Secondary Metabolite Profiling of Curcuma Species Grown at Different Locations Using GC/TOF and UPLC/Q-TOF MS
Abstract
:1. Introduction
2. Results and Discussion
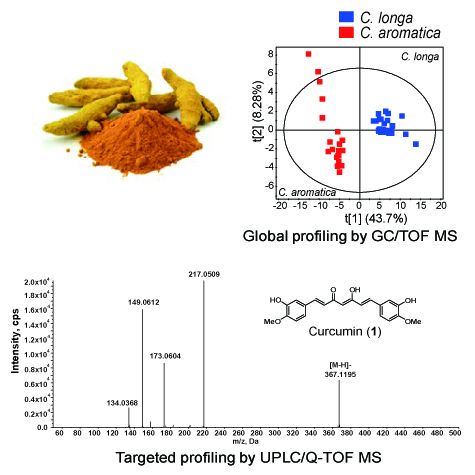
2.1. GC/TOF MS Analysis of Non-Polar Extracts in Curcuma Extracts

| RT # | Identified Metabolite | Formula | Accurate Mass | Main Fragments m/z (% bp) § | |
|---|---|---|---|---|---|
| 1 | 5.98 | Eucalyptol a | C10H18O | 154.1358 | 108.0945 (100); 154.1366 (96.7); 81.0707 (85.6) |
| 2 | 6.81 | Terpinolene †b | C10H16 | 136.1252 | 121.1021 (100); 136.1261 (80.7); 93.0701 (80.2) |
| 3 | 7.01 | Bergamol c | C12H20O2 | 196.1463 | 93.0704 (100); 69.0709 (44.0); 79.0534 (27.1) |
| I.S.-1 | 7.91 | 2,5-Dimethylphenol (IS) | C8H10O | 122.0732 | 107.0501 (100); 122.0732 (95.6); 121.0664 (39.8) |
| 4 | 12.44 | δ-Elemene b | C15H24 | 204.1878 | 121.1027 (100); 136.1266 (60.4); 93.0707 (55.8) |
| 5 | 13.90 | β-Elemene ‡a | C15H24 | 204.1878 | 93.0705 (100); 81.0710 (81.3); 147.1180 (68.7) |
| 6 | 14.22 | U-1 | C15H24 | 204.1878 | 119.0862 (100); 93.0710 (92.0); 91.0545 (55.6) |
| 7 | 14.78 | β-Caryophyllene b | C15H24 | 204.1878 | 133.1020 (100); 91.0545 (84.8); 93.0713 (83.0) |
| 8 | 15.58 | (Z)-β-Farnesene b | C15H24 | 204.1878 | 69.0703 (100); 93.0712 (87.9); 133.1023 (59.6) |
| 9 | 15.63 | β-Cedrene c | C15H24 | 204.1878 | 161.1329 (100); 69.0708 (86.3); 91.0546 (79.0) |
| 10 | 15.77 | α-Caryophyllene b | C15H24 | 204.1878 | 93.0705 (100); 121.121.1014 (39.1); 80.0632 (26.9) |
| 11 | 16.41 | α-Curcumene b | C15H22 | 202.1722 | 119.0865 (100); 132.0939 (82.1); 105.0709 (60.4) |
| 12 | 16.47 | Isoledene b | C15H24 | 204.1878 | 161.1341 (100); 105.0706 (41.1); 204.1890 (31.0) |
| 13 | Germacrene D b | C15H24 | 204.1878 | 161.1333 (100); 105.0705 (37.5); 91.0543 (31.3) | |
| 14 | 16.81 | U-2 | C15H24 | 204.1878 | 119.0844 (100); 93.0699 (82.3); 91.0544 (51.8) |
| 15 | 17.15 | β-Bisabolene a | C15H24 | 204.1878 | 93.0701 (100); 69.0700 (76.99); 204.1880 (57.02) |
| 16 | 17.62 | β-Sesquiphellandrene b | C15H24 | 204.1878 | 69.0707 (100); 91.0558 (98.5); 93.0713 (93.2) |
| 17 | 17.68 | α-Patchoulene c | C15H24 | 204.1878 | 93.0706 (100); 107.0861 (98.7); 135.1187 (58.1) |
| 18 | 19.08 | β-Guaiene c | C15H24 | 204.1878 | 161.1333 (100); 105.0709 (43.6); 204.1878 (36.1) |
| 19 | 19.44 | Cedr-9-ene b | C15H24 | 204.1878 | 119.0863 (100); 93.0702 (64.2); 105.0707 (46.10) |
| 20 | 20.1 | U-3 | C15H24 | 204.1878 | 119.0864 (100); 93.0703 (90.5); 91.0544 (70.3) |
| 21 | 20.56 | U-4 | C15H24 | 204.1878 | 119.0868 (100); 93.0706 (81.1); 91.0544 (50.3) |
| 22 | 21.23 | Agaruspirol c | C15H26O | 222.1984 | 161.1336 (100); 204.1890 (98.1); 189.1658 (84.3) |
| 23 | 21.43 | ar-Turmerone b | C15H20O | 216.1514 | 119.0861 (100); 83.0494 (91.6); 216.1524 (49.3) |
| 24 | 21.6 | Turmerone b | C15H22O | 218.1671 | 105.0705 (100); 83.0499 (86.8); 120.0940 (59.0) |
| 25 | 22.47 | Curlone b | C15H22O | 218.1671 | 120.0937 (100); 83.0499 (27.5); 105.0705 (25.0) |
| 26 | 22.81 | U-5 | C15H24O2 | 236.1787 | 180.1146 (100); 167.1069 (70.7); 109.0656 (35.6) |
