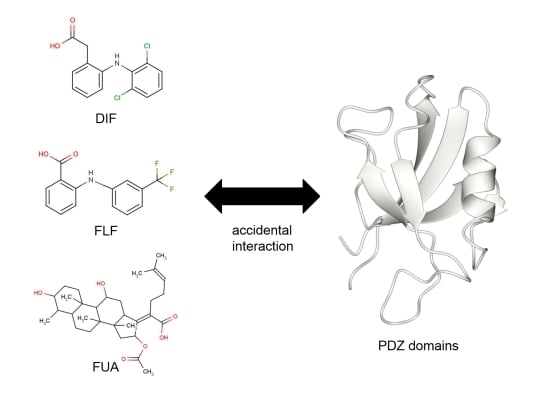
Accidental Interaction between PDZ Domains and Diclofenac Revealed by NMR-Assisted Virtual Screening
Abstract
:1. Introduction
2. Results and Discussion
2.1. eF-Seek Search for Non-peptide Ligands of PDZ Domains in the Human Genome
2.2. Target Selection
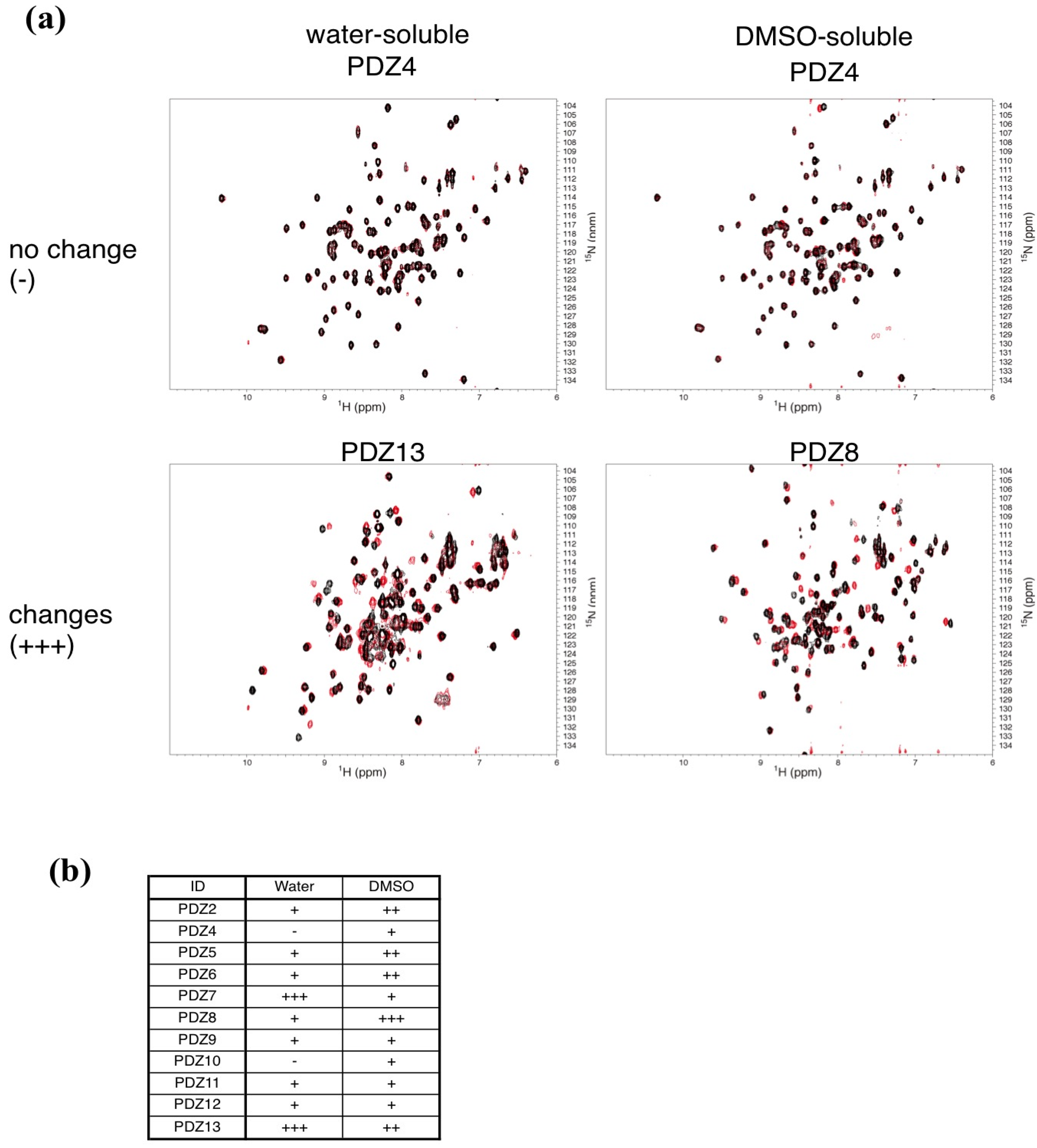
2.3. NMR Titration Experiments of PDZ Domains with the Predicted Ligands
| ID | Sample name | RefSeq ID | Length |
|---|---|---|---|
| PDZ1 * | NHE-RF1_PDZ1 | NP_004243 | 88 |
| PDZ2 | LNX2_PDZ2 | NP_699202 | 89 |
| PDZ3 * | NHE-RF4_PDZ3 | NP_079067 | 93 |
| PDZ4 | Stxbp4_PDZ1 | NP_848604 | 95 |
| PDZ5 | DVL2_PDZ | NP_004413 | 98 |
| PDZ6 | DVL1_PDZ | NP_004412 | 98 |
| PDZ7 | RHPN2_PDZ | NP_149094 | 99 |
| PDZ8 | Harmonin_PDZ2 | NP_710142 | 100 |
| PDZ9 | InaDL_PDZ8 | NP_795352 | 106 |
| PDZ10 | LIMK2_PDZ | NP_057952 | 107 |
| PDZ11 | Harmonin_PDZ3 | NP_710142 | 108 |
| PDZ12 | Neurabin-2_PDZ | NP_115984 | 110 |
| PDZ13 | InaDL_PDZ6 | NP_795352 | 113 |
| PDZ14 * | PAR-6beta_PDZ | NP_115910 | 127 |
| mZO-1 PDZ1 | Mouse ZO-1_PDZ1 | NP_009386 | 94 |
| Water-soluble | DMSO-soluble | ||||
|---|---|---|---|---|---|
| ID | Name | Structural formula | ID | Name | Structural formula |
| A2G | N-acetyl-2-deoxy-2-amino-galactose | 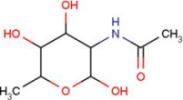 | MPB | 4-hydroxy-benzoic acid methyl ester | 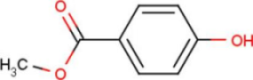 |
| DIF | 2-[(2,6-dichlorophenyl) amino] benzene-acetic acid |  | FLF | 2-[[3-(trifluoro-methyl) phenyl] amino] benzoic acid |  |
| FUA | fusidic acid | 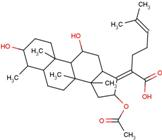 | MIL | milrinone |  |
| SIA | O-sialic acid | 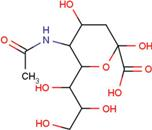 | MNA | 2-O-methyl-5-N-acetyl-α-D-neuraminic acid | 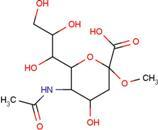 |
