Determining Chemical Reactivity Driving Biological Activity from SMILES Transformations: The Bonding Mechanism of Anti-HIV Pyrimidines
Abstract
:1. Introduction
- Computational toxicology [11];
- Integrative structure-property and structure-activity computational workflows [17];
- Interspecies toxicity analysis [18];
- QSAR-1: a defined endpoint;
- QSAR-2: an unambiguous algorithm;
- QSAR-3: a defined domain of applicability;
- QSAR-4: appropriate measures of goodness-of–fit, robustness and predictivity;
- QSAR-5: a mechanistic interpretation, if possible.
- Drug design oriented, which is generated through extensive database screening [39,40], similarity and domain considerations [41,42], producing QSAR models which should be then validated by internal [43,44], external and read-across techniques [45] so that finally the molecules or molecular fragments predicted as most active or inhibitive depending on the endpoint target can be selected;
- Mechanism oriented, which consists mainly in the identification of the fundamental types of interaction that happen at the chemical-to-biological scale so that the structural properties of a compound constitute the causes that can be related to the manifest and recorded effects at a biological site [46,47,48,49,50,51];
- Considering the descriptors of a QSAR model mainly with observable or physicochemical character, e.g., hydrophobicity for cellular wall transduction (the translation motion), the total energy for steric optimization (rotation motion), polarizability for molecular cloud deformation (vibrational motion) [57,58], or more recently, through the chemical reactivity indices (electronegativity, chemical hardness and related quantities) for gaining more insight into the subtle bonding description (binding movement)—leading to the so called chemical reactivity driven biological activity picture (which will be used also in the present work) [59];
- Considering the systematical collection of QSAR models of descriptors in the previous entry along with their basic statistics, e.g., correlation factors, to be then employed either in an algebraic formulation of descriptor-activity correlations, proved to be always superior to the basic statistical one, or to entering in Euclidian paths among the computed endpoints [60], thus involving the square form of the correlation factor, to produce and compare minimum distances toward the most comprehensive (superior in correlation) QSAR model (in turn presumed to be the closest in the QSAR pool of models to the real/recorded activity). This approach, consecrated as Spectral-SAR [57,58,61,62,63], provides the mechanistic interpretation of biological action in terms of the hierarchy of structural causes (descriptors) along the least computed path across available QSAR mode;
- Considering, more recently, the way of improving the previous entry by extensive use of the variational approach in all stages of Spectral-SAR, from screening (i.e., selecting the training set) from a set of toxicants, to assessing the minimum path by considering the molecular passage through cellular walls accompanied by the partial chemical bonds in molecules [64], according with the Simplified Molecular-Input Line-Entry System (SMILES) [65,66,67,68].
2. Results and Discussion
2.1. OECD-QSAR Principle 1: A Defined Endpoint
- (Eco-) toxicological studies, having various end-points (such as inhibition, activation, death, sterility, irritations, etc.) yet produced by a group of similar molecules, i.e., the case of congeneric studies;
- and carcinogenic studies, having essentially the same end-point as the exacerbated apoptosis that in principle diffuses in the organism no matter what the initial trigger point is, and may be initiated by highly structurally diverse molecules, being therefore classified as non-congeneric studies.
- the longest SMILES molecular chain (LoSMoC), when bonds are breaking on aromatic rings and moieties such that the resulting molecule displays a sort of 2D form of the original molecule along the “fractalic” chain, assumed to be the first stage in intermediary molecular defolding targeting the receptor. The maximum SMILES chains in LoSMoC are presumably responsible for best transport/transduction of ligand molecules through cellular (lipidic) walls, after which they may be released with a modified structure due to their further ionization resulting from interactions with cellular layers; accordingly, another SMILES form is generated and considered next, namely:
- the Branching SMILES (BraS), representing the second phase of molecular defolding and providing ligand bond breakages such that many “bays” are formed, yet with consistent “arms” linking the short molecular “skeleton” aiming to favor the binding with a receptor in its pockets. Accordingly, the branching is not necessary in the same points of molecules through a series, but the maximum branching combined with equilibrium of branches is to be obtained in the final BraS. For instance, a long branch adjacent to a short one will not make a strong enough “float” to bind in a receptor pocket; therefore, the branching principle is to have the float-clefs balanced among themselves. To this end branching up to fourth order is performed for the molecules in Table 1.
2.2. OECD-QSAR Principle 2: An Unambiguous Algorithm
2.2.1. Electronegativity and Its Principles
 principal SMILES chain;
principal SMILES chain;  secondary SMILES branch;
secondary SMILES branch;  tertiary SMILES branch;
tertiary SMILES branch;  quaternary SMILES branch; = double bond; # triple bond; /,\ directional bonds; ( ) branch; C, N, F, S, I — atoms present in the molecule; c, n — atoms place in an aromatic ring; C1/2/3, N1/2, c1/2/3, n2 — connectivity points.
quaternary SMILES branch; = double bond; # triple bond; /,\ directional bonds; ( ) branch; C, N, F, S, I — atoms present in the molecule; c, n — atoms place in an aromatic ring; C1/2/3, N1/2, c1/2/3, n2 — connectivity points.
| No. | Structure 2D | SMILES configurations | A | LogP | χ (eV) | η (eV) | π | ω (eV) | |
|---|---|---|---|---|---|---|---|---|---|
| IUPAC name MW AIDS code | LoSMoC | Code LoSMoC | ... LoSMoC ... | ||||||
| BraS | Code BraS | ... BraS ... | |||||||
| 1 |  [3-(2-Methyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 255.28 AIDS352092 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2ccc(C)c(C)c2 | 3.716698 | 0.91 | 23.107212 | 1.5817419 | 7.304356 | 168.78330 |
 | O=C1N(Cc(c(C)cc2)cc2)C(N(/C=C1\)CC#N)=O | 0.44 | 13.240955 | 2.8324015 | 2.3374078 | 30.949511 | |||
| 2 |  [3-(3-Methyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 255.28 AIDS352093 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2cccc(C)c2 | 5.173925 | 0.47 | 22.812517 | 1.5937610 | 7.156819 | 163.26505 |
 | O=C1N(Cc(cc(C)c2)cc2)C(N(/C=C1\)CC#N)=O | 0.44 | 13.043803 | 2.8273990 | 2.3066788 | 30.087865 | |||
| 3 |  [3-(4-Methyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 255.28 AIDS352094 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2ccc(C)cc2 | 4.023191 | 0.47 | 22.852718 | 1.5799314 | 7.232187 | 165.27512 |
 | O=C1N(Cc(ccc2C)cc2)C(N(/C=C1\)CC#N)=O | 0.88 | 13.149213 | 2.8323062 | 2.3212908 | 30.523148 | |||
| 4 |  [3-(2,4-Dimethyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 269.30 AIDS352888 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2ccc(C)cc2C | 3.943095 | 1.06 | 22.695343 | 1.4889604 | 7.621204 | 172.96584 |
 | O=C1N(Cc2c(cc(cc2)C)C)C(N(/C=C1\)CC#N)=O | 1.03 | 13.061603 | 2.7061581 | 2.4133112 | 31.521715 | |||
| 5 |  [3-(2,5-Dimethyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 269.30 AIDS352889 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2cc(C)ccc2C | 4.610833 | 1.06 | 22.961910 | 1.5967679 | 7.190121 | 165.09891 |
 | O=C1N(Cc(cc(C)c2)c(c2)C)C(N(/C=C1\)CC#N)=O | 0.6 | 13.344068 | 2.8843065 | 2.3132194 | 30.867758 | |||
| 6 |  [3-(2,6-Dimethyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 269.30 AIDS352890 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2c(C)cccc2C | 3.707743 | 1.06 | 22.914792 | 1.5375402 | 7.45177 | 170.75577 |
 | O=C1N(Cc(c(C)cc2)c(C)c2)C(N(/C=C1\)CC#N)=O | 0.6 | 13.174123 | 2.7474378 | 2.3975289 | 31.585343 | |||
| 7 |  [3-(3,5-Dimethyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 269.30 AIDS352095 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2cc(C)cc(C)c2 | 6.229147 | 0.63 | 22.322613 | 1.3441469 | 8.303636 | 185.35884 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)CC#N)=O | 1.03 | 12.688503 | 2.5160717 | 2.5214906 | 31.993942 | |||
| 8 |  [3-(3,4-Dimethyl-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 269.30 AIDS352891 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2ccc(C)c(C)c2 | 5.425968 | 0.63 | 22.513298 | 1.4966364 | 7.521298 | 169.32923 |
