stern-Volmer constant for the quenching of fluorescence by dynamic mechanism, V is a constant representing the static contributions to extinguish fluorescence and [Q] is the concentration of fluorescence extinguisher [
26].
By analyzing the data obtained, it was concluded that we must use another Stern-Volmer equation more suitable for our cases [
26,
27]. Therefore, we studied each representation separately and in accordance with these results Stern-Volmer representations and linear regressions were obtained using the Stern-Volmer expression according to Equation (2). Further, we studied the mechanisms of tryptophan fluorescence quenching by 2,6-DNP, DNOC 2,4-DNA and compared the mechanism of action of 2,4-DNP [
27].
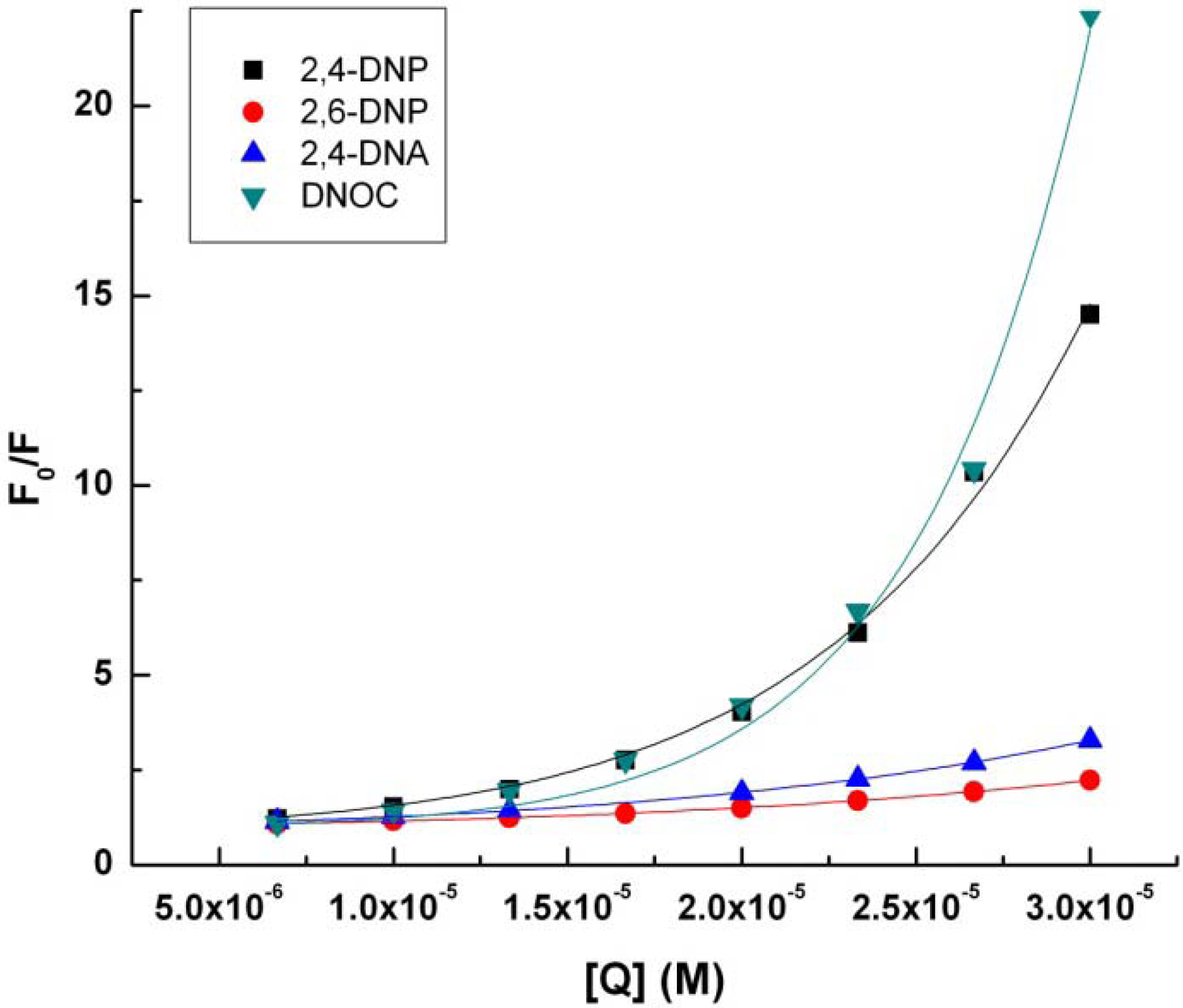
Data from the linear regression implementation of Stern-Volmer Equation (2) can be seen in
Table 2 and
Figure 3.