| NES | 2-(2-hydroxy-1,1-dihydroxymethyl-ethylamino)-ethanesulfonic acid | 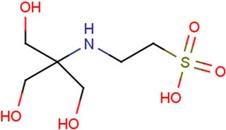 | TRP | tryptophan |  |
2.4. Determination of the Mouse Zo1-PDZ1 Binding Site of the 3 Ligands
| ID | A2G | DIF | FUA | SIA | NES | TRP | MPB | FLF | MIL | MNA |
|---|---|---|---|---|---|---|---|---|---|---|
| PDZ7 | − | ++ | ++ | − | − | − | − | + | − | − |
| PDZ8 | − | + | − | − | − | − | − | ++ | − | − |
| PDZ9 | − | + | − | − | − | − | − | + | − | − |
| PDZ11 | − | − | + | − | − | − | − | + | − | − |
| PDZ13 | n. e. | ++ | + | − | − | − | − | ++ | − | − |
| mZO-1 PDZ1 | n. e. | + | + | n. e. | n. e. | n. e. | n. e. | + | n. e. | n. e. |
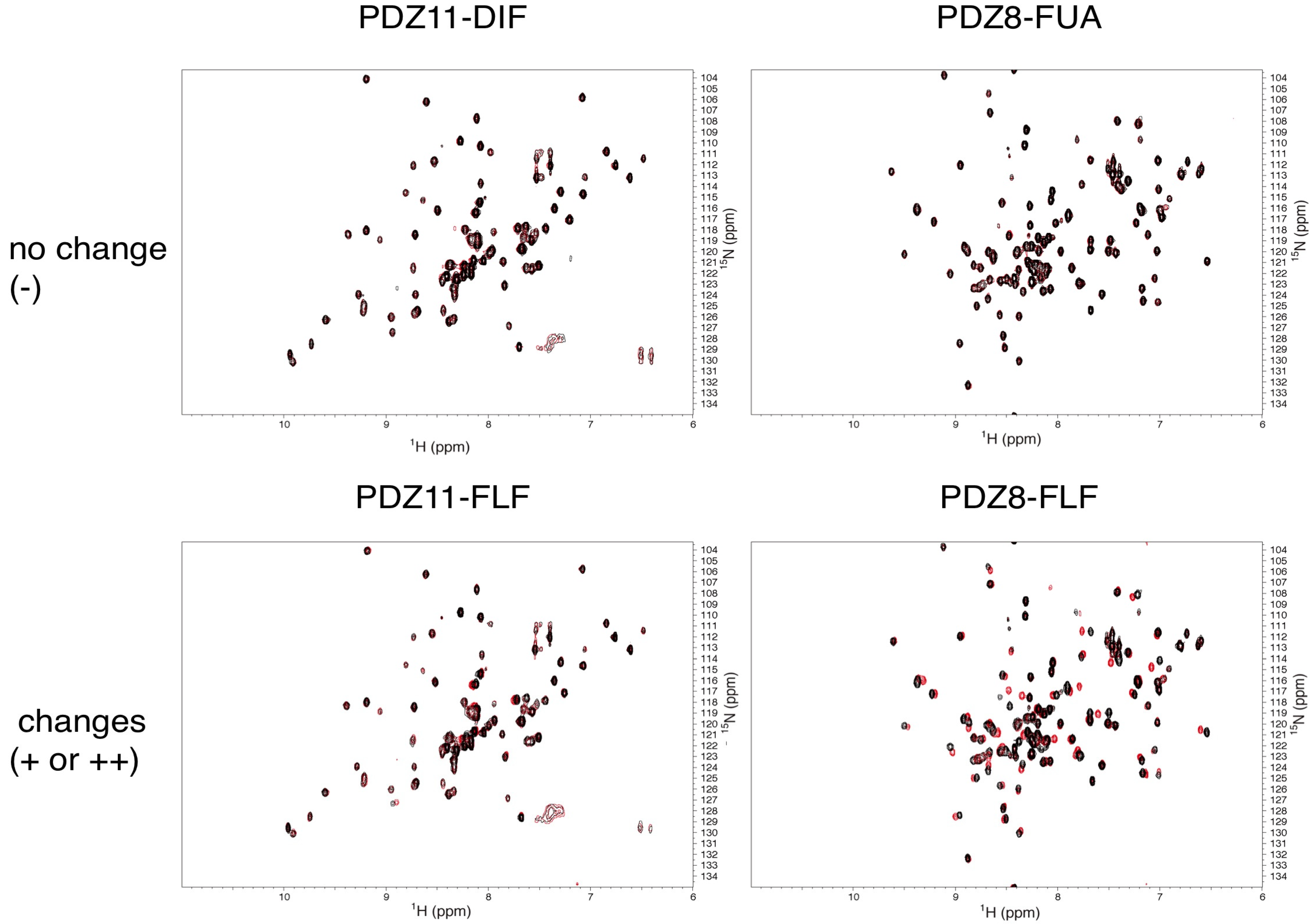

2.5. Critical Residues Involved in Accidental PDZ–Diclofenac Interactions
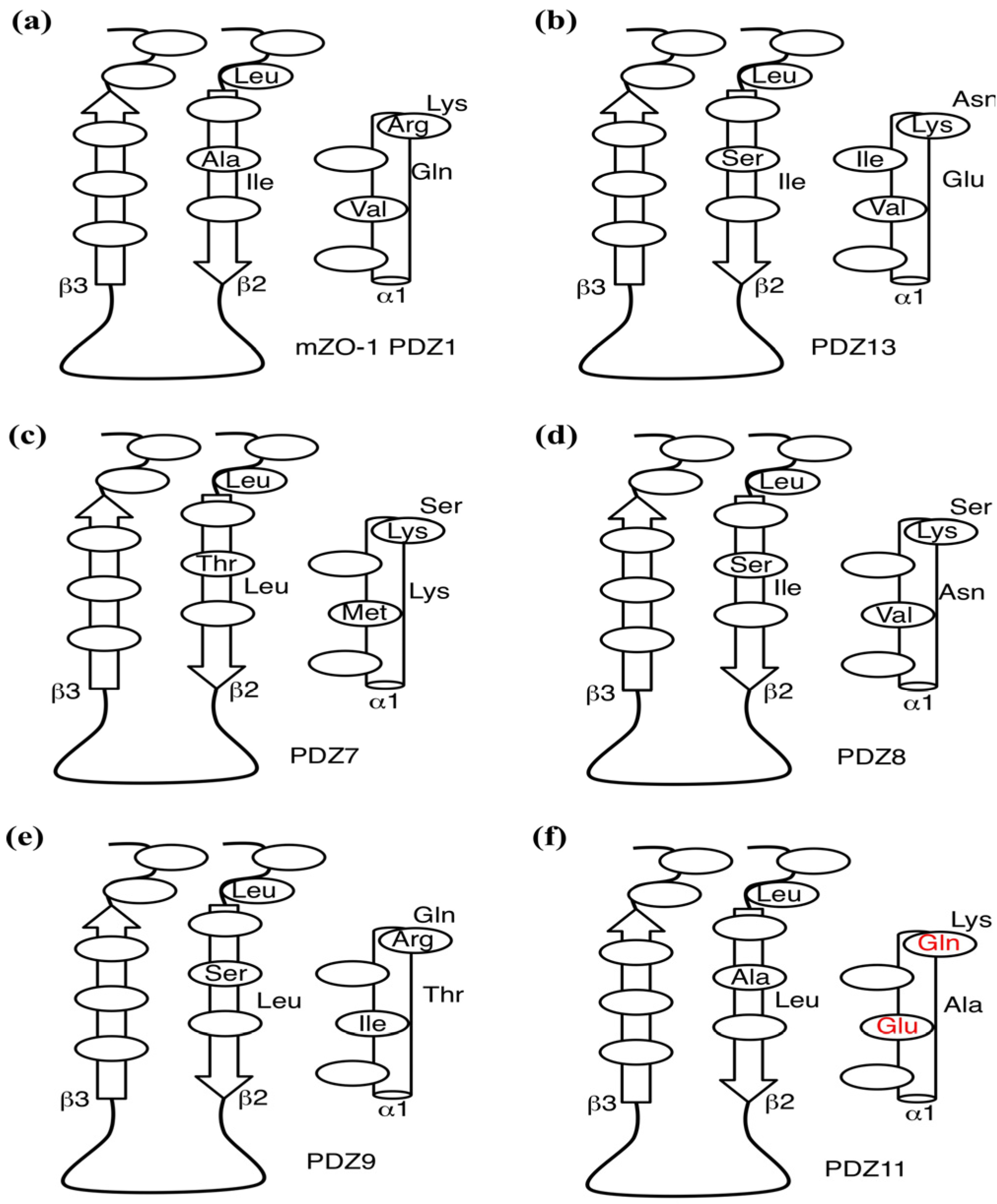
2.6. Pharmacological Implications of Accidental PDZ–Diclofenac Interactions

3. Experimental
3.1. Preparation of Protein Samples
3.2. NMR Experiments
4. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Brooijmans, N.; Kuntz, I.D. Molecular recognition and docking algorithms. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 335–373. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Protein-ligand docking: Current status and future challenges. Proteins 2006, 65, 15–26. [Google Scholar]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Goto, J.; Kataoka, R.; Muta, H.; Hirayama, N. ASEDock-docking based on alpha spheres and excluded volumes. J. Chem. Inf. Model. 2008, 48, 583–590. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Miller, M.D.; Kearsley, S.K.; Underwood, D.J.; Sheridan, R.P. FLOG: A system to select “quasi-flexible” ligands complementary to a receptor of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994, 8, 153–174. [Google Scholar] [CrossRef]