 | O=C1N(Cc(cc(c2C)C)cc2)C(N(/C=C1\)CC#N)=O | 1.03 | 12.964034 | 2.7262701 | 2.3776137 | 30.823468 | |||
| 9 |  [3-(2,4,6-trimethyl-benzyl)- 2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 283.33 AIDS352892 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2c(C)cc(C)cc2C | 3.716698 | 1.22 | 22.436637 | 1.3498377 | 8.310865 | 186.46785 |
 | O=C1N(Cc2c(cc(cc2C)C)C)C(N(/C=C1\)CC#N)=O | 1.62 | 12.848802 | 2.5836971 | 2.4865149 | 31.948740 | |||
| 10 |  [3-(3-cyanophenyl)methyl-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 266.26 AIDS352893 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2cccc(c2)C#N | 5.128427 | 0.04 | 22.981901 | 1.5807784 | 7.269172 | 167.05939 |
 | O=C1N(Cc(cc(C#N)c2)cc2)C(N(/C=C1\)CC#N)=O | 0.01 | 12.984607 | 2.7188679 | 2.3878703 | 31.00556 | |||
| 11 |  [3-(3,5-Dimethoxy-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 301.30 AIDS352897 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2cc(OC)cc(c2)OC | 5.248720 | -1.67 | 21.820275 | 1.0563595 | 10.32805 | 225.36097 |
 | O=C1N(Cc(cc2OC)cc(OC)c2)C(N(/C=C1\)CC#N)=O | -0.72 | 12.366078 | 2.2360288 | 2.7651875 | 34.194524 | |||
| 12 |  [3-(3,4,5-trimethoxy-benzyl)-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 331.33 AIDS352898 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2cc(OC)c(OC)c(c2)OC | 3.423658 | -2.66 | 21.365171 | 1.0625102 | 10.0541 | 214.80760 |
 | O=C1N(Cc2cc(c(OC)c(OC)c2)OC)C(N(/C=C1\)CC#N)=O | -2.26 | 12.143075 | 2.4593788 | 2.4687280 | 29.977950 | |||
| 13 |  |  | N#CCN1/C=C\C(=O)N(C1=O)Cc3c2ccccc2ccc3 | 5.268411 | 1.16 | 25.868615 | 1.4726275 | 8.78315 | 227.20792 |
| (3-Naphthalen-1-ylmethyl-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl)-acetonitrile 291.31
AIDS352899 |  | O=C1N(Cc(c(cc3)c(cc3)c2)cc2)C(N(/C=C1\)CC#N)=O | 0.25 | 14.682316 | 2.7628433 | 2.6571026 | 39.012422 | ||
| 14 |  (3-Naphthalen-2-ylmethyl-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl)-acetonitrile 291.31 AIDS352900 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc3cc2ccccc2cc3 | 4.435333 | 1.16 | 25.888824 | 1.3140309 | 9.850919 | 255.02871 |
 | O=C1N(Cc(cc(ccc3)c2c3)cc2)C(N(/C=C1\)CC#N)=O | 0.69 | 14.829177 | 2.6159392 | 2.8343888 | 42.031656 | |||
| 15 |  (3-Biphenyl-4-ylmethyl-2,4-dioxo-3,4-dihydro-2H-pyrimidin-1-yl)-acetonitrile 317.35 AIDS352901 |  | N#CCN1/C=C\C(=O)N(C1=O)Cc2ccc(cc2)c3ccccc3 | 4.236572 | 1.25 | 27.000458 | 1.2990428 | 10.39244 | 280.60074 |
 | O=C1N(Cc(c2)ccc(c(cc3)ccc3)c2)C(N(/C=C1\)CC#N)=O | 0.79 | 15.020930 | 2.3806514 | 3.1547941 | 47.387942 | |||
| 16 |  1-Benzyl-3-phenyl-1H-pyrimidine-2,4-dione 278.31 AIDS352902 |  | c1ccccc1CN2/C=C\C(=O)N(C2=O)c3ccccc3 | 3.665546 | 1.55 | 28.617336 | 1.4763650 | 9.691822 | 277.35413 |
 | O=C1N(c(cc2)ccc2)C(N(/C=C1\)Cc(ccc3)cc3)=O | 0.54 | 16.311764 | 2.7002385 | 3.0204302 | 49.268547 | |||
| 17 |  1,3-Dibenzyl-1H-pyrimidine-2,4-dione 292.34 AIDS352903 |  | c1ccccc1CN2/C=C\C(=O)N(C2=O)Cc3ccccc3 | 4.954677 | 1.53 | 27.627131 | 1.4262804 | 9.685028 | 267.56953 |
 | O=C1N(Cc(ccc2)cc2)C(N(/C=C1\)Cc(ccc3)cc3)=O | 1.06 | 15.538736 | 2.6492805 | 2.9326332 | 45.569415 | |||
| 18 |  1-Benzyl-3-(3,5-dimethyl-benzyl)-1H-pyrimidine-2,4-dione 320.39 AIDS352096 |  | c1ccccc1CN2/C=C\C(=O)N(C2=O)Cc3cc(C)cc(C)c3 | 6.630784 | 1.84 | 25.860489 | 0.7302591 | 17.70638 | 457.89563 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)Cc(ccc3)cc3)=O | 1.81 | 14.540931 | 1.5875011 | 4.5798175 | 66.594813 | |||
| 19 |  1-Benzyl-3-(4,6-dimethyl-pyridin-2-ylmethyl)-1H-pyrimidine-2,4-dione 321.38 AIDS352904 |  | c1ccccc1CN2/C=C\C(=O)N(C2=O)Cc3nc(C)cc(C)c3 | 5.136082 | 0.41 | 26.114347 | 0.8253111 | 15.82091 | 413.15277 |
 | O=C1N(Cc(cc(C)c2)nc2C)C(N(/C=C1\)Cc(ccc3)cc3)=O | 0.15 | 14.748792 | 1.7122755 | 4.3067812 | 63.519822 | |||
| 20 |  1-Benzyl-3-(3,5-dimethyl-benzyl)-5-methyl-1H-pyrimidine-2,4-dione334.42 AIDS352905 |  | c1ccccc1CN2/C=C\(C)C(=O)N(C2=O)Cc3cc(C)cc(C)c3 | 5.841637 | 2.12 | 25.007275 | 1.0403700 | 12.01845 | 300.54873 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\C)Cc(ccc3)cc3)=O | 2.39 | 14.063834 | 2.1272754 | 3.3055978 | 46.489379 | |||
| 21 |  1-Benzyl-3-(3,5-dimethyl-benzyl)-5-iodo-1H-pyrimidine-2,4-dione 446.29 AIDS352906 |  | c1ccccc1CN2/C=C\(I)C(=O)N(C2=O)Cc3cc(C)cc(C)c3 | 4.379863 | 2.48 | 25.393186 | 0.8931783 | 14.21507 | 360.96592 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\I)Cc(ccc3)cc3)=O | 2.53 | 13.656576 | 1.4894424 | 4.5844594 | 62.608023 | |||
| 22 |  1-(2,6-Difluoro-benzyl)-3-phenyl-1H-pyrimidine-2,4-dione 314.29 AIDS352907 |  | Fc1cccc(F)c1CN2/C=C\C(=O)N(C2=O)c3ccccc3 | 3.690369 | 1.08 | 28.610234 | 1.4786792 | 9.674253 | 276.78264 |
 | O=C1N(c(cc2)ccc2)C(N(/C=C1\)Cc(c(F)cc3)c(F)c3)=O | −0.66 | 16.175016 | 2.7665356 | 2.9233342 | 47.284980 | |||
| 23 |  1-(2,6-Difluoro-benzyl)-3-(3,5-dimethyl-benzyl)-1H-pyrimidine-2,4-dione 356.37 AIDS352908 |  | Fc1cccc(F)c1CN2/C=C\C(=O)N(C2=O)Cc3cc(C)cc(C)c3 | 6.939302 | 1.37 | 25.844444 | 0.7517152 | 17.19032 | 444.27415 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)Cc(c(F)cc3)c(F)c3)=O | 0.6 | 14.486247 | 1.5713578 | 4.6094680 | 66.773895 | |||
| 24 |  1-(2,6-Difluoro-benzyl)-3-(4,6-dimethyl-pyridin-2-ylmethyl)-1H-pyrimidine-2,4-dione357.36 AIDS352909 |  | Fc1cccc(F)c1CN2/C=C\C(=O)N(C2=O)Cc3nc(C)cc(C)c3 | 5.193820 | −0.06 | 26.085800 | 0.8406863 | 15.51458 | 404.71036 |
 | O=C1N(Cc(cc(C)c2)nc2C)C(N(/C=C1\)Cc(c(F)cc3)c(F)c3)=O | −1.05 | 14.690744 | 1.6779412 | 4.3776098 | 64.310348 | |||
| 25 |  1-(2,6-Difluoro-benzyl)-3-(2,6-dimethyl-pyridin-4-ylmethyl)-1H-pyrimidine-2,4-dione 357.36 AIDS352910 |  | Fc1cccc(F)c1CN2/C=C\C(=O)N(C2=O)Cc3cc(C)nc(C)c3 | 3.886056 | 0.57 | 26.493803 | 0.9063530 | 14.61561 | 387.22308 |
 | O=C1N(Cc(cc(C)n2)cc2C)C(N(/C=C1\)Cc(c(F)cc3)c(F)c3)=O | 0.77 | 14.950333 | 1.7825743 | 4.1934669 | 62.693730 | |||
| 26 |  1,3-Bis-(2,6-difluoro-benzyl)-1H-pyrimidine-2,4-dione 364.30 AIDS352911 |  | Fc1cccc(F)c1CN2/C=C\C(=O)N(C2=O)Cc3c(F)cccc3F | 4.379863 | 0.59 | 27.958833 | 1.5546911 | 8.991764 | 251.39924 |
 | O=C1N(Cc(c(F)cc2)c(F)c2)C(N(/C=C1\)Cc(c(F)cc3)c(F)c3)=O | −1.34 | 15.611849 | 2.8690618 | 2.7207236 | 42.475527 | |||
| 27 |  3-(3,5-Dimethyl-benzyl)-1-phenethyl-1H-pyrimidine-2,4-dione334.42 AIDS352912 |  | c1ccccc1CCN2/C=C\C(=O)N(C2=O)Cc3cc(C)cc(C)c3 | 5.206209 | 2.09 | 25.447501 | 0.8335692 | 15.26418 | 388.43520 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)CCc(cccc3)c3)=O | 2.06 | 14.410323 | 1.8477646 | 3.8993936 | 56.191522 | |||
| 28 |  3-(3,5-Dimethyl-benzyl)-1-prop-2-ynyl-1H-pyrimidine-2,4-dione 268.32 AIDS352913 |  | C#CCN1/C=C\C(=O)N(C1=O)Cc2cc(C)cc(C)c2 | 5.966576 | 0.77 | 21.628890 | 1.4603086 | 7.405589 | 160.17466 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)CC#C)=O | 1.18 | 12.392809 | 2.5046350 | 2.4739751 | 30.659502 | |||
| 29 |  1,3-Bis-(3,5-dimethyl-benzyl)-1H-pyrimidine-2,4-dione348.44 AIDS352914 |  | c1c(C)cc(C)cc1CN2/C=C\C(=O)N(C2=O)Cc3cc(C)cc(C)c3 | 6.283996 | 2.14 | 25.233546 | 0.8800182 | 14.33694 | 361.77196 |
 | O=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)Cc(cc(cc3C)C)c3)=O | 2.55 | 14.566107 | 1.9376961 | 3.7586149 | 54.748388 | |||
| 30 |  [3-(3,5-Dimethyl-benzyl)-2-oxo-4-thioxo-3,4-dihydro-2H-pyrimidin-1-yl]-acetonitrile 285.36 AIDS352915 |  | N#CCN1/C=C\C(=S)N(C1=O)Cc2cc(C)cc(C)c2 | 7.309803 | 1.28 | 21.897722 | 1.6182386 | 6.765913 | 148.15807 |
 | S=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)CC#N)=O | 1.68 | 12.764862 | 3.0237637 | 2.1107572 | 26.943525 | |||
| 31 |  1-Benzyl-3-(3,5-dimethyl-benzyl)-4-thioxo-3,4-dihydro-1H-pyrimidin-2-one 336.45 AIDS352916 |  | c1ccccc1CN2/C=C\C(=S)N(C2=O)Cc3cc(C)cc(C)c3 | 7.292429 | 2.49 | 25.217792 | 1.1471616 | 10.99139 | 277.17849 |
 | S=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)Cc(ccc3)cc3)=O | 2.45 | 14.289267 | 2.4197012 | 2.9526925 | 42.191813 | |||
| 32 |  1-(2,6-Difluoro-benzyl)-3-(3,5-dimethyl-benzyl)-4-thioxo-3,4-dihydro-1H-pyrimidin-2-one 372.43 AIDS352917 |  | Fc1cccc(F)c1CN2/C=C\C(=S)N(C2=O)Cc3cc(C)cc(C)c3 | 7.229147 | 2.02 | 25.321304 | 1.0761564 | 11.76469 | 297.89740 |
 | S=C1N(Cc(cc(C)c2)cc2C)C(N(/C=C1\)Cc(c(F)cc3)c(F)c3)=O | 1.25 | 14.434969 | 2.3806265 | 3.0317586 | 43.763344 | |||
2.2.2. Chemical Hardness and Its Principles
2.2.3. Chemical Power and Its Principle
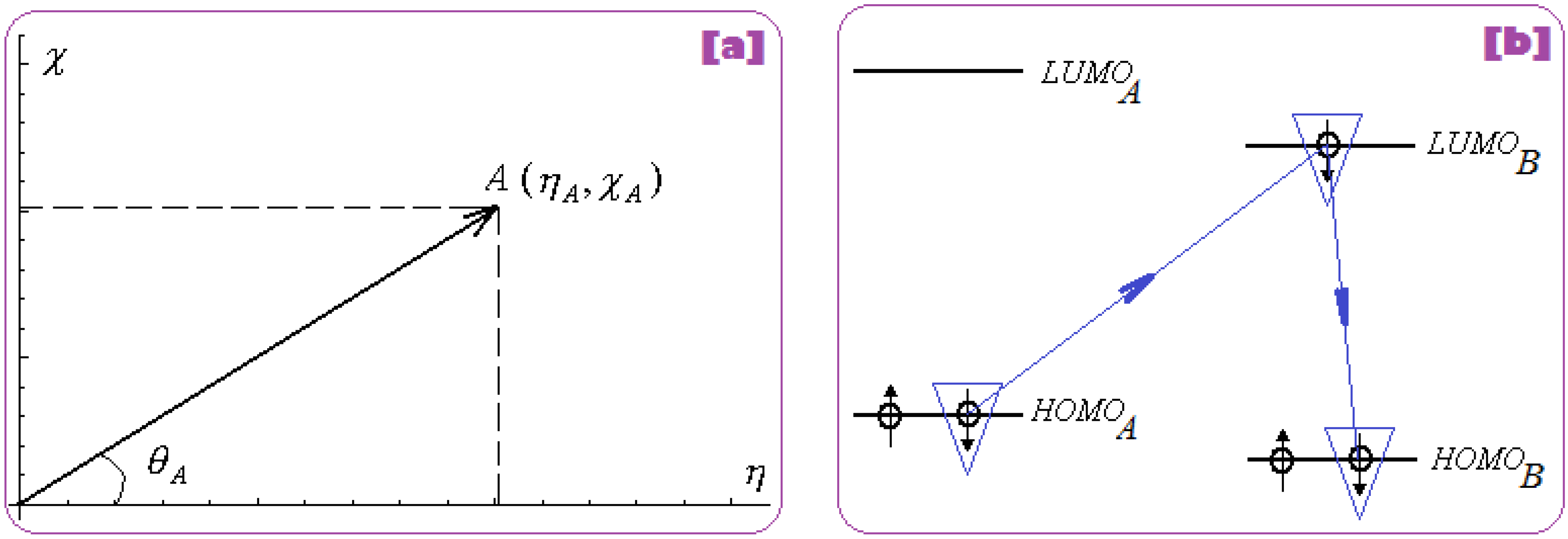
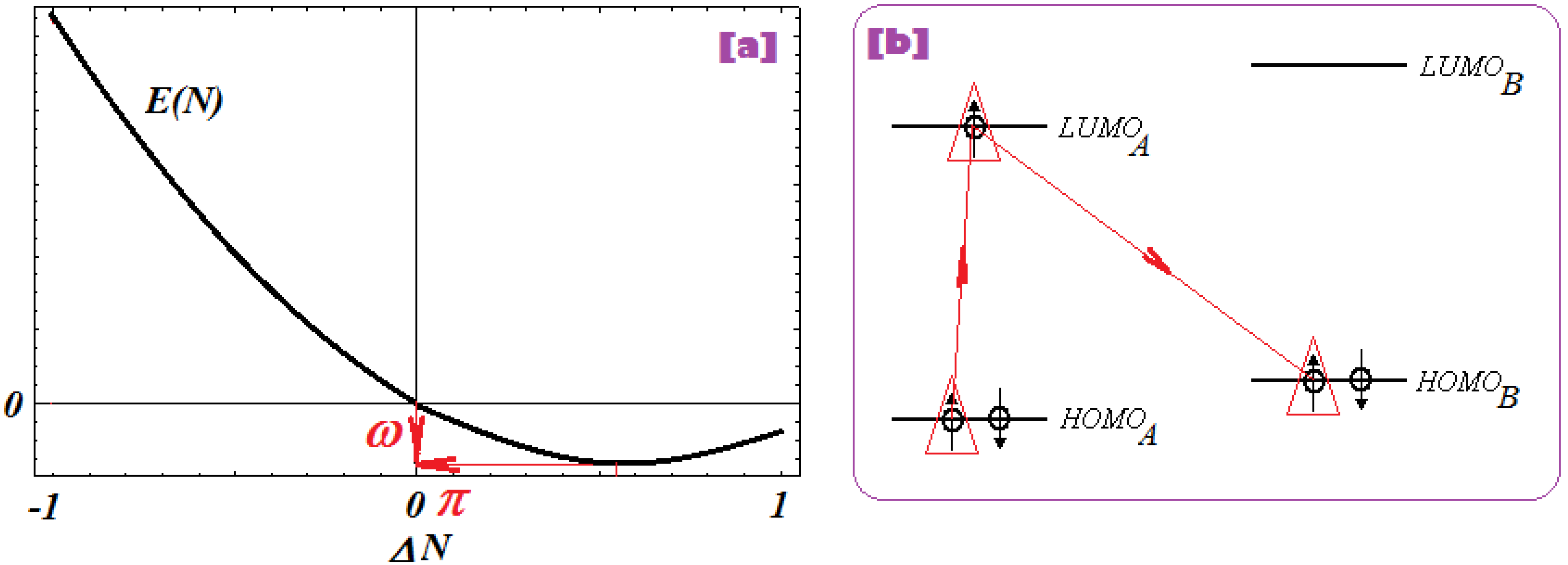
2.2.4. Electrophilicity and Its Principle
2.3. OECD-QSAR Principle 3: A Defined Domain of Applicability
- We consider only first orders of HOMO and LUMO for the LoSMoC molecules of Table 1;
- We consider all three orders of HOMO and LUMO for the BraS molecules of Table 1.