| 27 | 23.74 | U-6 | C15H24O2 | 2361791 | 180.1147 (100); 167.1079 (71.3); 109.0653 (33.4) |
| I.S.-2 | 34.93 | Methyl nonadecanoate (IS) | C20H40O2 | 312.3028 | 74.0361 (100); 87.0444 (87.5); 312.2987 (86.9) |

2.2. Quantitative Analysis of Curcuminoids in Curcuma Species Using UPLC/Q-TOF MS


| Samples | Calibration Parameters | C. aromatica Grown in Jeju-do | C. aromatica Grown in Jin-do | C. longa Grown in Jeju-do | C. longa Grown in Jin-do | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Linear Range (ng/mL) | R2 | a LOQ (ng/mL) | Mean ± SE (μg/mg) | b BQL (n) | Mean ± SE (μg/mg) | BQL (n) | Mean ± SE (μg/mg) | BQL(n) | Mean ± SE (μg/mg) | BQL (n) | |
| Curcumin (1) | 25–500 | 0.9989 | 0.5 | 1350.49 ± 72.66 | 0 | 239.24 ± 192.86 | 0 | 2591.43 ± 275.54 | 0 | 1240.38 ± 167.19 | 0 |
| Demethoxycurcumin (2) | 0.5–125 | 0.9999 | 0.5 | 276.60 ± 37.33 | 0 | 451.60 ± 26.79 | 0 | 15.49 ± 5.52 | 5 | 43.56 ± 0.00 | 9 |
| Bisdemethoxycurcumin (3) | 0.5–5 | 0.9944 | 0.5 | - | 10 | - | 10 | - | 10 | - | 10 |
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Plant Material
3.3. Procedures for Preparation of Sample Solution
3.3.1. Procedures for Preparation of Sample Solution for GC/TOF MS Analysis
3.3.2. Procedures for Preparation of Sample Solution for UPLC/Q-TOF MS Analysis
3.4. Instrumental Analysis
3.4.1. Instrumental Analysis of Non-Polar Extracts by GC/TOF MS
3.4.2. Instrumental Analysis of Curcuminoids by UPLC/Q-TOF MS
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wilson, B.; Abraham, G.; Manju, V.S.; Mathew, M.; Vimala, B.; Sundaresan, S.; Nambisan, B. Antimicrobial activity of Curcuma zedoaria and Curcuma malabarica tubers. J. Ethnopharmacol. 2005, 99, 147–151. [Google Scholar] [CrossRef]
- Lantz, R.C.; Chen, G.J.; Solyom, A.M.; Jolad, S.D.; Timmermann, B.N. The effect of turmeric extracts on inflammatory mediator production. Pytomedicine 2005, 12, 445–452. [Google Scholar] [CrossRef]
- Miquel, J.; Bernd, A.; Sempere, J.M.; Dı́az-Alperi, J.; Ramı́rez, A. The curcuma antioxidants: Pharmacological effects and prospects for future clinical use. A review. Arch. Gerontol. Geriatr. 2002, 34, 37–46. [Google Scholar] [CrossRef]
- Jayaprakasha, G.K.; Rao, L.J.; Sakariah, K.K. Antioxidant activities of curcumin, demethoxycurcumin and bisdemethoxycurcumin. Food Chem. 2006, 98, 720–724. [Google Scholar] [CrossRef]
- Tiyaboonchai, W.; Tungpradit, W.; Plianbangchang, P. Formulation and characterization of curcuminoids loaded solid lipid nanoparticles. Int. J. Pharm. 2007, 337, 299–306. [Google Scholar] [CrossRef]
- Lage, H.; Duarte, N.; Coburger, C.; Hilgeroth, A.; Ferreira, M.J.U. Antitumor activity of terpenoids against classical and atypical multidrug resistant cancer cells. Phytomedicine 2010, 17, 441–448. [Google Scholar] [CrossRef]
- Tolstikova, T.G.; Sorokina, I.V.; Tolstikov, G.A.; Tolstikov, A.G.; Flekhter, O.B. Biological activity and pharmacological prospects of lupane terpenoids: I. Natural lupane derivatives. Russ. J. Bioorg. Chem. 2006, 32, 37–49. [Google Scholar] [CrossRef]