Table 2.
Fluorescence extinction characteristics obtained using nonlinear regression Stern-Volmer Equation (2).
Table 2.
Fluorescence extinction characteristics obtained using nonlinear regression Stern-Volmer Equation (2).
| Compound | KSV (M−1) | V(M−1) | r2 |
|---|
| 2,4-DNP | 20117.21 ± 3482.26 | 104272.40 ± 6100.06 | 0.995 |
| 2,6-DNP | 9779.84 ± 241.19 | 47789.49 ± 918.83 | 0.999 |
| 2,4-DNA | 16339.86 ± 249.81 | 51308.83 ± 560.44 | 0.999 |
| DNOC | 4397.68 ± 1574.75 | 169008.33 ± 12208.50 | 0.992 |
Figure 3.
Stern-Volmer representation (2) for tryptophan (4.9 × 10−3 M) in presence of 2,4-DNP, 2,6-DNP, 2,4-DNA, DNOC.
Figure 3.
Stern-Volmer representation (2) for tryptophan (4.9 × 10−3 M) in presence of 2,4-DNP, 2,6-DNP, 2,4-DNA, DNOC.
KSV, the Stern-Volmer constant for the quenching of fluorescence by the dynamic mechanism increases in the following order: DNOC, 2,6-DNP, 2,4-DNA, 2,4-DNP. Surprisingly, DNOC fluorescence quenching by the dynamic mechanism was 80% less compared with 2,4-DNP, although both compounds are very strong. There is a big difference between the value of the constant KSV corresponding to 2,4-DNP compared to the other constants.
If we consider a static quenching mechanism of tryptophan fluorescence, as shown in the table above, V, the Stern-Volmer constant for the quenching of fluorescence by the static mechanism, increases in the following order: 2,6-DNP, 2,4-DNA, 2,4-DNP and DNOC.
If we consider the results in
Table 1,
Table 2, but also properties of a QSAR study of the 2,4-dinitro derivatives and their complexes with tryptophan, then a decreased tryptophan fluorescence quenching capacity through dynamic mechanism occurs with increasing volume (see the section on theoretical calculations). 2,4-DNP has a greater capacity for tryptophan fluorescence quenching that 2,6-DNP, probably due to the conjugation that arises from the position of the nitro group
para to the phenolic hydroxyl group.
2.2. Spectroscopic Studies on Some Dinitrophenol and Dinitrophenyl Ethers