| No. | HOMO1 | LUMO1 | HOMO2 | LUMO2 | HOMO3 | LUMO3 |
|---|---|---|---|---|---|---|
| ... LoSMoC ... | ||||||
| ... BraS ... | ||||||
| 1 | −24.49903 | −19.06514 | X | X | X | X |
| −24.48801 | −19.03451 | −24.88237 | −18.05411 | −25.10602 | −15.57611 | |
| 2 | −24.24188 | −18.7667 | X | X | X | X |
| −24.23715 | −18.75946 | −24.69547 | −17.93489 | −24.84179 | −15.32821 | |
| 3 | −24.25602 | −18.82835 | X | X | X | X |
| −24.2567 | −18.82977 | −24.58191 | −17.70927 | −24.85183 | −15.37954 | |
| 4 | −23.95141 | −18.83626 | X | X | X | X |
| −23.95204 | −18.83621 | −24.28008 | −17.60903 | −24.88104 | −15.38259 | |
| 5 | −24.38787 | −18.90236 | X | X | X | X |
| −24.38787 | −18.90236 | −24.42277 | −17.3528 | −24.90982 | −15.43411 | |
| 6 | −24.24172 | −18.95968 | X | X | X | X |
| −24.23569 | −18.95218 | −24.49526 | −17.86779 | −25.04196 | −15.49452 | |
| 7 | −23.35131 | −18.73365 | X | X | X | X |
| −23.35188 | −18.73767 | −24.22514 | −17.79723 | −24.54057 | −15.31291 | |
| 8 | −23.79299 | −18.65147 | X | X | X | X |
| −23.79239 | −18.65293 | −24.13254 | −17.38683 | −24.7122 | −15.20959 | |
| 9 | −23.46857 | −18.83136 | X | X | X | X |
| −23.46979 | −18.83921 | −24.28395 | −17.49789 | −24.46192 | −15.37871 | |
| 10 | −24.37925 | −18.94867 | X | X | X | X |
| −24.38 | −18.9506 | −24.99142 | −18.7636 | −25.14861 | −15.67584 | |
| 11 | −22.38345 | −18.75445 | X | X | X | X |
| −22.38345 | −18.75445 | −23.79029 | −17.31787 | −24.21747 | −15.29094 | |
| 12 | −21.96501 | −18.31488 | X | X | X | X |
| −21.96844 | −18.31149 | −23.85501 | −16.16856 | −23.89945 | −14.89846 | |
| 13 | −26.91465 | −21.85561 | X | X | X | X |
| −26.91465 | −21.85561 | −27.7802 | −20.69252 | −28.33738 | −18.9827 | |
| 14 | −26.66128 | −22.14708 | X | X | X | X |
| −26.66128 | −22.14708 | −27.47999 | −20.30796 | −27.875 | −19.37649 | |
| 15 | −27.68342 | −23.22071 | X | X | X | X |
| −27.68553 | −23.22033 | −28.91865 | −22.96564 | −28.9519 | −21.82943 | |
| 16 | −29.51216 | −24.44028 | X | X | X | X |
| −29.52823 | −24.42876 | −29.75423 | −23.06013 | −30.9813 | −22.86666 | |
| 17 | −28.49271 | −23.59289 | X | X | X | X |
| −28.47523 | −23.5581 | −29.36592 | −22.66404 | −30.06262 | −21.75203 | |
| 18 | −25.63183 | −23.12311 | X | X | X | X |
| −25.62548 | −23.11217 | −26.88207 | −22.20886 | −27.55654 | −20.01182 | |
| 19 | −26.0344 | −23.19914 | X | X | X | X |
| −26.03953 | −23.19181 | −27.0533 | −22.2293 | −27.84065 | −20.10159 | |
| 20 | −25.36022 | −21.78615 | X | X | X | X |
| −25.36493 | −21.78792 | −26.68329 | −20.71338 | −27.06831 | −19.37157 | |
| 21 | −25.47117 | −22.40276 | X | X | X | X |
| −24.47218 | −22.40179 | −26.77381 | −21.92655 | −27.2391 | −19.67952 | |
| 22 | −29.50944 | −24.42961 | X | X | X | X |
| −29.5088 | −24.42942 | −30.31708 | −23.21535 | −30.95683 | −22.84796 | |
| 23 | −25.65356 | −23.07113 | X | X | X | X |
| −25.6511 | −23.06654 | −26.89824 | −22.43524 | −27.60502 | −19.97251 | |
| 24 | −26.0339 | −23.14582 | X | X | X | X |
| −26.03578 | −23.15325 | −27.02319 | −22.45168 | −27.88159 | −20.08076 | |
| 25 | −26.5313 | −23.41763 | X | X | X | X |
| −26.55279 | −23.43647 | −27.38203 | −22.60093 | −28.51525 | −20.85345 | |
| 26 | −29.02596 | −23.685 | X | X | X | X |
| −28.90689 | −23.43443 | −29.78576 | −22.74551 | −29.8298 | −21.98785 | |
| 27 | −25.41998 | −22.55635 | X | X | X | X |
| −25.42157 | −22.55341 | −26.66657 | −21.11333 | −27.34483 | −19.49103 | |
| 28 | −22.8969 | −17.88018 | X | X | X | X |
| −22.90148 | −17.87531 | −22.96332 | −17.29096 | −24.06666 | −14.20252 | |
| 29 | −25.29808 | −22.27488 | X | X | X | X |
| −25.30234 | −22.27582 | −25.91112 | −20.09649 | −26.56577 | −19.35705 | |
| 30 | −23.42159 | −17.86232 | X | X | X | X |
| −23.42258 | −17.86581 | −23.58074 | −15.8222 | −23.9511 | −15.31823 | |
| 31 | −25.7421 | −21.80116 | X | X | X | X |
| −25.74458 | −21.80102 | −27.02937 | −20.01162 | −27.54453 | −19.79035 | |
| 32 | −25.71771 | −22.0207 | X | X | X | X |
| −25.71681 | −22.02284 | −27.02077 | −19.81276 | −27.47808 | −19.75182 | |
| Index | Criteria | CASE (i) | Case (ii) | ||
|---|---|---|---|---|---|
| Molecules | RQSAR | Molecules | RQSAR | ||
| V1 LoSMoC | Between 15–16 atoms LoSMoC | 1–4, 6–11, 28 | 0.90371960 (a) | 1–9, 28 | 0.92402295 (c) |
| V1 BraS | Main chain and secondary branch with maximum 14 atoms | 2–11, 13, 14, 16, 17, 22, 28 | 0.53158997 | 2, 3, 5–9, 13, 14, 16, 17, 22, 28, 29 | 0.70384894 |
| V2 LoSMoC | Between 18–21 atoms LoSMoC | 13–17, 19, 21, 22, 24, 26, 31, 32 | 0.75180080 | 15–18, 21–23, 27, 29, 31, 32 | 0. 95150144 (b) |
| V2 BraS | Main chain and secondary branch with minimum 14 atoms | 7, 11, 12, 15–17, 19, 22, 24–26, 28, 30–32 | 0.95109419 | 7, 15–17, 20, 21, 22, 27, 28, 29, 30–32 | 0.87354213 |
| V3 LoSMoC | At least one triple bond in the main chain LoSMoC | 1–7, 9–11, 13, 14, 28, 30 | 0.56411064 | 1–4, 6, 7, 9, 13, 15, 28, 30 | 0.49202776 |
| V3 BraS | Secondary and tertiary branches with maximum 14 atoms | 2–10, 13, 14, 28 | 0.62469181 | 1–7, 9, 13, 14, 28 | 0.75756597 |
| V4 LoSMoC | More than three branches in the main chain LoSMoC | 2–4, 6–11, 19, 21, 22, 24–26, 28, 30–32 | 0.43357261 | 2–4, 6, 7, 9, 15,20–23, 27–32 | 0.61510478 |
| V4 BraS | Secondary and tertiary branches with minimum 14 atoms | 11, 15–17, 19, 21–25, 31, 32 | 0.64694148 | 15–17, 20–23, 27, 29, 31, 32 | 0.94183439 |
| V5 LoSMoC | More than four branches in the main chain LoSMoC | 7–9, 11, 19, 21, 24–26, 28, 30–32 | 0.47454364 | 7, 8, 20, 23, 27–32 | 0.71500251 (d) |
| V5 BraS | Minimum 3 tertiary branches | 6, 11, 15–17, 19, 22–26, 31, 32 | 0.94899619 | 6, 15–17, 20–23, 27, 29, 31, 32 | 0.64718879 |
| V6 LoSMoC | Ramifications of LoSMoC main chain containing groups formed only carbon and hydrogen atoms (except common = O, C = O) | 2–4, 6–10, 19, 28, 30, 31 | 0.71050966 (b) | 2–4, 6, 7, 9, 15, 20,27–31 | 0.64508095 |
| V6 BraS | Minimum 1 quaternary branching | 1, 2, 4, 6–8, 10,13–15, 19, 21–25, 28, 30–32 | 0.48549586 | 1, 2, 4, 6–8,13, 14, 20–23, 27–29, 30–32 | 0.63906586 |
| V7 LoSMoC | Ramifications of LoSMoC main chain containing groups consisting of a single atom or –CH3 groups (except common = O, C = O) | 2–7, 9, 10, 19, 22,24–26, 28, 30–32 | 0.57636501 | 2–4, 6, 7, 9, 20–22,27– 32 | 0.61600596 (e) |
| V7 BraS | One of the secondary branches with minimum one triple bond | 1–7, 9–11, 13–15, 28 | 0.63904635 | 1–7, 9, 13–15, 28 | 0.73556023 (d) |
| V8 LoSMoC | At least one branch for the last 6 points main chain LoSMoC | 2–4, 6–11, 19, 23–25, 28, 30, 32 | 0. 51837657 | 2–4, 6, 7, 9, 20, 21, 27–32 | 0.69314160 (d) |
| V8 BraS | The secondary branch linked with C2 of pyrimidinic nucleus with minimum 2 heteroatoms | 1–6, 8–11, 13–15 | 0.58368204 | 1–6, 8, 9, 13–15 | 0.57765388 (f) |
| V9 LoSMoC | LoSMoC main chain contains after N3 atom of the pyrimidine nucleus (central main chain LoSMoC) a group –CH2– | 1–7, 9–11, 13–15, 19, 21, 24–26, 28, 30–32 | 0.37650771 | 1–8, 13–15, 20, 21, 27–32 | 0.63047473 |
| V9 BraS | The secondary branch linked with N3 of pyrimidinic nucleus contains only C and H atoms | 1–8, 10, 11, 13–17, 25, 26, 28, 30–32 | 0.63881109 | 1–8, 13–17, 20, 21, 27–29, 30–32 | 0.72514327 |
| V10 BraS | The secondary branch linked with N3 of pyrimidinic nucleus contains 4 Carbon atoms | 2–4, 6 8–10, 13, 14, 16, 19, 22, 24, 26 | 0.61480396 | 2–6, 8, 9, 13, 14, 16, 17, 22 | 0.53480139 |
| V11 BraS | The secondary branch linked with N3 of pyrimidinic nucleus contains 5–6 Carbon atoms | 7, 12, 15, 18, 21, 23, 25, 28, 30–32 | 0.66627959 | 7, 15, 18, 20, 21, 23, 28, 29, 30–32 | 0.59914507 |
| V12 BraS | The tertiary branching are formed by maximum 3 atoms of C and H | 2, 4–10, 13, 16, 19, 21–25, 28, 30–32 | 0.38470862 | 2, 4, 6–9, 13, 16–18, 20, 21, 28–32 | 0.61909773 |
| V13 BraS | The tertiary branches are formed only by C and H atoms | 2–10, 13, 14, 16, 17, 19, 28, 30, 31 | 0.56415743 | 2–9, 13–16, 20, 27–31 | 0.64691170 |
| V14 BraS | Quaternary branching are contains only one C atom or CH3 group | 1, 2, 5–7, 21–25, 28, 30–32 | 0.57731047 | 2, 5, 6, 20–23, 27, 28, 30–32 | 0.72850903 |
| V15 BraS | A single quaternary branching with maximum 2 atoms (C/O) and H | 1, 2, 5–7, 19, 21, 22, 28, 30, 31 | 0.93051865 | 1, 2, 5, 6, 20–22, 27, 28, 30, 31 | 0.90565106 |
- higher correlation factors;
- screening correlations having maxima of variables as descriptors;
- almost equal sets of compounds producing the precedent points;
- sets of compounds fulfilling the Topliss-Costello rule [152], or at least respecting the basic/independent descriptors of electronegativity and chemical hardness plus the hydrophobicity measure.
- the case (i)/V2 was chosen over V1 since it better fulfills the above criteria (e.g. being based on all variables and on 12 compounds and not on four variables and 11 compounds like V1);
- the case (ii)/V6 was chosen despite the fact versions V1 and V2 have lesser compounds in the set, and to be closer to the previous case, for molecular sets’ cardinals.
