Tethering in RNA: An RNA-Binding Fragment Discovery Tool
Abstract
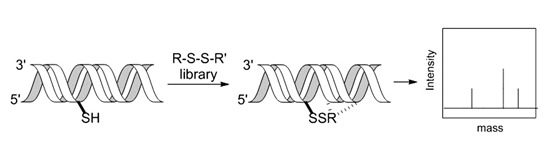
:1. Introduction
2. Results and Discussion
2.1. Synthesis of N6-(2-(triphenylmethylthio)ethyl)adenosine Phosphoramidite and Reactivity of Modified RNAs



2.2. RNA Tethering Screen with pre21 RNA 1 and pre21 RNA 2



3. Experimental Section
3.1. General Procedures




3.2. RNA Synthesis, Purification and S-Tr Deprotection
3.3. Mass Spectrometry Analysis of RNAs
3.4. Conjugation Reaction Procedure
3.5. Calculation of Reaction Product Abundances
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gallego, J.; Varani, G. Targeting RNA with Small-Molecule Drugs: Therapeutic Promise and Chemical Challenges. Acc. Chem. Res. 2001, 34, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Disney, M.D. Recent Advances in Developing Small Molecules Targeting RNA. ACS Chem. Biol. 2012, 7, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Griffey, R.H.; Sannes-Lowery, K.A.; Drades, J.J.; Venkatraman, M.; Swayze, E.E.; Hofstadler, S.A. Characterization of Low-Affinity Complexes between RNA and Small Molecule Using Electrospray Ionization Mass Spectrometry. J. Am. Chem. Soc. 2000, 122, 9933–9938. [Google Scholar] [CrossRef]
- Jimenez-Moreno, E.; Gomez-Pinto, I.; Corzana, F.; Santan, A.G.; Revuelta, J.; Bastida, A.; Jimenez-Barbero, J.; Gonzalez, C.; Asensio, J.L. Chemical Interrogation of Drug/RNA Complexes: From Chemical Reactivity to Drug Design. Angew. Chem. Int. Ed. 2013, 52, 3148–3151. [Google Scholar] [CrossRef]
- Hardy, J.A.; Lam, J.; Nguyen, J.; O’Brien, T.; Wells, J.A. Discovery of an allosteric site in the caspases. Proc. Natl. Acad. Sci. USA 2004, 101, 12461–12466. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Fucini, R.V.; Fahr, B.T.; Randal, M.; Lind, K.E.; Lam, M.B.; Lu, W.; Lu, Y.; Cary, D.R.; Romanowski, M.J.; et al. Fragment-based Discovery of Nonpeptidic BACE-1 Inhibitors Using Tethering. Biochemistry 2009, 48, 4488–4496. [Google Scholar] [CrossRef] [PubMed]
- Erlanson, D.A.; Braisted, A.C.; Raphael, M.; Stroud, R.M.; Gordon, E.M.; Wells, J.A. Site-directed ligand discovery. Proc. Natl. Acad. Sci. USA 2000, 97, 9367–9372. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Ferracin, M.; Prueit, R.L. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Chirayil, S.; Chirayil, R.; Luebke, K.J. Discovering ligands for a microRNA precursor with peptoid microarrays. Nucl. Acids Res. 2009, 37, 5486–5497. [Google Scholar] [CrossRef] [PubMed]
- Velagapudi, S.P.; Gallo, S.M.; Disney, M. Sequence-based Design of Bioactive Small Molecules that Target Precursor MicroRNAs. Nat. Chem. Biol. 2014, 10, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Bose, D.; Gopal, J.; Suryawanshi, H.; Agarwala, P.; Pore, S.K.; Banerjee, R.; Maiti, S. The Tuberculosis Drug Streptomycin as a Potential Cancer Therapeutic: Inhibition of miR-21 Function by Directly Targetting Its Precursor. Angew. Chem. Int. Ed. 2012, 51, 1019–1023. [Google Scholar] [CrossRef]
- Maiti, M.; Nauwelaerts, K.; Herdewijn, P. Pre-mircoRNA binding aminoglycosides and antitumor drugs as inhibitors of Dicer catalyzed microRNA processing. Bioorg. Med. Chem. Lett. 2012, 22, 1709–1711. [Google Scholar] [CrossRef] [PubMed]
- Allerson, C.; Swaine, L.; Verdine, G.L. A Chemical Method for Site-Specific Modification of RNA: The Convertible Nucleoside Approach. J. Am. Chem. Soc. 1997, 119, 7423–7433. [Google Scholar] [CrossRef]
- Porcher, S.; Myeyyappan, M.; Pitsch, S. Spontaneous Aminoacylation of a RNA Sequence Containing a 3'-Terminal 2'-Thioadenosine. Helvetica Chimica Acta 2005, 88, 2897–2910. [Google Scholar] [CrossRef]
- Geiermann, A.; Polacek, N.; Micura, R. Native Chemical Ligation of Hydrolysis-Resistant 3'-Peptidyl-RNA Mimics. J. Am. Chem. Soc. 2011, 133, 19068–19071. [Google Scholar] [CrossRef] [PubMed]
- Peacock, H.; Bachu, R.; Beal, P.A. Covalent stabilization of a small molecule-RNA complex. Bioorg. Med. Chem. Lett. 2011, 21, 5002–5005. [Google Scholar] [CrossRef] [PubMed]
- Burlingame, M.A.; Tom, C.T.; Renslo, A.R. Simple One-Pot Synthesis of Disulfide Fragments for Use in Disulfide-Exchange Screening. ACS Comb. Sci. 2011, 13, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Janda, L.; Nguyen, J.; Strekowski, L.; Wilson, W.D. The Interaction of Substituted 2-phenylquinoline Intercalators with Poly(A) • poly(U): Classical and Threading Intercalation Modes of RNA. Biopolymers 2004, 34, 61–73. [Google Scholar] [CrossRef]
- Battistutta, R.; de Moliner, E.; Sarno, S.; Zanotti, G.; Pinna, L.A. Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci. 2001, 10, 2200–2206. [Google Scholar] [CrossRef] [PubMed]
- Chirayil, S.; Wu, Q.; Amezcua, C.; Leubke, K.J. NMR Characterization of an Oligonucleotide Model of the MiR-21 Pre-Element. PLoS One 2014, 9, e108231. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yamato, K.; Ferguson, J.S.; Gong, B. Sequence-Specific Associaion in Aqueous Media by Integrating Hydrogen Bonding and Dynamic Covalent Interactions. J. Am. Chem. Soc. 2006, 128, 12628–12629. [Google Scholar] [CrossRef] [PubMed]
- Hamm, M.L.; Piccirilli, J.A. Incorporation of 2'-deoxy-2'-mercaptocytidine into Oligonucleotides via Phosphoramidite Chemistry. J. Org. Chem. 1997, 62, 3415–3420. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples are not available from authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, K.; Arkin, M.R.; Beal, P.A. Tethering in RNA: An RNA-Binding Fragment Discovery Tool. Molecules 2015, 20, 4148-4161. https://doi.org/10.3390/molecules20034148
Tran K, Arkin MR, Beal PA. Tethering in RNA: An RNA-Binding Fragment Discovery Tool. Molecules. 2015; 20(3):4148-4161. https://doi.org/10.3390/molecules20034148
Chicago/Turabian StyleTran, Kiet, Michelle R. Arkin, and Peter A. Beal. 2015. "Tethering in RNA: An RNA-Binding Fragment Discovery Tool" Molecules 20, no. 3: 4148-4161. https://doi.org/10.3390/molecules20034148