To elucidate the action of mono- and dinitro derivatives on the oxidative phosphorylation mechanism we studied the fluorescence quenching of tryptophan or other compounds in the presence these uncoupling agents.
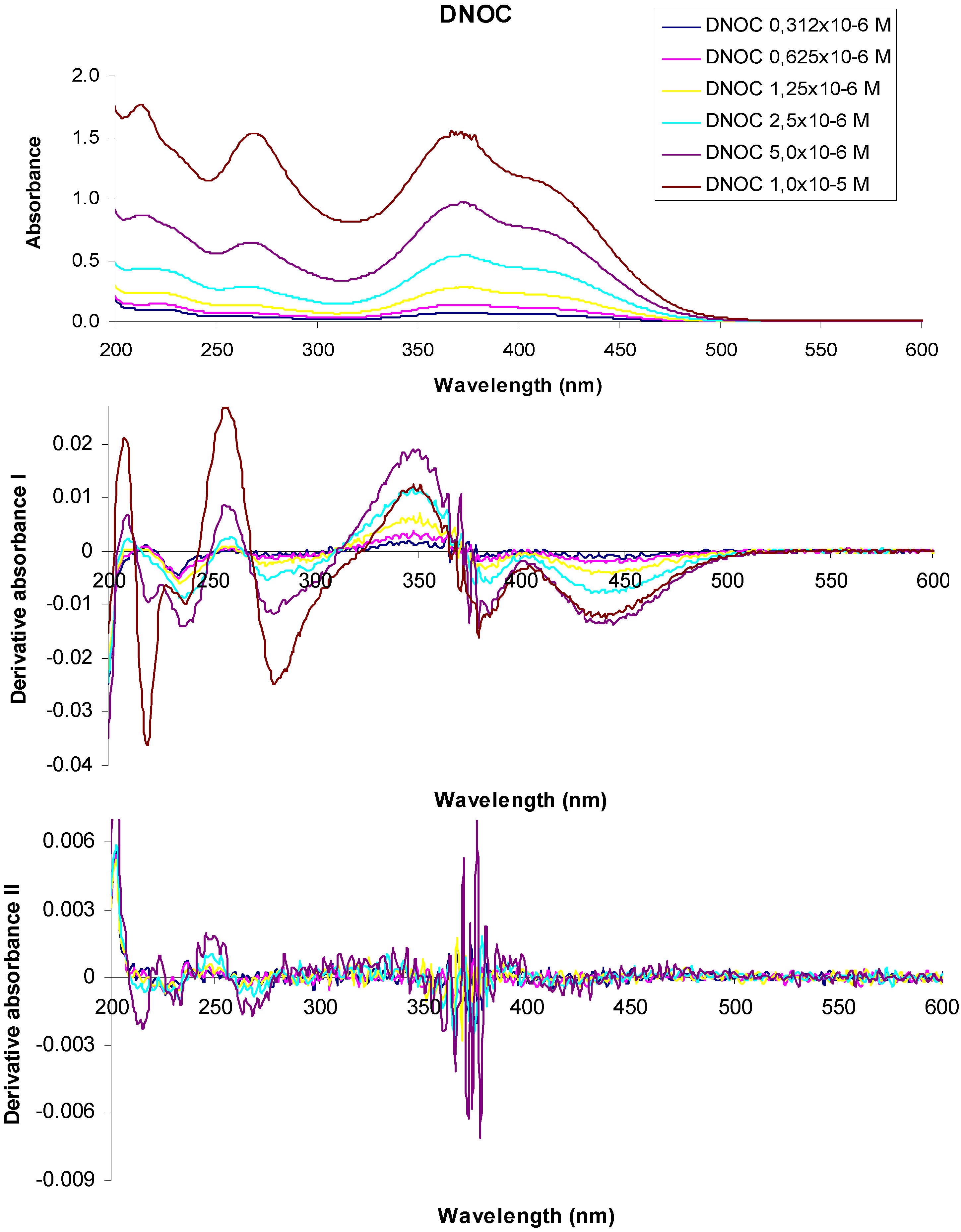
Figure 4 shows the absorption spectra and derivative curves of some representative dinitrophenol ethers and dinitrophenols.
Figure 4.
UV-VIS absorption spectra of 3,5-dinitro-o-cresol and their derivatives at various concentrations.
Figure 4.
UV-VIS absorption spectra of 3,5-dinitro-o-cresol and their derivatives at various concentrations.
In the UV-VIS absorption spectrum of a 1 × 10−5 M aqueous solution of 3,5-dinitro-o-cresol (DNOC), we observed three absorption maxima at 213 nm (A = 1.776), 268 nm (A = 1.537), 367 nm (A = 1.550) and two shoulders at 229 nm (A = 1.396) and 415 nm (A = 1.107). The UV-VIS absorption spectrum of a 5 × 10−6 M DNOC solution presented three absorption maxima at 213 nm (A = 0.868), 268 nm (A = 0.644), 372 nm (A = 0.975) and two shoulders at 230 nm (A = 0.752) and 416 nm (A = 0.720). Using 2.5 × 10−6 M DNOC concentration we observed three absorption maxima at 215 nm (A = 0.435), 266 nm (A = 0.285), 372 nm (A = 0.541) and a shoulder at 417 nm (A = 0.419). In the UV-VIS absorption spectrum of DNOC solutions of 1.25 × 10−6 M, 6.25 × 10−7 M and 3.12 × 10−7 M concentration, we observed three absorption maxima at 220 nm (A = 0.237), 264 nm (A = 0.139), 374 nm (A = 0.282) for the first solution, three absorption maxima at 220 nm (A = 0.144), 264 nm (A = 0.073), 377 nm (A = 0.141) in the case of the second solution and three absorption maxima from 222 nm (A = 0.105), 264 nm (A = 0.045), 376 nm (A = 0.080) in the case of the third solution, respectively.
The first order derivative of the 1 × 10−5 M DNOC solution spectra intersects the axis at wavelengths equal to 213 nm, 270 nm, 366 nm, at a concentration of 5 × 10−6 M, at 213 nm, 267 nm, 369 nm, at a concentration of 2.5 × 10−6 M, at 216 nm, 265 nm, 370 nm, at a concentration of 1.25 × 10−6 M at 217 nm, 264 nm, 369 nm, at a concentration of 6.25 × 10−7 M, at 212 nm, 263 nm, 368 nm and at a concentration of 3.12 × 10−7 M, at 215 nm, 264 nm, 368 nm respectively.