- the case (i)/V5 over variant V2 since it has a minimum of three tertiary branching instances, while being in the similar correlation range, so that it better fulfills the “spirit” of molecular branching;
- the case (ii)/V2 over versions V4 and V15 (with lesser compounds in the set), being nevertheless in the same range of higher correlations and having the same cardinal of molecules in the set as its companion case (i)/V5
2.4. OECD-QSAR Principle 4: Appropriate Measures of Goodness-of-Fit, Robustness and Predictivity
| No. | A(x) | LoSMoC | BraS | ||
|---|---|---|---|---|---|
| RCase V2/(i) | RCase V6/(ii) | RCase V5/(i) | RCase V2/(ii) | ||
| I1 | A(logP) | 0.36160241 | 0.43043863 | 0.45645057 | 0.51687516 |
| I2 | A(χ) | 0.70875308 | 0.04142206 | 0.32832072 | 0.63329686 |
| I3 | A(η) | 0.3850668 | 0.27082157 | 0.3694801 | 0.10466918 |
| I4 | A(π) | 0.20001171 | 0.23419593 | 0.23910446 | 0.36217604 |
| I5 | A(ω) | 0.0679732 | 0.21014 | 0.12316764 | 0.52996859 |
| II1 | A(logP, χ) | 0.72462236 | 0.54711991 | 0.54563771 | 0.68322871 |
| II2 | A(logP, η) | 0.53462981 | 0.45498598 | 0.58822038 | 0.78078563 |
| II3 | A(logP, π) | 0.4587341 | 0.47447182 | 0.53086816 | 0.8624387 |
| II4 | A(logP, ω ) | 0.40635079 | 0.49281211 | 0.48406183 | 0.85830581 |
| II5 | A(χ, η) | 0.72042921 | 0.34882836 | 0.44147923 | 0.65793015 |
| II6 | A(χ, π) | 0.72662887 | 0.32861178 | 0.42540934 | 0.67176394 |
| II7 | A(χ, ω) | 0.72663277 | 0.33323936 | 0.41607475 | 0.67165975 |
| II8 | A(η, π) | 0.74023092 | 0.31232276 | 0.46816571 | 0.69205634 |
| II9 | A(η, ω) | 0.74918964 | 0.3278778 | 0.47282745 | 0.6980058 |
| II10 | A(π, ω) | 0.72422189 | 0.31072122 | 0.4687647 | 0.66987725 |
| III1 | A(logP, χ, η ) | 0.72946153 | 0.54741756 | 0.62478127 | 0.83591477 |
| III2 | A(logP, χ, π) | 0.73229267 | 0.54735654 | 0.62197159 | 0.86508134 |
| III3 | A(logP, χ, ω) | 0.73214282 | 0.5471543 | 0.61493693 | 0.86624574 |
| III4 | A(logP, η, π) | 0.74609564 | 0.48854915 | 0.62416978 | 0.87096819 |
| III5 | A(logP, η, ω ) | 0.751297 | 0.51239927 | 0.63374038 | 0.86007179 |
| III6 | A(logP, π, ω) | 0.72648755 | 0.52806785 | 0.65025857 | 0.86552207 |
| III7 | A(χ, η, π) | 0.75053661 | 0.35028746 | 0.4752325 | 0.7019648 |
| III8 | A(χ, η, ω) | 0.74939285 | 0.34885082 | 0.52544907 | 0.70077495 |
| III9 | A(χ, π, ω ) | 0.72663285 | 0.35789332 | 0.83429197 | 0.67179626 |
| III10 | A(η, π, ω) | 0.74919138 | 0.33193549 | 0.47362344 | 0.70165085 |
| V | A(logP, χ, η, π, ω) | 0.7518008 | 0.64508095 | 0.94899619 | 0.87354213 |
| LoSMoC | BraS | ||||||
|---|---|---|---|---|---|---|---|
| Path | δV2/(i) | Path | δV6/(ii) | Path | δV5/(i) | Path | δV2/(ii) |
| I1-II1-III5-V | 0.363999003 | I1-II1-III1-V | 0.15216027 | I1-II1-III5-V | 0.339267818 | I1-II1-III3-V | 0.247430746 |
| I1-II1-III7-V | 0.363945917 | I1-II1-III2-V | 0.15219933 | I1-II1-III6-V | 0.328852605 γ | I1-II1-III4-V | 0.250851034 |
| I1-II1-III8-V | 0.363872037 | I1-II1-III3-V | 0.15232909 | I1-II1-III9-V | 0.323160465 β | I1-II1-III6-V | 0.246918396 |
| I1-II7-III5-V | 0.36586301 | I1-II2-III1-V | 0.13669055 | I1-II2-III5-V | 0.344705061 | I1-II2-III3-V | 0.277498475 |
| I1-II7-III7-V | 0.365814373 | I1-II2-III2-V | 0.13669292 | I1-II2-III6-V | 0.332349493 | I1-II2-III4-V | 0.27890546 |
| I1-II7-III8-V | 0.365747157 | I1-II2-III3-V | 0.13670114 | I1-II2-III9-V | 0.301780663 α | I1-II2-III6-V | 0.277296451 |
| I1-II8-III5-V | 0.378790523 | I1-II3-III1-V | 0.12960764 | I1-II3-III5-V | 0.339863056 | I1-II3-III3-V | 0.345661527 |
| I1-II8-III7-V | 0.378770846 | I1-II3-III2-V | 0.12961931 | I1-II3-III6-V | 0.330206319 | I1-II3-III4-V | 0.345678373 |
| I1-II8-III8-V | 0.378746997 | I1-II3-III3-V | 0.12965837 | I1-II3-III9-V | 0.332807819 | I1-II3-III6-V | 0.345670347 |
| I1-II9-III5-V | 0.387593286 | I1-II4-III1-V | 0.12810286 α | I1-II4-III5-V | 0.350074672 | I1-II4-III3-V | 0.341600891 |
| I1-II9-III7-V | 0.387591632 | I1-II4-III2-V | 0.1281234 β | I1-II4-III6-V | 0.342969246 | I1-II4-III4-V | 0.341675065 |
| I1-II9-III8-V | 0.387594763 | I1-II4-III3-V | 0.12819186 γ | I1-II4-III9-V | 0.369568114 | I1-II4-III6-V | 0.34160106 |
| I2-II1-III5-V | 0.031042298 | I2-II1-III1-V | 0.51504227 | I2-II1-III5-V | 0.392905816 | I2-II1-III3-V | 0.189846412 |
| I2-II1-III7-V | 0.030413493 | I2-II1-III2-V | 0.51505381 | I2-II1-III6-V | 0.383948387 | I2-II1-III4-V | 0.194283111 |
| I2-II1-III8-V | 0.029516257 β | I2-II1-III3-V | 0.51509217 | I2-II1-III9-V | 0.379084442 | I2-II1-III6-V | 0.18917817 |
| I2-II7-III5-V | 0.030467382 | I2-II2-III1-V | 0.43487567 | I2-II2-III5-V | 0.411103551 | I2-II2-III3-V | 0.170615371 β |
| I2-II7-III7-V | 0.029877668 γ | I2-II2-III2-V | 0.43487642 | I2-II2-III6-V | 0.40080012 | I2-II2-III4-V | 0.172894351 γ |
| I2-II7-III8-V | 0.029043119 α | I2-II2-III3-V | 0.434879 | I2-II2-III9-V | 0.37584055 | I2-II2-III6-V | 0.17028659 α |
| I2-II8-III5-V | 0.033370142 | I2-II3-III1-V | 0.44987922 | I2-II3-III5-V | 0.388579959 | I2-II3-III3-V | 0.229289585 |
| I2-II8-III7-V | 0.033146038 | I2-II3-III2-V | 0.44988258 | I2-II3-III6-V | 0.38016273 | I2-II3-III4-V | 0.22931498 |
| I2-II8-III8-V | 0.032872383 | I2-II3-III3-V | 0.44989384 | I2-II3-III9-V | 0.382424544 | I2-II3-III6-V | 0.229302881 |
| I2-II9-III5-V | 0.04049457 | I2-II4-III1-V | 0.46505147 | I2-II4-III5-V | 0.382158589 | I2-II4-III3-V | 0.225267191 |
| I2-II9-III7-V | 0.040478734 | I2-II4-III2-V | 0.46505713 | I2-II4-III6-V | 0.375660505 | I2-II4-III4-V | 0.225379654 |
| I2-II9-III8-V | 0.040508702 | I2-II4-III3-V | 0.46507599 | I2-II4-III9-V | 0.400091867 | I2-II4-III6-V | 0.225267448 |
| I3-II1-III5-V | 0.340602068 | I3-II1-III1-V | 0.29305119 | I3-II1-III5-V | 0.371725449 | I3-II1-III3-V | 0.606860446 |
| I3-II1-III7-V | 0.340545335 | I3-II1-III2-V | 0.29307147 | I3-II1-III6-V | 0.36224466 | I3-II1-III4-V | 0.608262992 |
| I3-II1-III8-V | 0.340466377 | I3-II1-III3-V | 0.29313888 | I3-II1-III9-V | 0.357085205 | I3-II1-III6-V | 0.606651729 |
| I3-II7-III5-V | 0.342455676 | I3-II2-III1-V | 0.22803128 | I3-II2-III5-V | 0.386400836 | I3-II2-III3-V | 0.681535121 |
| I3-II7-III7-V | 0.342403714 | I3-II2-III2-V | 0.2280327 | I3-II2-III6-V | 0.375420048 | I3-II2-III4-V | 0.682109209 |
| I3-II7-III8-V | 0.342331902 | I3-II2-III3-V | 0.22803762 | I3-II2-III9-V | 0.34864824 | I3-II2-III6-V | 0.681452889 |
| I3-II8-III5-V | 0.355336832 | I3-II3-III1-V | 0.23734499 | I3-II3-III5-V | 0.368802149 | I3-II3-III3-V | 0.75781421 |
| I3-II8-III7-V | 0.355315856 | I3-II3-III2-V | 0.23735136 | I3-II3-III6-V | 0.359922688 | I3-II3-III4-V | 0.757821894 |
| I3-II8-III8-V | 0.355290432 | I3-II3-III3-V | 0.23737269 | I3-II3-III9-V | 0.362310878 | I3-II3-III6-V | 0.757818233 |
| I3-II9-III5-V | 0.364129287 | I3-II4-III1-V | 0.24859544 | I3-II4-III5-V | 0.367313037 | I3-II4-III3-V | 0.753713772 |
| I3-II9-III7-V | 0.364127526 | I3-II4-III2-V | 0.24860602 | I3-II4-III6-V | 0.360547493 | I3-II4-III4-V | 0.753747392 |
| I3-II9-III8-V | 0.364130859 | I3-II4-III3-V | 0.24864131 | I3-II4-III9-V | 0.385936759 | I3-II4-III6-V | 0.753713849 |
| I4-II1-III5-V | 0.525288611 | I4-II1-III1-V | 0.32781038 | I4-II1-III5-V | 0.448453944 | I4-II1-III3-V | 0.369625875 |
| I4-II1-III7-V | 0.525251826 | I4-II1-III2-V | 0.32782851 | I4-II1-III6-V | 0.440627193 | I4-II1-III4-V | 0.371924125 |
| I4-II1-III8-V | 0.525200637 | I4-II1-III3-V | 0.32788877 | I4-II1-III9-V | 0.436395432 | I4-II1-III6-V | 0.369283099 |
| I4-II7-III5-V | 0.527198557 | I4-II2-III1-V | 0.25851495 | I4-II2-III5-V | 0.472588851 | I4-II2-III3-V | 0.42730628 |
| I4-II7-III7-V | 0.527164806 | I4-II2-III2-V | 0.2585162 | I4-II2-III6-V | 0.463653781 | I4-II2-III4-V | 0.428221331 |
| I4-II7-III8-V | 0.527118165 | I4-II2-III3-V | 0.25852055 | I4-II2-III9-V | 0.442255821 | I4-II2-III6-V | 0.42717511 |
| I4-II8-III5-V | 0.540332774 | I4-II3-III1-V | 0.26942851 | I4-II3-III5-V | 0.441695569 | I4-II3-III3-V | 0.500330351 |
| I4-II8-III7-V | 0.54031898 | I4-II3-III2-V | 0.26943412 | I4-II3-III6-V | 0.434308982 | I4-II3-III4-V | 0.500341989 |
| I4-II8-III8-V | 0.540302262 | I4-II3-III3-V | 0.26945292 | I4-II3-III9-V | 0.436290182 | I4-II3-III6-V | 0.500336444 |
| I4-II9-III5-V | 0.549182204 | I4-II4-III1-V | 0.281784 | I4-II4-III5-V | 0.426373085 | I4-II4-III3-V | 0.496246943 |
| I4-II9-III7-V | 0.549181037 | I4-II4-III2-V | 0.28179334 | I4-II4-III6-V | 0.420558718 | I4-II4-III4-V | 0.496298005 |
| I4-II9-III8-V | 0.549183247 | I4-II4-III3-V | 0.28182447 | I4-II4-III9-V | 0.44251816 | I4-II4-III6-V | 0.49624706 |
| I5-II1-III5-V | 0.657190923 | I5-II1-III1-V | 0.3508471 | I5-II1-III5-V | 0.534442949 | I5-II1-III3-V | 0.238824486 |
| I5-II1-III7-V | 0.657161522 | I5-II1-III2-V | 0.35086404 | I5-II1-III6-V | 0.52789265 | I5-II1-III4-V | 0.242366256 |
| I5-II1-III8-V | 0.657120609 | I5-II1-III3-V | 0.35092035 | I5-II1-III9-V | 0.524365617 | I5-II1-III6-V | 0.238293632 |
| I5-II7-III5-V | 0.65912139 | I5-II2-III1-V | 0.27934081 | I5-II2-III5-V | 0.563677521 | I5-II2-III3-V | 0.265077074 |
| I5-II7-III7-V | 0.659094395 | I5-II2-III2-V | 0.27934197 | I5-II2-III6-V | 0.556207653 | I5-II2-III4-V | 0.266549633 |
| I5-II7-III8-V | 0.659057091 | I5-II2-III3-V | 0.27934599 | I5-II2-III9-V | 0.53850008 | I5-II2-III6-V | 0.264865576 |
| I5-II8-III5-V | 0.672348982 | I5-II3-III1-V | 0.29108509 | I5-II3-III5-V | 0.52553652 | I5-II3-III3-V | 0.332571954 |
| I5-II8-III7-V | 0.672337897 | I5-II3-III2-V | 0.29109028 | I5-II3-III6-V | 0.519343768 | I5-II3-III4-V | 0.332589464 |
| I5-II8-III8-V | 0.672324461 | I5-II3-III3-V | 0.29110768 | I5-II3-III9-V | 0.521001709 | I5-II3-III6-V | 0.332581122 |
| I5-II9-III5-V | 0.681219886 | I5-II4-III1-V | 0.30401219 | I5-II4-III5-V | 0.502030388 | I5-II4-III3-V | 0.328514246 |
| I5-II9-III7-V | 0.681218945 | I5-II4-III2-V | 0.30402085 | I5-II4-III6-V | 0.497101738 | I5-II4-III4-V | 0.328591374 |
| I5-II9-III8-V | 0.681220726 | I5-II4-III3-V | 0.30404971 | I5-II4-III9-V | 0.515812781 | I5-II4-III6-V | 0.328514423 |
- whenever two descriptors are common for adjacent activities’ correlations—they will be considered as a single common influence in chemical causes for the observed biological activity.
- For case LoSMoC/V2/(i):
(α): I2-II7-III8-V δ[α]=0.029043119 (β): I2-II1-III8-V δ[β]=0.029516257 (γ): I2-II7-III7-V δ[γ]=0.029877668 - For case LoSMoC/V6/(ii):
(α): I1-II4-III1-V δ[α]=0.12810286 (β): I1-II4-III2-V δ[β]=0.1281234 (γ): I1-II4-III3-V δ[γ]=0.12819186 - For case BraS/V5/(i):
(α): I1-II2-III9-V δ[α]=0.301780663 (β): I1-II1-III9-V δ[β]=0.323160465 (γ): I1-II1-III6-V δ[γ]=0.328852605 - For case BraS/V2/(ii):
(α): I2-II2-III6-V δ[α]=0.17028659 (β): I2-II2-III3-V δ[β]=0.170615371 (γ): I2-II2-III4-V δ[γ]=0.172894351
- All the LoSMoC least path lengths are shorter than those of BraS, this way confirming that the chain based SMILES intermediates are prior to those displaying branching SMILES conformations, i.e., in accordance with the steps [A] →[B] of Figure 3 in pyrimidine-related uracil attack onreserve transcriptase;
- While passing from LoSMoC to BraS configurations in the chemical-biological interaction of uracil derivatives–reverse transcriptase binding phenomenology one notes the maintenance of the same criteria variants, namely V2 of Table 3:meaning that the chain-to-branching passage seems to require the same features of the principal chain and of the secondary branch alike;
- Looking now to the cases interchanged in the transformation of equation (22) one also notes that the passage from case (i) based on longest chain in the SMILES configuration to the case (ii) based on the pyrimidinic N3 atom’s neighbors, happens consistently. The mechanism of interaction is described as involving the trans-membrane transduction by means of the longest chain of SMILES configuration; it is followed by the bonding stage centered on the N3 atom of the pyrimidine ring nuclei as already proved to be specific for spirodiazine derivatives in their transformations towards recorded anti-inflammatory activities, anti-HIV activity included [126].