- McGann, M. FRED pose prediction and virtual screening accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. 1996, 9, 1–5. [Google Scholar] [CrossRef]
- Fukunishi, Y.; Mikami, Y.; Nakamura, H. Similarities among receptor pockets and among compounds: Analysis and application to in silico ligand screening. J. Mol. Graph. Model. 2005, 24, 34–45. [Google Scholar] [CrossRef]
- Ekins, S.; Williams, A.J.; Krasowski, M.D.; Freundlich, J.S. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov. Today 2011, 16, 298–310. [Google Scholar] [CrossRef]
- Dudley, J.T.; Deshpande, T.; Butte, A.J. Exploiting drug-disease relationships for computational drug repositioning. Brief. Bioinforma. 2011, 12, 303–311. [Google Scholar] [CrossRef]
- Dubus, E.; Ijjaali, I.; Barberan, O.; Petitet, F. Drug repositioning using in silico compound profiling. Future Med. Chem. 2009, 1, 1723–1736. [Google Scholar] [CrossRef]
- Xie, L.; Xie, L.; Bourne, P.E. Structure-based systems biology for analyzing off-target binding. Curr. Opin. Struct. Biol. 2011, 21, 189–199. [Google Scholar] [CrossRef]
- Lounkine, E.; Keiser, M.J.; Whitebread, S.; Mikhailov, D.; Hamon, J.; Jenkins, J.L.; Lavan, P.; Weber, E.; Doak, A.K.; Côté, S.; et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 2012, 486, 361–367. [Google Scholar]
- Kinoshita, K.; Murakami, Y.; Nakamura, H. eF-seek: Prediction of the functional sites of proteins by searching for similar electrostatic potential and molecular surface shape. Nucleic Acids Res. 2007, 35, W398–W402. [Google Scholar] [CrossRef]
- Kinoshita, K.; Nakamura, H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. Publ. Protein Soc. 2003, 12, 1589–1595. [Google Scholar] [CrossRef]
- Kinoshita, K.; Furui, J.; Nakamura, H. Identification of protein functions from a molecular surface database, eF-site. J. Struct. Funct. Genomics 2002, 2, 9–22. [Google Scholar] [CrossRef]