- Ulubelen, A. Cardioactive and antibacterial terpenoids from some Salvia species. Phytochemistry 2003, 64, 395–399. [Google Scholar] [CrossRef]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spec. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef]
- Jung, Y.; Ahn, Y.G.; Kim, H.K.; Moon, B.C.; Lee, A.Y.; Ryu, D.H.; Hwang, G.S. Characterization of dandelion species using 1H NMR- and GC-MS-based metabolite profiling. Analyst 2011, 136, 4222–4231. [Google Scholar] [CrossRef]
- Grata, E.; Boccard, J.; Guillarme, D.; Glauser, G.; Carrupt, P.A.; Farmer, E.E.; Wolfender, J.L.; Rudaz, S. UPLC-TOF-MS for plant metabolomics: A sequential approach for wound marker analysis in Arabidopsis thaliana. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 871, 261–270. [Google Scholar] [CrossRef]
- Kim, E.J.; Kwon, J.; Park, S.H.; Park, C.; Seo, Y.B.; Shin, H.K.; Kim, H.K.; Lee, K.S.; Choi, S.Y.; Ryu, D.H.; et al. Metabolite profiling of Angelica gigas from different geographical origins using 1H NMR and UPLC-MS analyses. J. Agric. Food Chem. 2011, 59, 8806–8815. [Google Scholar] [CrossRef]
- Raven, P.H.; Evert, R.F.; Eichhorm, S.E. Biology of Plants, 6th ed.; Freeman: New York, NY, USA, 1999; pp. 32–37. [Google Scholar]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Lei, Z.; Huhman, D.; Sumner, L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011, 268, 25435–25442. [Google Scholar]
- Halket, J.M.; Waterman, D.; Przyborowska, A.M.; Patel, R.K.P.; Fraser, P.D.; Bramley, P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005, 56, 219–243. [Google Scholar]
- Apbona, V.; Iglesias, D.J.; Talon, M.; Gomez-Cadenas, A. Plant phenotype demarcation using nontargeted LC-MS and GC-MS metabolite profiling. J. Agric. Food Chem. 2009, 57, 7338–7347. [Google Scholar]
- t’Kindt, R.; Morreel, K.; Deforce, D.; Boerjan, W.; Bocxlaer, J.V. Joint GC–MS and LC–MS platforms for comprehensive plant metabolomics: Repeatability and sample pre-treatment. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 3572–3580. [Google Scholar] [CrossRef]
- Jung, Y.; Lee, J.; Kim, H.K.; Moon, B.C.; Ji, Y.; Ryu, D.H.; Hwang, G.S. Metabolite profiling of Curcuma species grown in different regions using 1H NMR spectroscopy and multivariate analysis. Analyst 2012, 137, 5597–5606. [Google Scholar] [CrossRef]
- Herebian, D.; Choi, J.H.; Abd El-Aty, A.M.; Shim, J.H.; Spiteller, M. Metabolite analysis in Curcuma domestica using various GC-MS and LC-MS separation and detection techniques. Biomed. Chromatogr. 2009, 23, 951–965. [Google Scholar] [CrossRef]
- He, X.G.; Lin, L.Z.; Lian, L.Z.; Lindenmaier, M. Liquid chromatography electrospray mass spectrometric analysis of curcuminoids and sesquiterpenoids in turmeric (Curcuma longa). J. Chromatogr. A 1998, 818, 127–132. [Google Scholar] [CrossRef]
- Jiang, H.; Somogyi, A.; Jacobsen, N.E.; Timmermann, B.N.; Gang, D.R. Analysis of curcuminoids by positive and negative electrospray ionization and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 1001–1012. [Google Scholar] [CrossRef]