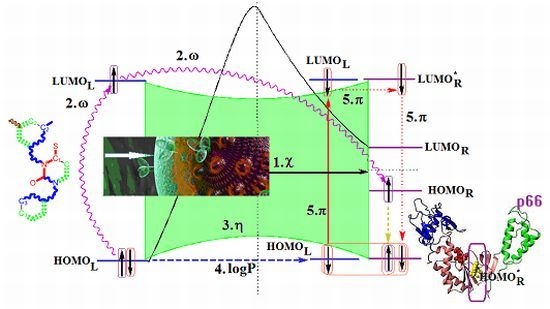
2.5. OECD-QSAR Principle 5: A Mechanistic Interpretation
- Transitivity chain rule, and
- Minimization of redundancies
- The development time is not the physical one but an internal one related with the reaction coordinates, so that the reactivity-driven-activity steps are phenomenological ordered through being interrelated and inter-conditioned during the entire physical time of the binding (on a nano-second scale);
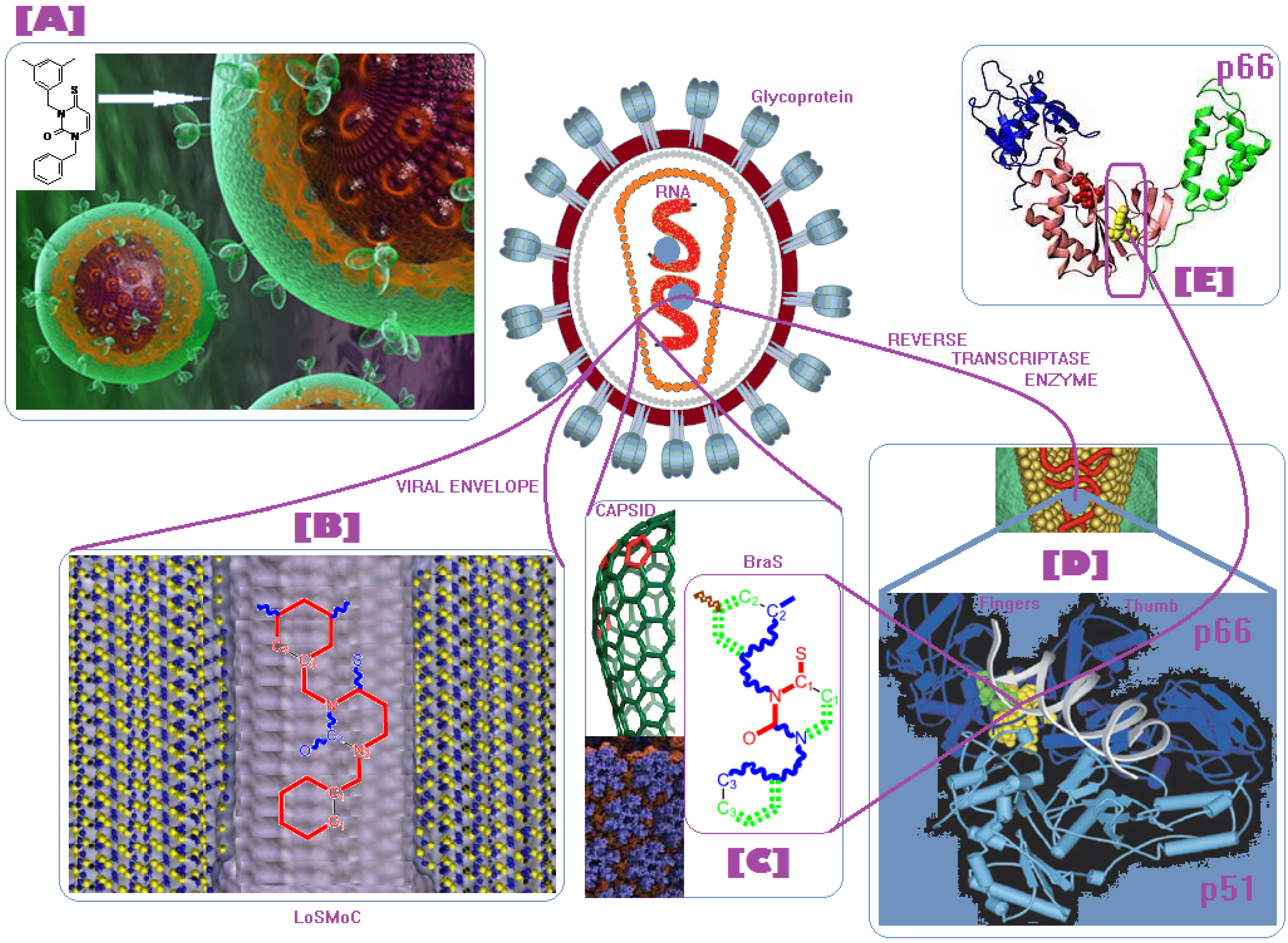
- The described interaction is spatially placed between the ligand (L) represented by the SMILES branched molecule resulted upon the HIV cell’s transduction (at least of the viral envelope) and the receptor–the palm region of the p66 region of the reverse transcriptase.
- The first step is triggered by electronegativity (χ) and of its principle of minimization difference between ligand (L) and receptor (R) HOMO-LUMO middle-levels, as provided by equation (3). In this stage the ligand and receptor are energetically aligned around a common electronegativity; it also associates with “preparation” of HOMO and LUMO states for ceding and accepting electrons by the accompanying interchanging charge;
- The second step accompanies the first one through the electrophilicity (ω) by putting into action the charge transfer by tunneling of the L-R barrier for one electron of the HOMOL level passing to the LUMOL and then down to the HOMOR state by means of the LLR mechanism, see Figure 2b; the minimization principle for electrophilicity, equation (11), further allows the relaxation of the transferred electron from the HOMOR to the HOMOR* level;
- The third step appears naturally “called” by the second one: the R to R* actually corresponds with the expansion of the HOMOR-LUMOR gap to HOMOR*-LUMOR* to be equal with HOMOL-LUMOL one, in accordance with the maximum hardness principle, equation (6), being this step driven by chemical hardness;
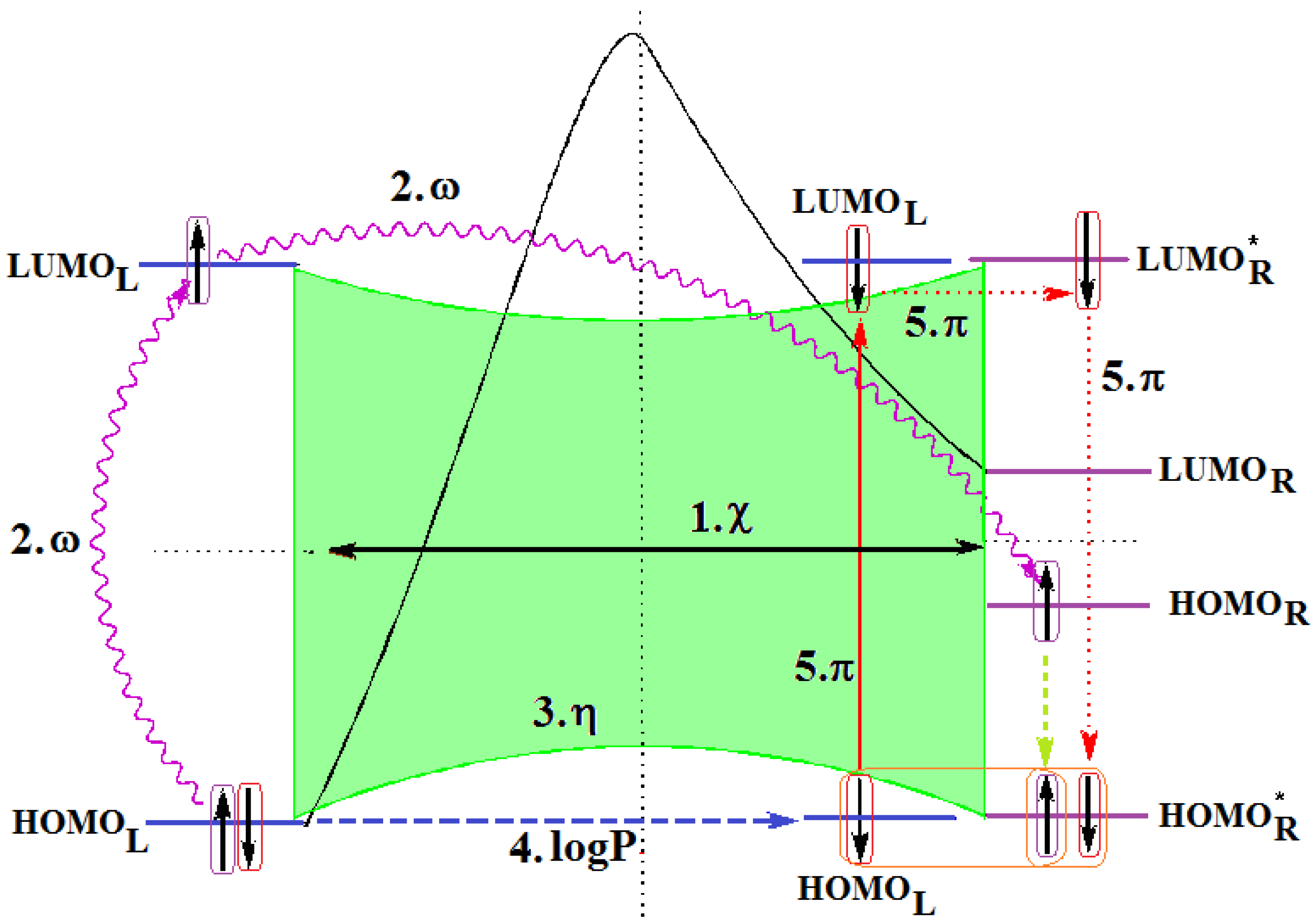
- The fourth step converts spatially the energetic HOMO-LUMO coupling of ligand-receptor by hydrophobicity/lipophilicity (logP) action eventually assuring also the capsid penetration; note that the previous charge transfer was realized through (quantum) tunneling, in accordance with electrophilicity driving action, thus being consistent with the earlier (second step) long-range action of the pyrimidines in the plasmidic region of HIV cell against its reverse transcriptase enzyme inside of the capsid, see Figure 3;
- The fifth and the last step is accomplished by chemical power (π) which assures the effective ligand-receptor binding (now also spatial in nature) by transferring the remaining electron of HOMOL to LUMOL and then to LUMOR* by means of the LRR mechanisms of Figure 1b; it nevertheless fulfils the minimization principle, equation (9), by undergoing the final LUMOR* to HOMOR* relaxation, when it pairs with the electron arrived from the electrophilicity step above.
- Spatially (the molecule is placed in the pocket of HIV’s reverse transcriptase);
- Energetically (all transitions compensate each other);
- By electronic pairing (assured by electrophilicity and chemical power actions);
- By bonding on the relaxed HOMOR* level
3. Conclusions
- For QSAR-OECD Principle 1 (a defined endpoint): considering SMILES longest chain (LoSMoC)- and branching (BraS)-based counterparts of envisaged molecules as the actual molecular ansatz for modeling the envisaged anti-HIV activity by the end-point of half maximal effective concentration (EC50, μM) antiviral activity of 1,3-disubstituted uracils against human immunodeficiency virus (HIV-1)—see Table 1;
- For QSAR-OECD Principle 2 (an unambiguous algorithm): implementing QSAR orthogonal descriptors with associate min-max principles of chemical reactivity: electronegativity and chemical hardness, and of their mixed forms under electrophilicity and chemical power indices; the first two descriptors were also considered with “branching” working forms for BraS molecules up to the third order in HOMO and LUMO, within Koopmans theorem and spectral like resolution frameworks; the last two descriptors are merely associated with chemical charge transfer at the molecular frontier (HOMO and LUMO). Together, they all assure the chemical reactivity-driving-biological activity and provide the molecular mechanism linking structural causes with recorded biological effects (anti-HIV in the present application), while being accompanied by the hydrophobicity/lipophilicity index (logP) modeling the transduction through cellular HIV membranes;
- For QSAR-OECD Principle 3 (a defined domain of applicability): selecting the appropriate QSAR correlation through the screening based on chain (LoSMoC) and branching (BraS) SMILES molecular structures; this stage allows further application of transitivity and minimum redundancy rules for the QSAR descriptors as they are present in the various multi-linear computed endpoints;
- For QSAR-OECD Principle 4 (appropriate measures of goodness-of–fit, robustness and predictivity): ordering the multi-descriptor dependencies with the help of spectral-path length hierarchy for chain (LoSMoC) and branching (BraS) SMILES molecular interaction, and globally in between them, with the aim of Euclidian path measure and of their systematic minimum search across all QSAR models and of their combinations;
- For QSAR-OECD Principle 5 (a mechanistic interpretation, if possible): constructing the molecular (orbital/frontier) diagram describing the mechanism of ligand-receptor interaction based on correlating the least alpha paths of LoSMoC and BraS QSAR analyses with the chemical reactivity descriptors’ electronic manifestations and principles.