- Motono, C.; Nakata, J.; Koike, R.; Shimizu, K.; Shirota, M.; Amemiya, T.; Tomii, K.; Nagano, N.; Sakaya, N.; Misoo, K.; et al. SAHG, a comprehensive database of predicted structures of all human proteins. Nucleic Acids Res. 2011, 39, D487–D493. [Google Scholar] [CrossRef]
- Harris, B.Z.; Lim, W.A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001, 114, 3219–3231. [Google Scholar]
- Sheng, M. Molecular organization of the postsynaptic specialization. Proc. Natl. Acad. Sci. USA 2001, 98, 7058–7061. [Google Scholar] [CrossRef]
- Letunic, I.; Copley, R.R.; Schmidt, S.; Ciccarelli, F.D.; Doerks, T.; Schultz, J.; Ponting, C.P.; Bork, P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004, 32, D142–D144. [Google Scholar] [CrossRef]
- Gaudet, S.; Branton, D.; Lue, R.A. Characterization of PDZ-binding kinase, a mitotic kinase. Proc. Natl. Acad. Sci. USA 2000, 97, 5167–5172. [Google Scholar] [CrossRef]
- Muders, M.H.; Vohra, P.K.; Dutta, S.K.; Wang, E.; Ikeda, Y.; Wang, L.; Udugamasooriya, D.G.; Memic, A.; Rupasinghe, C.N.; Rupashinghe, C.N.; et al. Targeting GIPC/synectin in pancreatic cancer inhibits tumor growth. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 4095–4103. [Google Scholar] [CrossRef]
- Jeong, K.W.; Kim, H.-Z.; Kim, S.; Kim, Y.S.; Choe, J. Human papillomavirus type 16 E6 protein interacts with cystic fibrosis transmembrane regulator-associated ligand and promotes E6-associated protein-mediated ubiquitination and proteasomal degradation. Oncogene 2007, 26, 487–499. [Google Scholar] [CrossRef]
- Fallon, L.; Moreau, F.; Croft, B.G.; Labib, N.; Gu, W.-J.; Fon, E.A. Parkin and CASK/LIN-2 associate via a PDZ-mediated interaction and are co-localized in lipid rafts and postsynaptic densities in brain. J. Biol. Chem. 2002, 277, 486–491. [Google Scholar]
