Rapid Identification of Antioxidant Compounds of Genista saharae Coss. & Dur. by Combination of DPPH Scavenging Assay and HPTLC-MS
Abstract
:1. Introduction
2. Results and Discussion
| Sample | Part | Harvesting period | Extraction Yield (%) | % Flavonoids content (SD) | % Total phenolic content (SD) |
|---|---|---|---|---|---|
| 1 | Roots | August 2009 | 16.31 | 2,441(0,012) | 9,328(0,217) |
| 2 | Roots | March 2010 | 8.78 | 0,212(0,014) | 5,490(0,198) |
| 3 | Stems | August 2009 | 10.93 | 0,213(0,022) | 5,799(0,148) |
| 4 | Stems | March 2010 | 15.07 | 0,924(0,012) | 6,809(0,106) |
| 5 | Flowers | March 2010 | 18.00 | 2,614(0,028) | 9,067(0,144) |
| 6 | Fruit pods | August 2009 | 6.69 | 1,273(0,012) | 5,062(0,163) |
| 7 | Fruit Seeds | August 2009 | 7.70 | 0,518(0,016) | 1,996(0,163) |

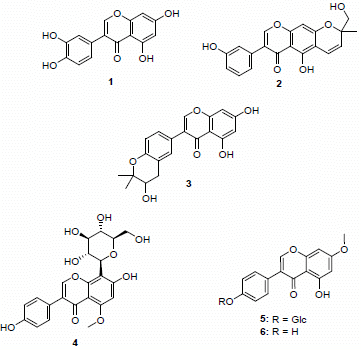
| Spot numbers | m/z values [M−H]− | Putative structures |
|---|---|---|
| A1 | 299.2 | - |
| A2 | 285.1 | 3',4',5,7-Tetrahydroxyisoflavone (1) |
| A3 | 351.2 | Hydroxyalpinumisoflavone (2) |
| A4 | 465.2 | - |
| A5 | 285.1/353.1 | Tetrahydroxyisoflavone (1)/ficuisoflavone (3) |
| B1 | - | - |
| B2 | 445.2 | 5-Methoxy-4',7-trihydroxy-8-glucopyranosyl (4) |
| B3 | 445.2 | 4',5-Dihydroxy-7-methoxyisoflavone-4'-O-β-d-glucopyranoside (5) |
| B4 | 283.1 | 4',5-Dihydroxy-7-methoxyisoflavone (6) |
| B5 | 445.2/575.6/325.2 | - |

2.1. Identification of Compound 4
2.2. Radical Scavenging Capacity (DPPH)
3. Experimental
3.1. General
3.2. Plant Material
3.3. Extraction
3.4. Total Polyphenol and Flavonoids Dosage
3.4.1. Total Phenolic Content

3.4.2. Flavonoids Content

3.5. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) Radical Scavenging Capacity Assay