In the second order derivative of the spectra of a 1 × 10−5 M DNOC solution, the minimum is observed more at 215 nm, 234 nm, 267 nm, 365 nm, 370 nm, 379 nm, of which the most important are at 215 nm, 267 nm, 370 nm. If the concentration is 5 × 10−6 M, the minimum is observed more at 215 nm, 230 nm, 233 nm, 263 nm, 268 nm, 361 nm, 366 nm, 372 nm, 374 nm, 376 nm, 378 nm, among which the most important is at 215 nm, 267 nm, 378 nm; If the concentration is 2.5 × 10−6 M, at 216 nm, 233 nm, 263 nm, 268 nm, 360 nm, 365 nm, 369 nm, 374 nm, 376 nm, 378 nm, of the most important is at 215 nm, 267 nm, 378 nm. For the the concentration of 1.25 × 10−6 M, at 377 nm and when the concentration is 6.25 × 10−7 M, the minimum is observed at 376 nm.
Radiant energy absorbed by one mole of substance, E, was calculated using the following equation:
where E is the energy, λ
max is the wavelength at which maximum power is calculated [
28]. Studying
Table 3, it is noted that 2,4-DNP has the largest absorbed radiant energy.
Table 3.
Average absorbance and radiant energy absorbed by one mole of substance calculated for 1 M solutions of nitro compound.
Table 3.
Average absorbance and radiant energy absorbed by one mole of substance calculated for 1 M solutions of nitro compound.
| Name of substance | λmax | Average absorbance at concentration of M × 10−5 | E kcal/mol |
|---|
| 2,4-DNP | 212 | 2.55 | 135.02 |
| 259 | 2.10 | 110.56 |
| 359 | 2.88 | 79.76 |
| 2,6-DNP | 222 | 2.58 | 128.99 |
| 255 | 0.89 | 112.29 |
| 432 | 0.87 | 66.39 |
| 2,4-DNA | 213 | 1.74 | 134.44 |
| 260 | 1.21 | 110.13 |
| 299 | 1.26 | 95.77 |
| DNOC | 213 (2.5−10.0 ×10−6 M) | 1.74 | 134.44 |
| 268 (2.5−10.0 ×10−6 M) | 1.31 | 108.47 |
| 372 (2.5−10.0 ×10−6 M) | 1.88 | 76.98 |
2.3. Theoretical Studies
We looked for possible correlations between these parameters and optical properties of the biological compounds investigated. We studied the molecular parameters of dinitrophenol and dinitrophenyl ethers parameters compared with other compounds or their analogues showing similar properties.
According to PM3 and fluorescence quenching methods the volume, refractivity and polarizability can be correlated with the quenching constants. The tryptophan conformers were analyzed in the first step, then the most stable variants were chosen. Only the 3,5-dinitro-
o-cresol:tryptophan complex is discussed as an example (
Figure 5). Some molecular properties of dinitro-derivative:tryptophan complexes calculated by the MM+ and PM3 methods are presented for comparison in
Table 4. Variation of energy complexes calculated by the PM3 methods are presented for in
Table 5. For a deeper study of the properties of molecular complexes with dinitro-derivatives and tryptophan we studied the most stable structures.
If we take into account the electronic chemical potential of 2,4-DNP compared with that of 2,4-DNA, respectively, at the DNOC site it is noted that 2,4-DNP has lower electronic chemical potential (
Table 6).
Table 4.
QSAR properties of complexes calculated by MM+ and PM3 methods.
Table 4.
QSAR properties of complexes calculated by MM+ and PM3 methods.
| Complex Methods | 2.4-DNP:Trip | DNOC:Trip | DNAN:Trip |
| MM+ | PM3 | MM+ | PM3 | MM+ | PM3 |
| Volume Å3 | 919.12 | 1037.42 | 965.51 | 1038.70 | 960.24 | 1084.00 |
| Refractivity Å3 | 95.09 | 95.09 | 99.57 | 99.57 | 99.03 | 99.99 |
| Polarizability Å3 | 36.82 | 36.82 | 38.66 | 38.66 | 38.66 | 38.66 |
Table 5.
Variation of energy complexes PM3.
Table 5.
Variation of energy complexes PM3.
| Complex | ΔTotal Energy | ΔBinding Energy | ΔHeat of Formation |
|---|
| kcal/mol | kcal/mol | kcal/mol |
|---|
| 2,4-DNP:Tryp | −24.79 | −30.78 | −24.79 |
| 2,4-DNOC:Tryp | −28.85 | −27.85 | −28.85 |
| 2,4-DNA:Tryp | −38.05 | −38.05 | −38.06 |
Figure 5.