- Jayaprakasha, G.K.; Rao, L.J.M.; Sakariah, K.K. Improved HPLC method for the determination of curcumin, demethoxycurcumin, and bisdemethoxycurcumin. J. Agric. Food Chem. 2002, 50, 3668–3672. [Google Scholar] [CrossRef]
- Yan, J.; Chen, G.; Tong, S.; Feng, Y.; Sheng, L.; Lou, J. Preparative isolation and purification of germacrone and curdione from the essential oil of the rhizomes of Curcuma wenyujin by high-speed counter-current chromatography. J. Chromatogr. A 2005, 1070, 207–210. [Google Scholar] [CrossRef]
- Devaraj, S.; Esfahani, A.S.; Ismail, S.; Ramanathan, S.; Yam, M.F. Evaluation of the antinociceptive activity and acute oral toxicity of standardized ethanolic extract of the rhizome of Curcuma xanthorrhiza Roxb. Molecules 2010, 15, 2925–2934. [Google Scholar] [CrossRef]
- Raina, V.K.; Srivastava, S.K. Rhizome and leaf oil composition of Curcuma longa from the lower Himalayan region of northern India. J. Essent. Oil Res. 2005, 17, 556–559. [Google Scholar] [CrossRef]
- Meckes, M.; Calzada, F. Terpenoids isolated from Psidium guajava hexane extract with depressant activity on central nervous system. Phytother. Res. 1996, 10, 600–603. [Google Scholar] [CrossRef]
- Fraenkel, G.S. The raison d’être of secondary plant substances. Science 1959, 129, 1466–1470. [Google Scholar]
- Pichersky, E.; Gang, D.R. Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 2000, 5, 439–445. [Google Scholar] [CrossRef]
- Brenes-Arguedas, T.; Coley, P.D. Phenotypic variation and spatial structure of secondary chemistry in a natural population of a tropical tree species. OIKOS 2005, 108, 410–420. [Google Scholar] [CrossRef]
- Tsai, S.Y.; Huang, S.J.; Chyau, C.C.; Tsai, C.H.; Weng, C.C.; Mau, J.L. Composition and antioxidant properties of essential oils from Curcuma rhizome. Asian J. Arts Sci. 2011, 2, 57–66. [Google Scholar]
- Ritwiz, S.B.; Kusha, S.B.; Ranjeet, D.; Ganesh, N. Curcuma longa leaves exhibits a potential antioxidant, antibacterial and immunomodulating properties. Int. J. Phytomed. 2011, 3, 270–278. [Google Scholar]
- Bergdahl, M.; Bergdahl, J. Burning mouth syndrome: Prevalence and associated factors. J. Oral Pathol. Med. 1999, 28, 350–354. [Google Scholar] [CrossRef]
- Peana, A.T.; D’Aquila, P.S.; Panin, F.; Serra, G.; Pippia, P.; Moretti, M.D.L. Anti-inflammatory activity of linalool and linalyl acetate constituents of essential oils. Phytomedicine 2002, 9, 721–726. [Google Scholar] [CrossRef]
- Bosak, T.; Losick, R.M.; Pearson, A. A polycyclic terpenoid that alleviates oxidative stress. Proc. Natl. Acad. Sci. USA 2008, 105, 6725–6729. [Google Scholar]
- Singh, G.; Kapoor, I.P.S.; Singh, P.; de Heluani, C.S.; de Lampasona, M.P.; Catalan, C.A.N. Comparative study of chemical composition and antioxidant activity of fresh and dry rhizomes of turmeric (Curcuma longa Linn.). Food Chem. Toxicol. 2010, 48, 1026–1031. [Google Scholar] [CrossRef]
- Singh, S.; Sankar, B.; Rajesh, S.; Sahoo, K.; Subudhi, E.; Nayak, S. Chemical composition of turmeric oil (Curcuma longa L. cv. Roma) and its antimicrobial activity against eye infecting pathogens. J. Essent. Oil Res. 2011, 23, 11–18. [Google Scholar]