- Aarts, M.; Liu, Y.; Liu, L.; Besshoh, S.; Arundine, M.; Gurd, J.W.; Wang, Y.-T.; Salter, M.W.; Tymianski, M. Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science 2002, 298, 846–850. [Google Scholar] [CrossRef]
- Tomita, S.; Ozaki, T.; Taru, H.; Oguchi, S.; Takeda, S.; Yagi, Y.; Sakiyama, S.; Kirino, Y.; Suzuki, T. Interaction of a neuron-specific protein containing PDZ domains with Alzheimer’s amyloid precursor protein. J. Biol. Chem. 1999, 274, 2243–2254. [Google Scholar] [CrossRef]
- Dev, K.K. Making protein interactions druggable: Targeting PDZ domains. Nat. Rev. Drug Discov. 2004, 3, 1047–1056. [Google Scholar] [CrossRef]
- Bach, A.; Stuhr-Hansen, N.; Thorsen, T.S.; Bork, N.; Moreira, I.S.; Frydenvang, K.; Padrah, S.; Christensen, S.B.; Madsen, K.L.; Weinstein, H.; et al. Structure-activity relationships of a small-molecule inhibitor of the PDZ domain of PICK1. Org. Biomol. Chem. 2010, 8, 4281–4288. [Google Scholar] [CrossRef]
- Fujii, N.; Haresco, J.J.; Novak, K.A.P.; Stokoe, D.; Kuntz, I.D.; Guy, R.K. A selective irreversible inhibitor targeting a PDZ protein interaction domain. J. Am. Chem. Soc. 2003, 125, 12074–12075. [Google Scholar] [CrossRef]
- Boucherle, B.; Vogrig, A.; Deokar, H.; Bouzidi, N.; Ripoche, I.; Thomas, I.; Marin, P.; Ducki, S. Synthesis and evaluation of bidentate ligands designed to interact with PDZ domains. Bioorg. Med. Chem. 2011, 19, 4346–4354. [Google Scholar] [CrossRef]
- Goda, N.; Tenno, T.; Takasu, H.; Hiroaki, H.; Shirakawa, M. The PRESAT-vector: Asymmetric T-vector for high-throughput screening of soluble protein domains for structural proteomics. Protein Sci. Publ. Protein Soc. 2004, 13, 652–658. [Google Scholar] [CrossRef]
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68. [Google Scholar] [CrossRef]