Supplementary Materials
Acknowledgments
Conflicts of Interest
References and Notes
- Péry, A.R.R.; Brochot, C.; Zeman, F.A.; Mombelli, E.; Desmots, S.; Pavan, M.; Fioravanzo, E.; Zaldivar, J.M. Prediction of dose-hepatotoxic response in humans based on toxicokinetic/ toxicodynamic modeling with or without in vivo data: A case study with acetaminophen. Toxicol. Lett. 2013, 220, 26–34. [Google Scholar] [CrossRef]
- Diaz Ochoa, J.G.; Bucher, J.; Péry, A.R.R.; Zaldivar, J.M.; Niclas, J.; Mauch, K. A multi-scale modeling framework for individualized, spatiotemporal prediction of drug effects and toxicological risk. Front. Pharmacol. 2013, 3, 204. [Google Scholar]
- Worth, A.P.; Lapenna, S.; Serafimova, R. QSAR and metabolic assessment tools in the assessment of genotoxicity. Method. Mol. Biol. 2012, 930, 125–162. [Google Scholar]
- Tonnelier, A.; Coecke, S.; Zaldivar, J.M. Screening of chemicals for human bioaccumulative potential with a physiologically based toxicokinetic model. Arch. Toxicol. 2012, 86, 393–403. [Google Scholar] [CrossRef]
- Burello, E.; Worth, A.P. QSAR modeling of nanomaterials. WIREs Nanomed. Nanobiotechnol. 2011, 3, 298–306. [Google Scholar] [CrossRef]
- Burello, E.; Worth, A.P. A theoretical framework for predicting the oxidative stress potential of oxide nanoparticles. Nanotoxicology 2011, 5, 228–235. [Google Scholar]
- Burello, E.; Worth, A.P. Computational nanotoxicology: Predicting toxicity of nanoparticles. Nat. Nanotechnol. 2011, 6, 138–139. [Google Scholar] [CrossRef]
- Lo Piparo, E.; Worth, A.; Manibusan, M.; Yang, C.; Schilter, B.; Mazzatorta, P.; Jacobs, M.N.; Steinkellner, H.; Mohimont, L. Use of computational tools in the field of food safety. Regul. Toxicol. Pharm. 2011, 60, 354–362. [Google Scholar] [CrossRef]
- Raevsky, O.A.; Liplavskaya, E.A.; Yarkova, A.V.; Raevskaya, O.E.; Worth, A.P. Linear and nonlinear QSAR models of acute intravenous toxicity of organic chemicals for mice. Biochem. (Moscow) Suppl. Ser. B Biomed. Chem. 2011, 5, 213–225. [Google Scholar] [CrossRef]
- Worth, A.P.; Mostrag-Szlichtyng, A. Towards a common regulatory framework for computational toxicology: current status and future perspectives. In New Horizons in Predictive Toxicology: Methods and Applications; Wilson, A.G.E., Ed.; The Royal Society of Chemistry: Cambridge, UK, 2011; pp. 38–69. [Google Scholar]
- Mostrag-Szlichtyng, A.; Zaldivar, J.M.; Worth, A.P. Computational toxicology at the European Commission`s Joint Research Centre. Expert Opin. Drug Met. 2010, 6, 785–792. [Google Scholar] [CrossRef]
- Novellino, A.; Zaldivar, J.M. Recurrence quantification analysis of spontaneous electrophysiological activity during development: Characterization of in vitro neuronal networks cultured on multi electrode array chips. Adv. Artificial Intelligence 2010, 2010. [Google Scholar] [CrossRef]
- Raevsky, O.A.; Grigor'ev, V.J.; Modina, E.A.; Worth, A.P. Prediction of acute toxicity to mice by the Arithmetic Mean Toxicity (AMT) modelling approach. SAR QSAR Environ. Res. 2010, 21, 265–275. [Google Scholar] [CrossRef]
- Bacelar, F.S.; Dueri, S.; Hernandez-Garcia, E.; Zaldivar, J.M. Joint effects of nutrients and contaminants on the dynamics of a food chain in marine ecosystems. Math. Biosci. 2009, 218, 24–32. [Google Scholar] [CrossRef]
- Poater, A.; Gallegos, A.; Sola, M.; Cavallo, L.; Worth, A.P. Computational methods to predict the reactivity of nanoparticles through structure-property relationships. Expert Opin. Drug Del. 2009, 7, 1–11. [Google Scholar]
- Poater, A.; Gallegos, A.; Carbo-Dorca, R.; Poater, J.; Sola, M.; Cavallo, L.; Worth, A.P. Modeling the structure-property relationships of nanoneedles: A journey toward nanomedicine. J. Comput. Chem. 2009, 30, 275–284. [Google Scholar] [CrossRef]
- Bassan, A.; Worth, A.P. The integrated use of models for the properties and effects of chemicals by means of a structured workflow. QSAR Comb. Sci. 2008, 27, 6–20. [Google Scholar] [CrossRef]
- Kahn, I.; Maran, U.; Benfenati, E.; Netzeva, T.I.; Schultz, T.W.; Cronin, M.T.D. Comparative quantitative structure-activity-activity relationships for toxicity to Tetrahymena pyriformis and Pimephales promelas. Altern. Lab. Anim. ATLA 2007, 35, 15–24. [Google Scholar]
- Gallegos, A. Mini-review on chemical similarity and prediction of toxicity. Curr. Comp.-Aid. Drug. 2006, 2, 105–122. [Google Scholar] [CrossRef]
- Patlewicz, G. Computational methods to predict drug safety. Curr. Comp.-Aid. Drug. 2006, 2, 151–168. [Google Scholar] [CrossRef]
- Dimitrov, S.; Dimitrova, G.; Pavlov, T.; Dimitrova, N.; Patlewicz, G.; Niemela, J.; Mekenyan, O. A stepwise approach for defining the applicability domain of SAR and QSAR models. J. Chem. Inf. Model. 2005, 45, 839–849. [Google Scholar] [CrossRef]
- Gallegos, A.; Carbó-Dorca, R.; Lodier, F.; Cancès, E.; Savin, A. Maximal probability domains in linear molecules. J. Comput. Chem. 2005, 26, 455–460. [Google Scholar]
- Gallegos, A.; Gironés, X. Topological quantum similarity indices based on fitted densities: Theoretical background and QSPR application. J. Chem. Inf. Model. 2005, 45, 321–326. [Google Scholar] [CrossRef]
- Gallegos, A.; Gironés, X. Topological quantum similarity measures: Applications in QSAR. J. Mol. Struct. (THEOCHEM) 2005, 727, 97–106. [Google Scholar] [CrossRef]
- Netzeva, T.I.; Aptula, A.O.; Benfenati, E.; Cronin, M.T.D.; Gini, G.; Lessigiarska, I.; Maran, U.; Vracko, M.; Schuurmann, G. Description of the electronic structure of organic chemicals using semiempirical and ab initio methods for development of toxicological QSARs. J. Chem. Inf. Model. 2005, 45, 106–114. [Google Scholar] [CrossRef]
- Ponec, R.; Bultinck, P.; Gallegos, A. Multicenter bond indices as a new means for the quantitative characterization of homoaromaticity. J. Phys. Chem. A 2005, 109, 6606–6609. [Google Scholar] [CrossRef]
- Schultz, T.W.; Netzeva, T.I.; Roberts, D.W.; Cronin, M.T.D. Structure-Toxicity relationships for the effects to Tetrahymena pyriformis of aliphatic, α,β-carbonyl-containing, unsaturated chemicals. Chem. Res. Toxicol. 2005, 18, 330–341. [Google Scholar] [CrossRef]
- Lessigiarska, I.; Worth, A.P.; Sokull-Klüttgen, B.; Jeram, S.; Dearden, J.C.; Netzeva, T.I.; Cronin, M.T.D. QSAR investigation of a large data set for fish, algae and Daphnia toxicity. SAR QSAR Environ. Res. 2004, 15, 413–431. [Google Scholar] [CrossRef]
- Cronin, M.T.D.; Dearden, J.C.; Walker, J.D.; Worth, A.P. Quantitative structure-activity relationships for human health effects: Commonalities with other endpoints. Environ. Toxicol. Chem. 2003, 22, 1829–1843. [Google Scholar] [CrossRef]
- Worth, A.P.; Cronin, M.T.D. The use of discriminant analysis, logistic regression and classification tree analysis in the development of classification models for human health effects. J. Mol. Struct. (Theochem) 2003, 622, 97–111. [Google Scholar] [CrossRef]
- Cronin, M.T.D.; Dearden, J.C.; Duffy, J.C.; Edwards, R.; Manga, N.; Worth, A.P.; Worgan, A.D.P. The importance of hydrophobicity and electrophilicity descriptors in mechanistically-based QSARs for toxicological endpoints. SAR QSAR Environ. Res. 2002, 13, 167–176. [Google Scholar] [CrossRef]
- Cronin, M.T.D.; Aptula, A.O.; Dearden, J.C.; Duffy, J.C.; Netzeva, T.I.; Patel, H.; Rowe, P.H.; Schultz, T.W.; Worth, A.P.; Voutzoulidis, K.; et al. Structure-based classification of antibacterial activity. J. Chem. Inf. Comp. Sci. 2002, 42, 869–878. [Google Scholar] [CrossRef]
- Worth, A.P.; van Leeuwen, C.J.; Hartung, T. The prospects for using (Q)SARs in a changing political environment — high expectations and a key role for the Commission's Joint Research Centre. SAR QSAR Environ. Res. 2004, 15, 331–343. [Google Scholar] [CrossRef]
- Worth, A.P.; Cronin, M.T.D.; van Leeuwen, C.J. A framework for promoting the acceptance and regulatory use of (Quantitative) Structure-Activity Relationships. In Predicting Chemical Toxicity and Fate; Cronin, M.T.D., Livingstone, D., Eds.; CRC Press: Boca Raton, FL, USA, 2004; pp. 427–438. [Google Scholar]
- OECD. Report from the Expert Group on (Quantitative) Structure-Activity Relationships [(Q)SARs] on the Principles for the Validation of (Q)SARs. Series on Testing and Assessment, No. 49; OECD: Paris, France, 2004. Available online: http://www.oecd.org/document/30/0,2340,en_2649_34365_1916638_1_1_1_1,00.html (accessed on 3 March 2011).
- OECD. Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment. Series on Testing and Assessment, No. 34; OECD: Paris, France, 2005. Available online: http://www.oecd.org/document/30/0,2340,en_2649_34365_1916638_1_1_1_1,00.html (accessed on 3 March 2011).
- OECD. Report on the Regulatory Uses and Applications in OECD Member Countries of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models in the Assessment of New and Existing Chemicals, Series on Testing and Assessment, No. 58. OECD: Paris, France, 2006. Available online: http://www.oecd.org/document/30/0,2340,en_2649_34365_1916638_1_1_1_1,00.html (accessed on 3 March 2011).
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. Series on Testing and Assessment, No. 69. OECD: Paris, France, 2006; 154. Available online: http://www.oecd.org/document/30/0,2340,en_2649_34365_1916638_1_1_1_1,00.html (accessed on 3 March 2011).
- Patlewicz, G.; Jeliazkova, N.; Gallegos, A.; Worth, A.P. Toxmatch—A new software tool to aid in the development and evaluation of chemically similar groups. SAR QSAR Environ. Res. 2008, 19, 397–412. [Google Scholar] [CrossRef]
- Todeschini, R.; Ballabio, D.; Consonni, V.; Mauri, A.; Pavan, M. CAIMAN (Classification And Influence Matrix Analysis): A new approach to the classification based on leverage-scaled functions. Chemom. Intell. Lab. 2007, 87, 3–17. [Google Scholar] [CrossRef]
- Patlewicz, G.; Jeliazkova, N.; Safford, R.J.; Worth, A.P.; Aleksiev, B. An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. SAR QSAR Environ. Res. 2008, 19, 495–524. [Google Scholar] [CrossRef]
- Gallegos, A.; Poater, A.; Jeliazkova, N.; Patlewicz, G.; Worth, A.P. Toxmatch—A chemical classification and activity prediction tool based on similarity measures. Regul. Toxicol. Pharm. 2008, 52, 77–84. [Google Scholar] [CrossRef]
- Patlewicz, G.; Aptula, A.O.; Uriarte, E.; Roberts, D.W.; Kern, P.S.; Gerberick, G.F.; Kimber, I.; Dearman, R.J.; Ryan, C.A.; Basketter, D.A. An evaluation of selected global (Q)SARs/expert systems for the prediction of skin sensitisation potential. SAR QSAR Environ. Res. 2007, 18, 515–541. [Google Scholar] [CrossRef]
- Jaworska, J.; Nikolova-Jeliazkova, N; Aldenberg, T. QSAR applicability domain estimation by projection of the training set descriptor space: a review. Altern. Lab. Anim. 2005, 33, 445–459. [Google Scholar]
- Jeliazkova, N.; Jaworska, J.; Worth, A.P. Open source tools for read across and category formation. In In Silico Toxicology. Principles and Applications; Cronin, M.T.D., Madden, J., Eds.; Royal Society of Chemistry: Cambridge, UK, 2010; pp. 408–445. [Google Scholar]
- Pavan, M.; Todeschini, R. Multi-criteria decision making methods. In Comprehensive Chemometrics; Brown, S., Walczak, B., Tauler, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 591–629. [Google Scholar]
- Scientific data ranking methods: theory and applications. Data handling. In Science and Technology, 1st ed.; Pavan, M.; Todeschini, R. (Eds.) Elsevier: Oxford, UK, 2008; Volume 27.
- Roberts, D.W.; Aptula, A.O.; Patlewicz, G.; Pease, C. Chemical reactivity indices and mechanism-based read-across for non-animal based assessment of skin sensitisation potential. J. Appl. Toxicol. 2008, 28, 443–454. [Google Scholar] [CrossRef]
- Spycher, S.; Netzeva, T.I.; Worth, A.P.; Escher, B.I. Mode of action-based classification and prediction of activity of uncouplers for the screening of chemical inventories. SAR QSAR Environ. Res. 2008, 19, 433–463. [Google Scholar] [CrossRef]
- Benigni, R.; Bossa, C.; Netzeva, T.I.; Rodomonte, A.; Tsakovska, I. Mechanistic QSAR of aromatic amines: New models for discriminating between homocyclic mutagens and nonmutagens, and validation of models for carcinogens. Environ. Mol. Mutagen. 2007, 48, 754–771. [Google Scholar] [CrossRef]
- Vracko, M.; Bandelj, V.; Barbieri, P.; Benfenati, E.; Chaudhry, Q.; Cronin, M.T.D.; Devillers, J.; Gallegos, A.; Gini, G.; Gramatica, P.; et al. Validation of counter propagation neural network models for predictive toxicology according to the OECD principles: A case study. SAR QSAR Environ. Res. 2006, 17, 265–284. [Google Scholar] [CrossRef]
- Dirac, P.A.M. The Principles of Quantum Mechanics, 2nd ed.; Clarendon Press: Oxford, UK, 1947. [Google Scholar]
- QSAR & SPECTRAL-SAR in Computational Ecotoxicology; Putz, M.V. (Ed.) Apple Academics: Ontario, Canada, 2012.