3.6. HPTLC-DPPH Scavenging Assay
3.7. HPTLC-MS
3.8. Purification of Compound 4
 = +17; (c 0.1, CH3OH); ESIMS m/z 445.2 [M−H]−; 1H-NMR as reported in [20].
= +17; (c 0.1, CH3OH); ESIMS m/z 445.2 [M−H]−; 1H-NMR as reported in [20].4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Johansen, J.; Harris, A.; Rychly, D.; Ergul, A. Oxidative stress and the use of antioxidants in diabetes: Linking basic science to clinical practice. Cardiovasc. Diabetol. 2005, 4, 5. [Google Scholar] [CrossRef]
- Sies, H.; Hollman, P.C.H.; Grune, T.; Stahl, W.; Biesalski, H.K.; Williamson, G. Protection by flavanol-rich foods against vascular dysfunction and oxidative damage: 27th hohenheim consensus conference. Adv. Nutr. 2012, 3, 217–221. [Google Scholar] [CrossRef]
- Unnikrishnan, M.K.; Veerapur, V.; Nayak, Y.; Mudgal, P.P.; Mathew, G. Antidiabetic, antihyperlipidemic and antioxidant effects of the flavonoids. In Polyphenols in Human Health and Disease; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Academic Press: San Diego, CA, USA, 2014; Chapter 13; pp. 143–161. [Google Scholar]
- Quézel, P.; Santa, S. Nouvelle flore d'algérie et des régions désertiques méridionales; Editions du Centre National de la Recherche Scientifique: Paris, France, 1963; Volume 1–2. (In French) [Google Scholar]
- Bisby, F.A.; Buckingham, J.; Harborne, J.B.; Southon, I.W.; Zarucchi, J.L. Phytochemical Dictionary of the Leguminosae; Springer: Heildelberg, Germany, 1994; Volume 2. [Google Scholar]
- Serrilli, A.M.; Graziosi, V.; Ballero, M.; Foddis, C.; Serafini, M.; Poli, F.; Scartezzini, P.; Bianco, A. Endemic sardinian plants: The case of Genista cadasonensis valsecchi. Nat. Prod. Res. 2010, 24, 942–947. [Google Scholar] [CrossRef]
- Orhan, I.E.; Tosun, F.; Tamer, U.; Duran, A.; Alan, B.; Kok, A.F. Quantification of genistein and daidzein in two endemic genista species and their antioxidant activity. J. Serb. Chem. Soc. 2011, 76, 35–42. [Google Scholar]
- Rigano, D.; Cardile, V.; Formisano, C.; Maldini, M.T.; Piacente, S.; Bevilacqua, J.; Russo, A.; Senatore, F. Genista sessilifolia DC. and Genista tinctoria l. Inhibit UV light and nitric oxide-induced DNA damage and human melanoma cell growth. Chem. Biol. Interact. 2009, 180, 211–219. [Google Scholar] [CrossRef]
- Rauter, A.P.; Martins, A.; Lopes, R.; Ferreira, J.; Serralheiro, L.M.; Araújo, M.-E.; Borges, C.; Justino, J.; Silva, F.V.; Goulart, M.; et al. Bioactivity studies and chemical profile of the antidiabetic plant Genista tenera. J. Ethnoph. 2009, 122, 384–393. [Google Scholar] [CrossRef]
- Maire, R. Flore de l'afrique du nord; Éditions Paul Lechevalier: Paris, France, 1967; Volume 13. (In French) [Google Scholar]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef]
- Zhang, J.; Subramanian, S.; Stacey, G.; Yu, O. Flavones and flavonols play distinct critical roles during nodulation of medicago truncatula by Sinorhizobium meliloti. Plant J. 2009, 57, 171–183. [Google Scholar] [CrossRef]
- Sahgal, G.; Ramanathan, S.; Sasidharan, S.; Mordi, M.N.; Ismail, S.; Mansor, S.M. In vitro antioxidant and xanthine oxidase inhibitory activities of methanolic Swietenia mahagoni seed extracts. Molecules 2009, 14, 4476–4485. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, J.-S.; Yang, B.; Lv, G.-P.; Li, S.-P. Free radical scavenging activity and characterization of sesquiterpenoids in four species of curcuma using a TLC bioautography assay and GC-MS analysis. Molecules 2010, 15, 7547–7557. [Google Scholar] [CrossRef]
- Dictionnary of Natural Products. Available online: http://dnp.Chemnetbase.Com/ (accessed on 16 December 2013).
- Ozimina, I.I.; Bandyukova, V.A. Compactin-a novel glycoside of orobol from Genista compacta. Khim. Prir. Soedin. 1985, 507–510. [Google Scholar]
- Pistelli, L.; Bertoli, A.; Giachi, I.; Manunta, A. Flavonoids from Genista ephedroides. J. Nat. Prod. 1998, 61, 1404–1406. [Google Scholar] [CrossRef]
- Pistelli, L.; Giachi, I.; Potenza, D.; Morelli, I. A new isoflavone from Genista corsica. J. Nat. Prod. 2000, 63, 504–506. [Google Scholar] [CrossRef]
- Reich, E.; Schibli, A. High-Performance Thin-Layer Chromatography for the Analysis of Medicinal Plants; Thieme: Stuttgart, Germany, 2006. [Google Scholar]
- Mekkiou, R.; Touahar, H.; Dijoux-Franca, M.G.; Mariotte, A.M.; Benayache, S.; Benayache, F. A new isoflavone from Genista saharae (fabaceae). Biochem. Systemat. Ecol. 2005, 33, 635–638. [Google Scholar] [CrossRef]
- Nakov, N.; Tolkachev, O.N.; Akhtardzhiev, K. Flavonoid content of Genista carinalis Grsb. Pharmazie 1978, 33, 463. [Google Scholar]
- Sato, S.; Hiroe, K.; Kumazawa, T.; Jun-ichi, O. Total synthesis of two isoflavone C-glycosides: Genistein and orobol 8-C-β-D-glucopyranosides. Carbohydr. Res. 2006, 341, 1091–1095. [Google Scholar] [CrossRef]
- Tannins in herbal drugs. In European Pharmacopoeia Online 8.0 General Methods; European Directorate for the Quality of Medicines: Brussels, Belgium, 2014.
- Monographs: Sophorae japonicae flos (Sophora japonica, Fabaceae), Carthami flos (safflower, Carthamus tinctorius L., Asteraceae). In European Pharmacopoeia Online 8.0; European Directorate for the Quality of Medicines: Brussels, Belgium, 2014.
- Sample Availability: Samples of compound 4 are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Meriane, D.; Genta-Jouve, G.; Kaabeche, M.; Michel, S.; Boutefnouchet, S. Rapid Identification of Antioxidant Compounds of Genista saharae Coss. & Dur. by Combination of DPPH Scavenging Assay and HPTLC-MS. Molecules 2014, 19, 4369-4379. https://doi.org/10.3390/molecules19044369
Meriane D, Genta-Jouve G, Kaabeche M, Michel S, Boutefnouchet S. Rapid Identification of Antioxidant Compounds of Genista saharae Coss. & Dur. by Combination of DPPH Scavenging Assay and HPTLC-MS. Molecules. 2014; 19(4):4369-4379. https://doi.org/10.3390/molecules19044369
Chicago/Turabian StyleMeriane, Djamila, Grégory Genta-Jouve, Mohamed Kaabeche, Sylvie Michel, and Sabrina Boutefnouchet. 2014. "Rapid Identification of Antioxidant Compounds of Genista saharae Coss. & Dur. by Combination of DPPH Scavenging Assay and HPTLC-MS" Molecules 19, no. 4: 4369-4379. https://doi.org/10.3390/molecules19044369
APA StyleMeriane, D., Genta-Jouve, G., Kaabeche, M., Michel, S., & Boutefnouchet, S. (2014). Rapid Identification of Antioxidant Compounds of Genista saharae Coss. & Dur. by Combination of DPPH Scavenging Assay and HPTLC-MS. Molecules, 19(4), 4369-4379. https://doi.org/10.3390/molecules19044369