- Hiroaki, H. Recent applications of isotopic labeling for protein NMR in drug discovery. Expert Opin. Drug Discov. 2013, 8, 523–536. [Google Scholar] [CrossRef]
- Schanda, P.; Kupce, E.; Brutscher, B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 2005, 33, 199–211. [Google Scholar] [CrossRef]
- Zhang, Y.; Yeh, S.; Appleton, B.A.; Held, H.A.; Kausalya, P.J.; Phua, D.C.Y.; Wong, W.L.; Lasky, L.A.; Wiesmann, C.; Hunziker, W.; et al. Convergent and divergent ligand specificity among PDZ domains of the LAP and zonula occludens (ZO) families. J. Biol. Chem. 2006, 281, 22299–22311. [Google Scholar] [CrossRef]
- Appleton, B.A.; Zhang, Y.; Wu, P.; Yin, J.P.; Hunziker, W.; Skelton, N.J.; Sidhu, S.S.; Wiesmann, C. Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity. J. Biol. Chem. 2006, 281, 22312–22320. [Google Scholar]
- Umetsu, Y.; Goda, N.; Taniguchi, R.; Satomura, K.; Ikegami, T.; Furuse, M.; Hiroaki, H. 1H, 13C, and 15N resonance assignment of the first PDZ domain of mouse ZO-1. Biomol. NMR Assign. 2011, 5, 207–210. [Google Scholar] [CrossRef]
- Bach, A.; Clausen, B.H.; Møller, M.; Vestergaard, B.; Chi, C.N.; Round, A.; Sørensen, P.L.; Nissen, K.B.; Kastrup, J.S.; Gajhede, M.; et al. A high-affinity, dimeric inhibitor of PSD-95 bivalently interacts with PDZ1-2 and protects against ischemic brain damage. Proc. Natl. Acad. Sci. USA 2012, 109, 3317–3322. [Google Scholar] [CrossRef]
- Grandy, D.; Shan, J.; Zhang, X.; Rao, S.; Akunuru, S.; Li, H.; Zhang, Y.; Alpatov, I.; Zhang, X.A.; Lang, R.A.; et al. Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled. J. Biol. Chem. 2009, 284, 16256–16263. [Google Scholar] [CrossRef]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.-H.; Reva, B.; Held, H.A.; Appleton, B.A; Evangelista, M.; Wu, Y.; et al. A specificity map for the PDZ domain family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef]