- Putz, M.V. Chemical orthogonal spaces; Mathematical Chemistry Monographs; Publisher: Kragujevac, Serbia, 2012; Volume 14. [Google Scholar]
- Putz, M.V. Chemical Orthogonal Spaces (COSs): From structure to reactivity to biological activity. Int. J. Chem. Model. 2013, 5. in press. [Google Scholar]
- Putz, M.V.; Putz, A.M.; Lazea, M.; Ienciu, L.; Chiriac, A. Quantum-SAR extension of the Spectral-SAR algorithm. Application to polyphenolic anticancer bioactivity. Int. J. Mol. Sci. 2009, 10, 1193–1214. [Google Scholar] [CrossRef]
- Putz, M.V.; Lacrămă, A.M. Introducing spectral structure activity relationship (S-SAR) Analysis. application to ecotoxicology. Int. J. Mol. Sci. 2007, 8, 363–391. [Google Scholar] [CrossRef]
- Putz, M.V.; Putz, A.M. Timisoara Spectral–Structure Activity Relationship (Spectral-SAR) Algorithm: From statistical and algebraic fundamentals to quantum consequences. Series, Chemistry Research and Applications; In Quantum Frontiers of Atoms and Molecules; Putz, M.V., Ed.; NOVA Science Publishers, Inc.: New York, NY, USA, 2011; pp. 539–580. [Google Scholar]
- Putz, M.V.; Putz, A.M. DFT chemical reactivity driven by biological activity: Applications for the toxicological fate of chlorinated PAHs. Struct. Bond. 2013, 150, 181–232. [Google Scholar] [CrossRef]
- Putz, M.V.; Putz, A.M.; Lazea, M.; Chiriac, A. Spectral vs. statistic approach of Structure-Activity Relationship. Application on ecotoxicity of aliphatic amines. J. Theor. Comput. Chem. 2009, 8, 1235–1251. [Google Scholar] [CrossRef]
- Lacrămă, A.M.; Putz, M.V.; Ostafe, V. A Spectral-SAR model for the anionic-cationic interaction in ionic liquids: application to Vibrio fischeri ecotoxicity. Int. J. Mol. Sci. 2007, 8, 842–863. [Google Scholar] [CrossRef]
- Putz, M.V.; Lacrămă, A.M.; Ostafe, V. Spectral-SAR ecotoxicology of ionic liquids. The Daphnia magna Case. Int. J. Ecology (former Res. Lett. Ecology) 2007, 2007. [Google Scholar] [CrossRef]
- Putz, M.V.; Putz, A.M.; Ostafe, V.; Chiriac, A. Spectral-SAR Ecotoxicology of Ionic Liquids-Acetylcholine Interaction on E. Electricus Species. Int. J. Chem. Model. 2010, 2, 85–96. [Google Scholar]
- Putz, M.V.; Dudaş, N.A. Variational principles for mechanistic quantitative structure–activity relationship (QSAR) studies: Application on uracil derivatives’ anti-HIV action. Struct. Chem. 2013. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Weininger, D.; Weininger, A.; Weininger, J.L. SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Model. 1989, 29, 97–101. [Google Scholar] [CrossRef]
- Weininger, D. SMILES. 3. DEPICT. Graphical depiction of chemical structures. J. Chem. Inf. Model. 1990, 30, 237–243. [Google Scholar] [CrossRef]
- Helson, H.E. Structure Diagram Generation. In Reviews in Computational Chemistry; Lipkowitz, K.B., Boyd, D.B., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; Volume 13, pp. 313–398. [Google Scholar]
- Maruyama, T.; Kozai, S.; Yamasaki, T.; Witvrouw, M.; Pannecouque, C.; Balzarini, J.; Snoeck, R.; Andrei, G.; De Clercq, E. Synthesis and antiviral activity of 1,3-disubstituted uracils against HIV-1 and HCMV. Antivir. Chem. Chemoth. 2003, 14, 271–279. [Google Scholar]
- Putz, M.V.; Putz, A.M.; Barou, R. Spectral-SAR realization of OECD-QSAR principles. Int. J. Chem. Model. 2011, 3, 173–190. [Google Scholar]
- Putz, M.V.; Duda-Seiman, C.; Duda-Seiman, D.M.; Putz, A.M. Turning SPECTRAL-SAR into 3D-QSAR analysis. Application on H+K+-ATPase inhibitory activity. Int. J. Chem. Model. 2008, 1, 45–62. [Google Scholar]
- Garg, R.; Gupta, S.P.; Gao, H.; Babu, M.S.; Debnath, A.K.; Hansch, C. QSAR studies on anti HIV-1 drugs. Chem. Rev. 1999, 99, 3525–3601. [Google Scholar] [CrossRef]
- Mehellou, Y.; De Clercq, E. Twenty-six years of anti-HIV drug discovery: Where do we stand and where do we go? J. Med. Chem. 2010, 53, 521–538. [Google Scholar] [CrossRef]
- Esposito, F.; Corona, A.; Tramontano, E. HIV-1 reverse transcriptase still remains a new drug target: structure, function, classical inhibitors, and new inhibitors with innovative mechanisms of actions. Mol. Biol. Int. 2012, 2012. [Google Scholar] [CrossRef]
- Quashie, P.K.; Sloan, R.D.; Wainberg, M.A. Novel therapeutic strategies targeting HIV integrase. BMC Medicine 2012, 10, 34. [Google Scholar]
- Kaufmann, G.R.; Cooper, D.A. Antiretroviral therapy of HIV-1 infection: established treatment strategies and new therapeutic options. Curr. Opin. Microbiol. 2000, 3, 508–514. [Google Scholar] [CrossRef]
- De Clercq, E. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. of Antimicrob. Agents 2009, 33, 307–320. [Google Scholar] [CrossRef]
- Antiviral strategies. In Handbook of Experimental Pharmacology; Krausslich, H.G.; Bartenschlager, R. (Eds.) Springer-Verlag: Berlin-Heidelberg, Germany, 2010; Volume 189.
- Benigni, R.; Netzeva, T.I.; Benfenati, E.; Bossa, C.; Franke, R.; Helma, C.; Hulzebos, E.; Marchant, C.; Richard, A.; Woo, Y.T.; et al. The expanding role of predictive toxicology: An update on the (Q)SAR models for mutagens and carcinogens. J. Environ. Sci. Health C 2007, 25, 53–97. [Google Scholar] [CrossRef]
- Benigni, R.; Bossa, C.; Jeliazkova, N.; Netzeva, T.; Worth, A. The Benigni/Bossa Rules for Mutagenicity and CarcInogenicity—A Module of Toxtree; European Commission report EUR 23241 EN. IdeaConsult Ltd.: Sofia, Bulgaria, 2008. Available online: http://toxtree.sourceforge.net/carc.html (accessed on 3 August 2011).
- Lessigiariska, I.; Worth, A.P.; Cronin, M.T.D. Structure-Activity Relationships for Pharmacotoxicological Endpoints; Lambert Academic Publishing: Saarbrucken, Germany, 2010. [Google Scholar]
- Putz, M.V. Residual-QSAR. Implications for Genotoxic Carcinogenesis. Chem. Cent. J. 2011, 5, 29–40. [Google Scholar] [CrossRef]
- Tarko, L.; Putz, M.V. On Quantitative Structure-Toxicity Relationships (QSTR) using high chemical diversity molecules group. J. Theor. Comput. Chem. 2012, 11, 265–272. [Google Scholar] [CrossRef]
- Chemical Identifier Resolver beta 4. Available online: http://cactus.nci.nih.gov/chemical/structure (accessed on 23 February 2013).
- Moldoveanu, C.C.; Jones, P.G.; Mangalagiu, I.I. Spiroheterocyclic compounds: Old stories with new outcomes. Tetrahedron Lett. 2009, 50, 7205–7208. [Google Scholar] [CrossRef]
- Gammon, D.B.; Snoeck, R.; Fiten, P.; Krecmerova, M.; Holy, A.; De Clercq, E.; Opdenakker, G.; Evans, D.H.; Andrei, G. Mechanism of antiviral drug resistance of Vaccinia virus: Identification of residues in the viral DNA polymerase conferring differential resistance to Antipoxvirus drugs. J. Virol. 2008, 82, 12520–12534. [Google Scholar] [CrossRef]
- Fan, S.Y.; Zheng, Z.B.; Mi, C.L.; Zhou, X.B.; Yan, H.; Gong, Z.H.; Li, S. Synthesis and evaluation of novel chloropyridazine derivatives as potent human rhinovirus (HRV) capsid-binding inhibitors. Bioorg. Med. Chem. 2009, 17, 621–624. [Google Scholar] [CrossRef]
- De Clercq, E. New approaches toward anti-HIV chemotherapy. J. Med. Chem. 2005, 48, 1297–1313. [Google Scholar] [CrossRef]
- Muhanji, C.I.; Hunter, R. Current developments in the synthesis and biological activity of HIV-1 double-drug inhibitors. Curr. Med. Chem. 2007, 14, 1207–1222. [Google Scholar] [CrossRef]
- Butnariu, R.; Mangalagiu, I.I. New pyridazine derivatives: Synthesis, Chemistry and biological activity. Bioorg. Med. Chem. 2009, 17, 2823–2829. [Google Scholar] [CrossRef]
- Balan, A.M.; Florea, O.; Moldoveanu, C.; Zbancioc, G.; Iurea, D.; Mangalagiu, I.I. Diazinium salts with dihydroxyacetophenone skeleton: syntheses and antimicrobial activity. Eur. J. Med. Chem. 2009, 44, 2275–2279. [Google Scholar] [CrossRef]
- Butnariu, R.; Caprosu, M.; Bejan, V.; Ungureanu, M.; Poiata, A.; Tuchilus, C.; Florescu, M.; Mangalagiu, I.I. Pyridazine and phthalazine derivatives with potential antimicrobial activity. J. Heterocycl. Chem. 2007, 44, 1149–1152. [Google Scholar] [CrossRef]
- Parr, R.G.; Yang, W. Density-Functional Theory Of Atoms And Molecules; Oxford University Press: New York, NY, USA, 1989. [Google Scholar]
- Koopmans, T. Uber die Zuordnung von Wellen Funktionen und Eigenwerter zu den einzelnen Elektronen eines Atom. Physica 1934, 1, 104–113. [Google Scholar] [CrossRef]
- Parr, R.G.; Donnelly, R.A.; Levy, M.; Palke, W.E. Electronegativity: The density functional viewpoint. J. Chem. Phys. 1978, 68, 3801–3808. [Google Scholar] [CrossRef]
- Mortier, W.J.; Genechten, K.V.; Gasteiger, J. Electronegativity equalization: Application and parametrization. J. Am. Chem. Soc. 1985, 107, 829–835. [Google Scholar] [CrossRef]
- Sanderson, R.T. Principles of electronegativity Part I. General nature. J. Chem. Edu. 1988, 65, 112–119. [Google Scholar] [CrossRef]
- Putz, M.V. Chemical action concept and principle. MATCH Commun. Math. Comput. Chem. 2011, 66, 35–63. [Google Scholar]
- Tachibana, A. Density functional rationale of chemical reaction coordinate. Int. J. Quantum Chem. 1987, 21, 181–190. [Google Scholar] [CrossRef]
- Tachibana, A.; Parr, R.G. On the redistribution of electrons for chemical reaction systems. Int. J. Quantum. Chem. 1992, 41, 527–555. [Google Scholar] [CrossRef]
- Tachibana, A.; Nakamura, K.; Sakata, K.; Morisaki, T. Application of the regional density functional theory: The chemical potential inequality in the HeH+ system. Int. J. Quantum Chem. 1999, 74, 669–679. [Google Scholar] [CrossRef]
- Parr, R.G.; Yang, W. Density functional approach to the frontier electron theory of chemical reactivity. J. Am. Chem. Soc. 1984, 106, 4049–4050. [Google Scholar] [CrossRef]
- Yang, W.; Parr, R.G.; Pucci, R. Electron density, Kohn-Sham frontier orbitals, and Fukui functionS. J. Chem. Phys. 1984, 81, 2862–2863. [Google Scholar] [CrossRef]
- Putz, M.V. Contributions within Density Functional Theory with Applications in Chemical Reactivity Theory and Electronegativity; Dissertation.com: Parkland, FL, USA, 2003. [Google Scholar]
- Pearson, R.G. Chemical Hardness; Wiley-VCH: Weinheim, Germany, 1997. [Google Scholar]
- Putz, M.V. Electronegativity and chemical hardness: different patterns in quantum chemistry. Curr. Phys. Chem. 2011, 1, 111–139. [Google Scholar] [CrossRef]
- Putz, M.V. Electronegativity: Quantum observable. Int. J. Quantum Chem. 2009, 109, 733–738. [Google Scholar] [CrossRef]
- Putz, M.V. Chemical hardness: Quantum observable? Studia Univ. Babeş-Bolyai-Ser. Chem. 2010, 55, 47–50. [Google Scholar]
- Chattaraj, P.K.; Schleyer, P.V.R. An ab initio study resulting in a greater understanding of the HSAB principle. J. Am. Chem. Soc. 1994, 116, 1067–1071. [Google Scholar] [CrossRef]
- Chattaraj, P.K.; Maiti, B. HSAB principle applied to the time evolution of chemical reactions. J. Am. Chem. Soc. 2003, 125, 2705–2710. [Google Scholar] [CrossRef]
- Putz, M.V.; Russo, N.; Sicilia, E. On the application of the HSAB principle through the use of improved computational schemes for chemical hardness evaluation. J. Comput. Chem. 2004, 25, 994–1003. [Google Scholar] [CrossRef]
- Pearson, R.G. Hard and soft acids and bases—the evolution of a chemical concept. Coord. Chem. Rev. 1990, 100, 403–425. [Google Scholar] [CrossRef]
- Pearson, R.G. Absolute electronegativity and absolute hardness of Lewis acids and bases. J. Am. Chem. Soc. 1985, 107, 6801–6806. [Google Scholar] [CrossRef]
- Chattaraj, P.K.; Lee, H.; Parr, R.G. Principle of maximum hardness. J. Am. Chem. Soc. 1991, 113, 1854–1855. [Google Scholar] [CrossRef]
- Chattaraj, P.K.; Liu, G.H.; Parr, R.G. The maximum hardness principle in the Gyftpoulos-Hatsopoulos three-level model for an atomic or molecular species and its positive and negative ions. Chem. Phys. Lett. 1995, 237, 171–176. [Google Scholar] [CrossRef]
- Putz, M.V. Maximum hardness index of quantum acid-base bonding. MATCH Commun. Math. Comput. Chem. 2008, 60, 845–868. [Google Scholar]
- Ayers, P.W.; Parr, R.G. Variational principles for describing chemical reactions: The Fukui function and chemical hardness revisited. J. Am. Chem. Soc. 2000, 122, 2010–2018. [Google Scholar] [CrossRef]