DNOC:Tryptophan structures optimized by the PM3 method.
Figure 5.
DNOC:Tryptophan structures optimized by the PM3 method.
Table 6.
Energy data calculated theoretically PM3.
Table 6.
Energy data calculated theoretically PM3.
| Name of substance | EHOMO | ELUMO | ΔE | μ=-χM | η | Δ
N |
|---|
| eV | eV | eV | eV | eV |
|---|
| 2,4-DNP | −10.79 | −1.88 | −8.91 | −6.33 | −4.46 | 0.22 |
| 2,6-DNP | −10.66 | −1.99 | −8.67 | −6.33 | −4.33 | 0.22 |
| 2,4-DNA | −10.81 | −1.69 | −9.12 | −6.25 | −4.56 | 0.21 |
| DNOC | −10.67 | −1.75 | −8.92 | −6.21 | −4.46 | 0.21 |
If the most stable complexes were obtained the results are as summarized in the following table:
Comparing the data summarized in
Table 6,
Table 7 we observe that for the studied compounds the electronegativity values differ greatly from those of the corresponding complexes. The electronic chemical potential value of the 2,4-DNP-tryptophan complex
versus the electronic chemical potential values corresponding to the other complexes is higher for the derivatives, therefore, the electronegativity is the smallest. The electronic chemical potential could make important contributions to the elucidation of the mechanism of toxicity of 2,4-DNP community. We sought possible correlations between these parameters and the optical or biological properties of the investigated compounds [
29,
30]. Analysis was carried out about the physicochemical properties of mono-complexes and dinitro-derivatives with tryptophan using Pearson’s reactions [
31].
Table 7.
Theoretical energy data calculated using the HyperChem program PM3 method.
Table 7.
Theoretical energy data calculated using the HyperChem program PM3 method.
| Name of substance | EHOMO | ELUMO | ΔE | μ=-χM | η | Δ
N |
|---|
| eV | eV | eV | eV | eV |
|---|
| 2,4-DNP:Tryp | −8.55 | −1.40 | −7.15 | −4.98 | −3.57 | 0.22 |
| 2,6-DNP:Tryp | −8.67 | −1.94 | −6.74 | −5.31 | −3.37 | 0.22 |
| 2,4-DNA:Tryp | −8.81 | −1.56 | −7.24 | −5.18 | −3.62 | 0.21 |
| DNOC:Tryp | −8.58 | −1.76 | −6.82 | −5.17 | −3.41 | 0.21 |
2.4. Toxicological Studies
We studied the biological activity of some dinitrophenols on the germination of wheat seeds. The results were statistically interpreted [
32]. We thus studied the biological effects of DNOC, 2,4-DNP and dinocap on the germination of wheat seeds of the variety Gasparom compared to a control sample of 1 × 10
−3 M Tris solution at pH 7 and 22 °C temperature. Treatment solutions had the same concentration as blank Tris (
Figure 6). The toxicity of DNOC and 2,4-DNP at a concentration of 3 × 10
−3 M is 100%. At a concentration of 3 × 10
−4 M they inhibited germination—dinocap by 19%, 2,4-DNP by 87% and DNOC by 99%. Comparing the results of these treatments, it appears that at the same concentration DNOC is the most toxic and dinocap the least toxic. In the case of DNOC no plantlets were obtained (
Figure 6).
Figure 6.
Effect of 4,6-dinitro-o-cresol (DNOC), 2,4-dinitrophenol (2,4-DNP) and dinocap on wheat seed germination.
Figure 6.
Effect of 4,6-dinitro-o-cresol (DNOC), 2,4-dinitrophenol (2,4-DNP) and dinocap on wheat seed germination.
If we consider the total height of the plantlets in a lot, the plantlets’ mass in a lot and the average mass of plantlets, 2,4-DNP reduced the total height of the plantlets by 97.3%, the plantlets’ mass in a lot by 96.4% and the average mass of plantlets by 61.5% and dinocap reduced the total height of the plantlets in a lot by 76.9%, the plantlets’ mass in a lot by 36.3% and the average mass of plantlets by 54.7%. According to the Tukey test differences were significant compared to the blank, with one exception, germination of sample 6. The toxicity of 3 × 10−4 M concentrations increases in the following order: mixture of Tris with dinocap is less toxic than the mixture of Tris with DNP, which is less toxic than the mixture of Tris with DNOC.