- Mayasundari, A.; Ferreira, A.M.; He, L.; Mahindroo, N.; Bashford, D.; Fujii, N. Rational design of the first small-molecule antagonists of NHERF1/EBP50 PDZ domains. Bioorg. Med. Chem. Lett. 2008, 18, 942–945. [Google Scholar]
- Ciucci, A.G. A review of spontaneously reported adverse drug reactions with diclofenac sodium (Voltarol). Rheumatol. Rehabil. 1979, Suppl. 2, 116–121. [Google Scholar]
- Catalano, M.A. Worldwide safety experience with diclofenac. Am. J. Med. 1986, 80, 81–87. [Google Scholar] [CrossRef]
- Graham, D.J. COX-2 inhibitors, other NSAIDs, and cardiovascular risk: The seduction of common sens. JAMA J. Am. Med. Assoc. 2006, 296, 1653–1656. [Google Scholar] [CrossRef]
- Todd, P.A.; Sorkin, E.M. Diclofenac sodium. A reappraisal of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy. Drugs 1988, 35, 244–285. [Google Scholar] [CrossRef]
- O’brien, W.M. Adverse reactions to nonsteroidal anti-inflammatory drugs diclofenac compared with other nonsteroidal anti-inflammatory drugs. Am. J. Med. 1986, 80, 70–80. [Google Scholar] [CrossRef]
- Jing, H.; Na, T.; Zhang, W.; Wu, G.; Liu, C.; Peng, J.-B. Concerted actions of NHERF2 and WNK4 in regulating TRPV5. Biochem. Biophys. Res. Commun. 2011, 404, 979–984. [Google Scholar] [CrossRef]
- Hruska-Hageman, A.M.; Wemmie, J.A.; Price, M.P.; Welsh, M.J. Interaction of the synaptic protein PICK1 (protein interacting with C kinase 1) with the non-voltage gated sodium channels BNC1 (brain Na+ channel 1) and ASIC (acid-sensing ion channel). Biochem. J. 2002, 361, 443–450. [Google Scholar] [CrossRef]
- Anzai, N.; Deval, E.; Schaefer, L.; Friend, V.; Lazdunski, M.; Lingueglia, E. The multivalent PDZ domain-containing protein CIPP is a partner of acid-sensing ion channel 3 in sensory neurons. J. Biol. Chem. 2002, 277, 16655–16661. [Google Scholar]