- Mineva, T.; Sicilia, E.; Russo, N. Density functional approach to hardness evaluation and its use in the study of the maximum hardness principle. J. Am. Chem. Soc. 1998, 120, 9053–9058. [Google Scholar] [CrossRef]
- Torrent-Sucarrat, M.; Solà, M. Gas-phase structures, rotational barriers, and conformational properties of hydroxyl and mercapto derivatives of cyclohexa-2,5-dienone and cyclohexa-2,5-dienthione. J. Phys. Chem. A 2006, 110, 8901–8911. [Google Scholar] [CrossRef]
- Parr, R.G.; Szentpaly, L.V.; Liu, S. Electrophilicity Index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Pérez, P.; Toro-Labbé, A.; Aizman, A.; Contreras, R. Comparison between experimental and theoretical scales of electrophilicity in benzhydryl cations. J. Org. Chem. 2002, 67, 4747–2752. [Google Scholar] [CrossRef]
- Chamorro, E.; Chattaraj, P.K.; Fuentealba, P. Variation of the electrophilicity index along the reaction path. J. Phys. Chem. A 2003, 107, 7068–7072. [Google Scholar] [CrossRef]
- Parthasarathi, R.; Elango, M.; Subramanian, V.; Chattaraj, P.K. Variation of electrophilicity during molecular vibrations and internal rotations. Theor. Chem. Acc. 2005, 113, 257–266. [Google Scholar] [CrossRef]
- Domingo, L.R.; Asensio, A.; Arroyo, P. Density functional theory study of the Lewis acid-catalyzed Diels–Alder reaction of nitroalkenes with vinyl ethers using aluminum derivatives. J. Phys. Org. Chem. 2002, 15, 660–666. [Google Scholar] [CrossRef]
- Domingo, L.R.; Aurell, M.J.; Pérez, P.; Contreras, R. Quantitative characterization of the global electrophilicity power of common diene/dienophile pairs in Diels-Alder reactions. Tetrahedron 2002, 58, 4417–4423. [Google Scholar] [CrossRef]
- Pérez, P.; Toro-Labbé, A.; Contreras, R. Solvent effects on electrophilicity. J. Am. Chem. Soc. 2001, 123, 5527–5531. [Google Scholar] [CrossRef]
- Meneses, L.; Fuentealba, P.; Contreras, R. On the variations of electronic chemical potential and chemical hardness induced by solvent effects. Chem. Phys. Lett. 2006, 433, 54–57. [Google Scholar] [CrossRef]
- Parthasarathi, R.; Subramanian, V.; Chattaraj, P.K. Effect of electric field on the global and local reactivity indices. Chem. Phys. Lett. 2003, 382, 48–56. [Google Scholar] [CrossRef]
- Parthasarathi, R.; Padmanabhan, J.; Subramanian, V.; Maiti, B.; Chattaraj, P.K. Chemical reactivity profiles of two selected polychlorinated biphenyls. J. Phys. Chem. A 2003, 107, 10346–10352. [Google Scholar] [CrossRef]
- Parthasarathi, R.; Subramanian, V.; Roy, D.R.; Chattaraj, P.K. Electrophilicity index as a possible descriptor of biological activity. Bioorg. Med. Chem. 2004, 12, 5533–5543. [Google Scholar] [CrossRef]
- Padmanabhan, J.; Parthasarathi, R.; Subramanian, V.; Chattaraj, P.K. Molecular structure, reactivity, and toxicity of the complete series of chlorinated benzenes. J. Phys. Chem. A. 2005, 109, 11043–11049. [Google Scholar] [CrossRef]
- Padmanabhan, J.; Parthasarathi, R.; Subramanian, V.; Chattaraj, P.K. Chemical reactivity indices for the complete series of chlorinated benzenes: solvent effect. J. Phys. Chem. A 2006, 110, 2739–2745. [Google Scholar] [CrossRef]
- Padmanabhan, J.; Parthasarathi, R.; Subramanian, V.; Chattaraj, P.K. Group philicity and electrophilicity as possible descriptors for modeling ecotoxicity applied to chlorophenols. Chem. Res. Toxicol. 2006, 19, 356–364. [Google Scholar] [CrossRef]
- Rong, C.; Lian, S.; Yin, D.; Zhong, A.; Zhang, R.; Liu, S. Effective simulation of biological systems: choice of density functional and basis set for heme-containing complexes. Chem. Phys. Lett. 2007, 434, 149–154. [Google Scholar] [CrossRef]
- Roy, D.R.; Pal, N.; Mitra, A.; Bultinck, P.; Parthasarathi, R.; Subramanian, V.; Chattaraj, P.K. An atom counting strategy towards analyzing the biological activity of sex hormones. Eur. J. Med. Chem. 2007, 42, 1365–1369. [Google Scholar] [CrossRef]
- Schultz, T.W.; Hewitt, M.; Netzeva, T.I.; Cronin, M.T.D. Assessing applicability domains of toxicological QSARs: Definition, Confidence in predicted values, And the role of mechanisms of action. QSAR Comb. Sci. 2007, 26, 238–254. [Google Scholar] [CrossRef]
- Netzeva, T.I.; Gallegos, A.; Worth, A.P. Comparison of the applicability domain of a QSAR for estrogenicity with a large chemical inventory. Environ. Toxicol. Chem. 2006, 25, 1223–1230. [Google Scholar] [CrossRef]
- Roberts, D.W.; Aptula, A.O.; Patlewicz, G. Mechanistic applicability domains for non-animal based prediction of toxicological endpoints. QSAR analysis of the Schiff base applicability domain for skin sensitization. Chem. Res. Toxicol. 2006, 19, 1228–1233. [Google Scholar] [CrossRef]
- Aptula, A.O.; Patlewicz, G.; Roberts, D.W. Skin sensitization: Reaction mechanistic applicability domains for Structure-Activity Relationships. Chem. Res. Toxicol. 2005, 18, 1420–1426. [Google Scholar] [CrossRef]
- Program Package, HyperChem 7.01, Hypercube, Inc.: Gainesville, FL, USA, 2002.
- Putz, M.V. On absolute aromaticity within electronegativity and chemical hardness reactivity pictures. MATCH Commun. Math. Comput. Chem. 2010, 64, 391–418. [Google Scholar]
- Lele, S.K. Compact finite difference schemes with spectral-like resolution. J. Comput. Phys. 1992, 103, 16–42. [Google Scholar] [CrossRef]
- ViiV Healthcare Group of Companies. The biology of HIV, 2012. Available online: http://www.apositivelife.com/forasos/biology-of-hiv.html (accessed on 4 May 2013).
- Genetic Engineering & Biotechnology News. Pfizer Inks Deal with K.U. Leuven for HIV drugs with new mechanism of action. 2010. Available online: http://www.genengnews.com/gen-news-highlights/pfizer-inks-deal-with-k-u-leuven-for-hiv-drugs-with-new-mechanism-of-action/81243931/ (accessed on 5 April 2013).
- Aksimentiev, A.; Heng, J.B.; Timp, G.; Schulten, K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys. J. 2004, 87, 2086–2097. [Google Scholar] [CrossRef]
- Heng, J.B.; Ho, C.; Kim, T.; Timp, R.; Aksimentiev, A.; Grinkova, Y.V.; Sligar, S.; Schulten, K.; Timp, G. Sizing DNA using a nanometer-diameter pore. Biophys. J. 2004, 87, 2905–2911. [Google Scholar] [CrossRef]
- Perilla, J.R.; Zhao, G.; Chandler, D.; Gronenborn, A.; Zhang, P.; Schulten, K. Refinement of Atomic Models of HIV-1 Oligomers. Available online: http://www.ks.uiuc.edu/ (accessed on 5 April 2013).
- Ganser-Pornillos, B.K.; Yeager, M.; Sundquist, W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008, 18, 203–217. [Google Scholar] [CrossRef]
- Picado, M.J. Avances en la diseminación del VIH. La Ciencia y sus Demonios. 30 April 2012. Available online: http://lacienciaysusdemonios.com/2012/04/30/avances-en-la-diseminacion-del-vih/#more-24038 (accessed on 5 April 2013).
- C.H.A.N.G.E.—Counteracting HIV/AIDS through new gene enhancement. Available online: http://dev.nsta.org/evwebs/577/Present_Technology_Page.html (accessed on 5 April 2013).
- Madrid, M.; Jacobo-Molina, A.; Ding, J.; Arnold, E. Major subdomain rearrangement in HIV-1 reverse transcriptase simulated by molecular dynamics. Proteins Struct. Funct. Bioinf. 1999, 35, 332–337. [Google Scholar] [CrossRef]
- Topliss, J.G.; Costello, J.D. Chance correlation in structure-activity studies using multiple regression analysis. J. Med. Chem. 1972, 15, 1066–1069. [Google Scholar] [CrossRef]
- Puzyn, T.; Mostrag-Szlichtyng, A.; Gajewicz, A.; Skrzynski, M.; Worth, A.P. Investigating the influence of data splitting on the predictive ability of QSAR/QSPR models. Struct. Chem. 2011, 22, 795–804. [Google Scholar] [CrossRef] [Green Version]
- Pavan, M.; Netzeva, T.I.; Worth, A.P. Validation of a QSAR Model for Acute Toxicity. SAR QSAR Environ. Res. 2006, 17, 147–171. [Google Scholar] [CrossRef]
- Putz, M.V.; Lazea, M.; Putz, A.M.; Seiman-Duda, C. Introducing Catastrophe-QSAR. Application on Modeling Molecular Mechanisms of Pyridinone Derivative-Type HIV Non-Nucleoside Reverse Transcriptase Inhibitors. Int. J. Mol. Sci. 2011, 12, 9533–9569. [Google Scholar] [CrossRef]
- Balaban, A.T. Chemical Applications of Graph Theory; Academic Press: London, UK, 1976. [Google Scholar]
- Balaban, A.T. Highly discriminating distance-based topological index. Chem. Phys. Lett. 1982, 89, 399–404. [Google Scholar] [CrossRef]
- Basak, S.C.; Mills, D.R.; Balaban, A.T.; Gute, B.D. Prediction of mutagenicity of aromatic and heteroaromatic amines from structure: a hierarchical QSAR approach. J. Chem. Inf. Comput. Sci. 2011, 41, 671–678. [Google Scholar]
- Balaban, A.T. From chemical topology to 3D geometry. J. Chem. Inf. Comput. Sci. 1997, 37, 645–650. [Google Scholar] [CrossRef]
- Lewis, G.N. The atom and the molecule. J. Am. Chem. Soc. 1916, 38, 762–785. [Google Scholar] [CrossRef]
- Pauling, L.; Wheland, G.W. The nature of the chemical bond. V. The quantum-mechanical calculation of the resonance energy of benzene and naphthalene and the hydrocarbon free radicals. J. Chem. Phys. 1933, 1, 362–375. [Google Scholar] [CrossRef]
- Pauling, L.; Sherman, J. The nature of the chemical bond. VI. The calculation from thermochemical data of the energy of resonance of molecules among several electronic structures. J. Chem. Phys. 1933, 1, 606–618. [Google Scholar] [CrossRef]
- Pauling, L.; Wilson, E.B. Introduction to Quantum Mechanics with Applications to Chemistry; McGraw-Hill: New York, NY, USA, 1935. [Google Scholar]
- Wheland, G.W. The Theory of Resonance and its Application to Organic Chemistry; Wiley: New York, NY, USA, 1944. [Google Scholar]
- Heitler, W.; London, F. Wechselwirkung neutraler Atome und homöopolare Bindung nach der Quantenmechanik. Z. Phys. 1927, 44, 455–472. [Google Scholar] [CrossRef]
- Hückel, E. Quantentheoretische Beiträge zum Benzolproblem. I. Z. Physik 1931, 71, 204–286. [Google Scholar] [CrossRef]
- Hückel, E. Quantentheoretische Beiträge zum Benzolproblem. II. Z. Physik 1931, 72, 310–337. [Google Scholar] [CrossRef]
- Hoffmann, R. An extended Hückel theory. I. Hydrocarbons. J. Chem. Phys. 1963, 39, 1397–1412. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules - A Quantum Theory; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Bader, R.F.W. Principle of stationary action and the definition of a proper open system. Phys. Rev. B 1994, 49, 13348–13356. [Google Scholar] [CrossRef]
- Bader, R.F.W. A bond path: a universal indicator of bonded interactions. J. Phys. Chem. A 1998, 102, 7314–7323. [Google Scholar] [CrossRef]
- Gillespie, R.J.; Nyholm, R.S. Inorganic stereochemistry. Quart. Rev. Chem. Soc. 1957, 11, 339–380. [Google Scholar] [CrossRef]
- Gillespie, R.J.; Hargittai, I. The VSEPR Model of Molecular Geometry; Allyn and Bacon: Boston, MA, USA, 1991. [Google Scholar]
- Kohn, W.; Becke, A.D.; Parr, R.G. Density functional theory of electronic structure. J. Phys. Chem. 1996, 100, 12974–12980. [Google Scholar] [CrossRef]
- Putz, M.V. Chemical action and chemical bonding. J. Mol. Structure THEOCHEM 2009, 900, 64–70. [Google Scholar] [CrossRef]
- Putz, M.V. Quantum parabolic effects of electronegativity and chemical hardness on carbon π-systems. In Carbon Bonding And Structures: Advances in Physics and Chemistry; Putz, M.V., Ed.; Springer Verlag: London, UK, 2011; pp. 1–32. [Google Scholar]
- Putz, M.V. Quantum Theory: Density, Condensation, and Bonding; Apple Academics: Ontario, New Jersey, NJ, USA, 2012. [Google Scholar]
- Sample Availability: Not available.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Putz, M.V.; Dudaş, N.A. Determining Chemical Reactivity Driving Biological Activity from SMILES Transformations: The Bonding Mechanism of Anti-HIV Pyrimidines. Molecules 2013, 18, 9061-9116. https://doi.org/10.3390/molecules18089061
Putz MV, Dudaş NA. Determining Chemical Reactivity Driving Biological Activity from SMILES Transformations: The Bonding Mechanism of Anti-HIV Pyrimidines. Molecules. 2013; 18(8):9061-9116. https://doi.org/10.3390/molecules18089061
Chicago/Turabian StylePutz, Mihai V., and Nicoleta A. Dudaş. 2013. "Determining Chemical Reactivity Driving Biological Activity from SMILES Transformations: The Bonding Mechanism of Anti-HIV Pyrimidines" Molecules 18, no. 8: 9061-9116. https://doi.org/10.3390/molecules18089061