- De Abreu Gonzaga, W.; Weber, A.D.; Giacomelli, S.R.; Simionatto, E.; Dalcol, I.I.; Dessoy, E.C.; Morel, A.F. Composition and antibacterial activity of the essential oils from Zanthoxylum rhoifolium. Planta Med. 2003, 69, 773–775. [Google Scholar]
- El-Baroty, G.S.; El-Baky, H.H.A.; Farag, R.S.; Saleh, M.A. Characterization of antioxidant and antimicrobial compounds of cinnamon and ginger essential oils. Afr. J. Biochem. Res. 2010, 4, 167–174. [Google Scholar]
- Prakash, P.; Misra, A.; Surin, W.R.; Jain, M.; Bhatta, R.S.; Pal, R.; Raj, K.; Barthwal, M.K.; Dikshit, M. Anti-platelet effects of Curcuma oil in experimental models of myocardial ischemia-reperfusion and thrombosis. Thromb. Res. 2011, 127, 111–118. [Google Scholar]
- Sukari, M.A.; Wah, T.S.; Saad, S.M.; Rashid, N.Y.; Rahmani, M.; Lajis, N.H.; Hin, T.Y. Bioactive sesquiterpenes from Curcuma ochrorhiza and Curcuma heyneana. Nat. Prod. Res. 2014, 24, 838–845. [Google Scholar]
- Ramanathan, R.; Jemal, M.; Ramagiri, S.; Xia, Y.Q.; Humpreys, W.G.; Olah, T.; Korfmacher, W.A. It is time for a paradigm shift in drug discovery bioanalysis: From SRM to HRMS. J. Mass Spectrom. 2011, 46, 595–601. [Google Scholar]
- Jiang, H.; Timmermann, B.N.; Gang, D.R. Use of liquid chromatography–electrospray ionization tandem mass spectrometry to identify diarylheptanoids in turmeric (Curcuma longa L.) rhizome. J. Chromatogr. A 2006, 1111, 21–31. [Google Scholar] [CrossRef]
- Hayakawa, H.; Kobayashi, T.; Minaniya, Y.; Ito, K.; Miyasaki, A.; Fukuda, T.; Yamamoto, Y. Development of a molecular marker to identify a candidate line of turmeric (Curcuma longa L.) with a high curcumin content. Am. J. Plant Sci. 2011, 2, 15–26. [Google Scholar]
- Sasikumar, B. Genetic resources of Curcuma: Diversity, characterization and utilization. Plant Genet. Resour. 2005, 3, 230–251. [Google Scholar]
- Hou, X.-L.; Hayashi-Nakamura, E.; Takatani-Nakase, T.; Tanaka, K.; Takahashi, K.; Komatsu, K.; Takahashi, K. Curdione plays an important role in the inhibitory effect of Curcuma aromatica on CYP3A4 in Caco-2 cells. Evid. Based Complement. Alternat. Med. 2011, 2011. [Google Scholar] [CrossRef]
- Sample Availability: Not available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Jung, Y.; Shin, J.-H.; Kim, H.K.; Moon, B.C.; Ryu, D.H.; Hwang, G.-S. Secondary Metabolite Profiling of Curcuma Species Grown at Different Locations Using GC/TOF and UPLC/Q-TOF MS. Molecules 2014, 19, 9535-9551. https://doi.org/10.3390/molecules19079535
Lee J, Jung Y, Shin J-H, Kim HK, Moon BC, Ryu DH, Hwang G-S. Secondary Metabolite Profiling of Curcuma Species Grown at Different Locations Using GC/TOF and UPLC/Q-TOF MS. Molecules. 2014; 19(7):9535-9551. https://doi.org/10.3390/molecules19079535
Chicago/Turabian StyleLee, Jueun, Youngae Jung, Jeoung-Hwa Shin, Ho Kyoung Kim, Byeong Cheol Moon, Do Hyun Ryu, and Geum-Sook Hwang. 2014. "Secondary Metabolite Profiling of Curcuma Species Grown at Different Locations Using GC/TOF and UPLC/Q-TOF MS" Molecules 19, no. 7: 9535-9551. https://doi.org/10.3390/molecules19079535