- Pichon, X.; Wattiez, A.S.; Becamel, C.; Ehrlich, I.; Bockaert, J.; Eschalier, A.; Marin, P.; Courteix, C. Disrupting 5-HT(2A) receptor/PDZ protein interactions reduces hyperalgesia and enhances SSRI efficacy in neuropathic pain. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 1462–1470. [Google Scholar] [CrossRef]
- Tao, F.; Johns, R.A. Effect of disrupting N-methyl-d-aspartate receptor-postsynaptic density protein-95 interactions on the threshold for halothane anesthesia in mice. Anesthesiology 2008, 108, 882–887. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar]
- Goddard, T.D.; Kneller, D.G. Sparky 3, 2004; University of California: San Francisco, CA, USA, 2004. [Google Scholar]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E.M. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr. Section D Biol. Crystallogr. 2011, 67, 386–394. [Google Scholar]
- Sample Availability: Expression plasmids of the PDZ domains as well as PRESAT-vector (pGEX-6P-PRESAT) are available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tenno, T.; Goda, N.; Umetsu, Y.; Ota, M.; Kinoshita, K.; Hiroaki, H. Accidental Interaction between PDZ Domains and Diclofenac Revealed by NMR-Assisted Virtual Screening. Molecules 2013, 18, 9567-9581. https://doi.org/10.3390/molecules18089567
Tenno T, Goda N, Umetsu Y, Ota M, Kinoshita K, Hiroaki H. Accidental Interaction between PDZ Domains and Diclofenac Revealed by NMR-Assisted Virtual Screening. Molecules. 2013; 18(8):9567-9581. https://doi.org/10.3390/molecules18089567
Chicago/Turabian StyleTenno, Takeshi, Natsuko Goda, Yoshitaka Umetsu, Motonori Ota, Kengo Kinoshita, and Hidekazu Hiroaki. 2013. "Accidental Interaction between PDZ Domains and Diclofenac Revealed by NMR-Assisted Virtual Screening" Molecules 18, no. 8: 9567-9581. https://doi.org/10.3390/molecules18089567